Impact of G449 Single-Point Mutation on Glucansucrase URE 13-300 Enzyme Properties and Polysaccharide Structure
Abstract
1. Introduction
2. Results
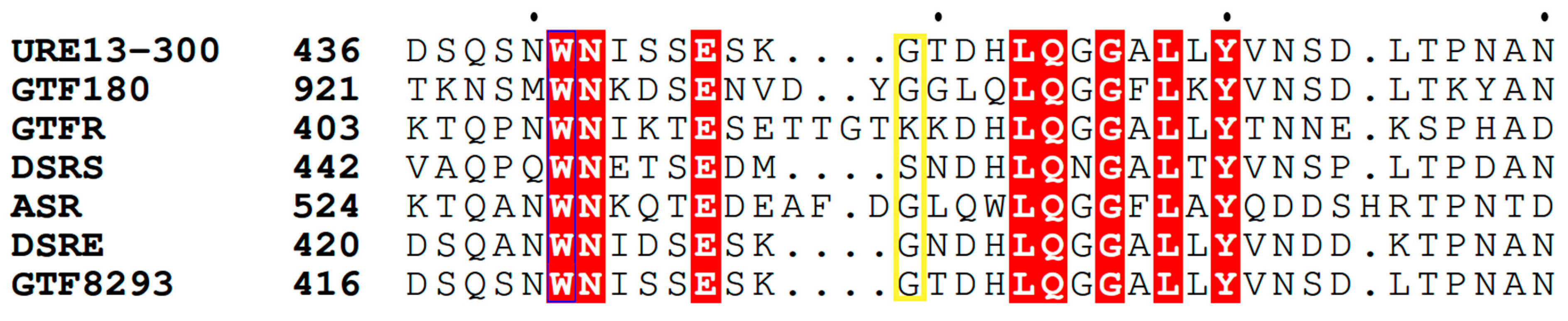
2.1. Design of the Site-Directed Mutagenesis and Expression of the Mutated Enzyme U13M1
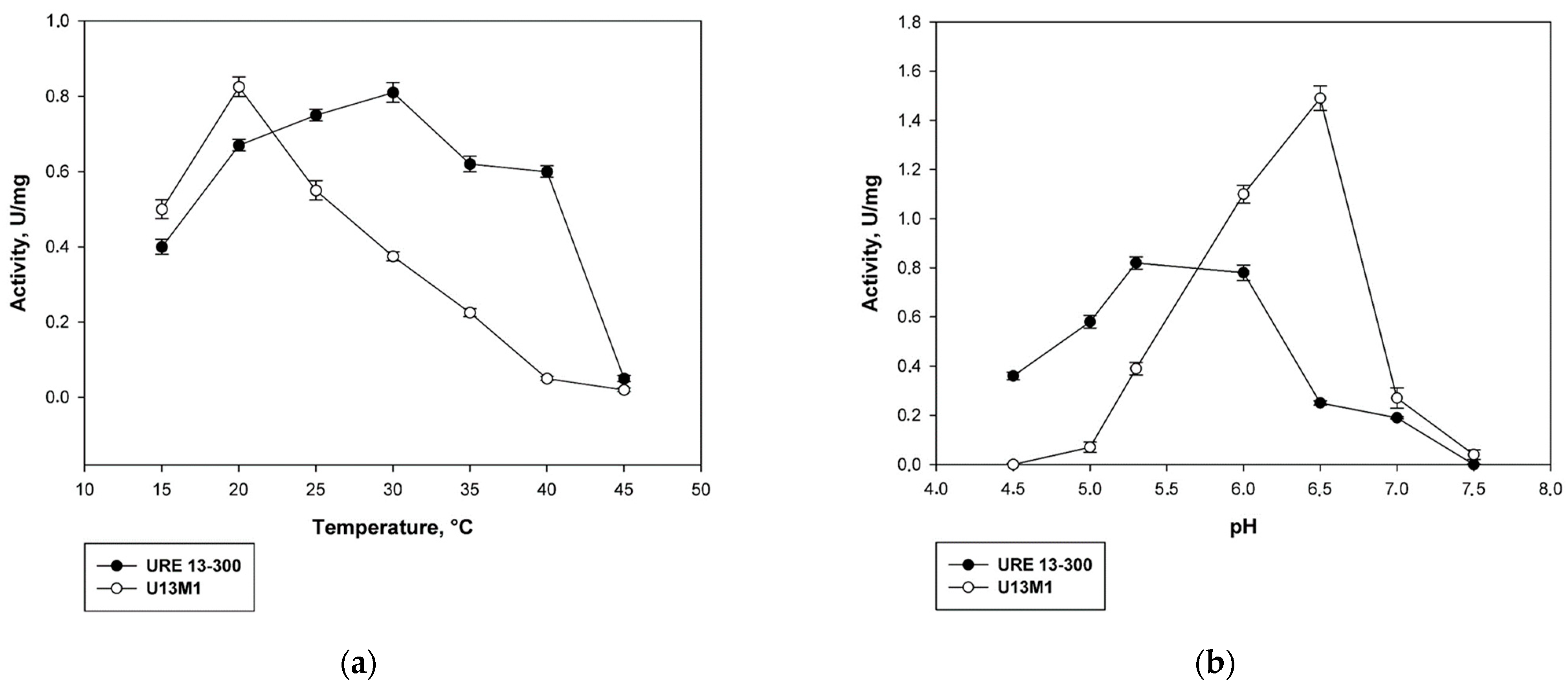
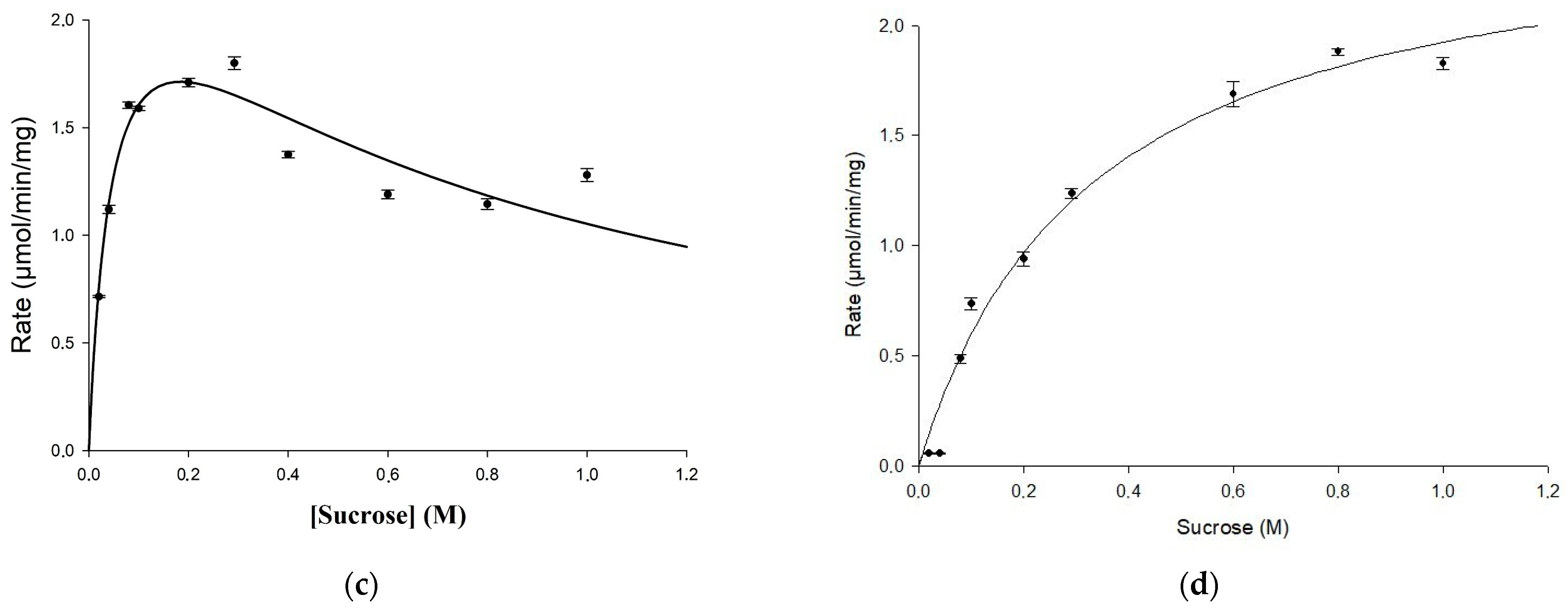
2.2. Biochemical Characterization of the Mutant Enzyme U13M1
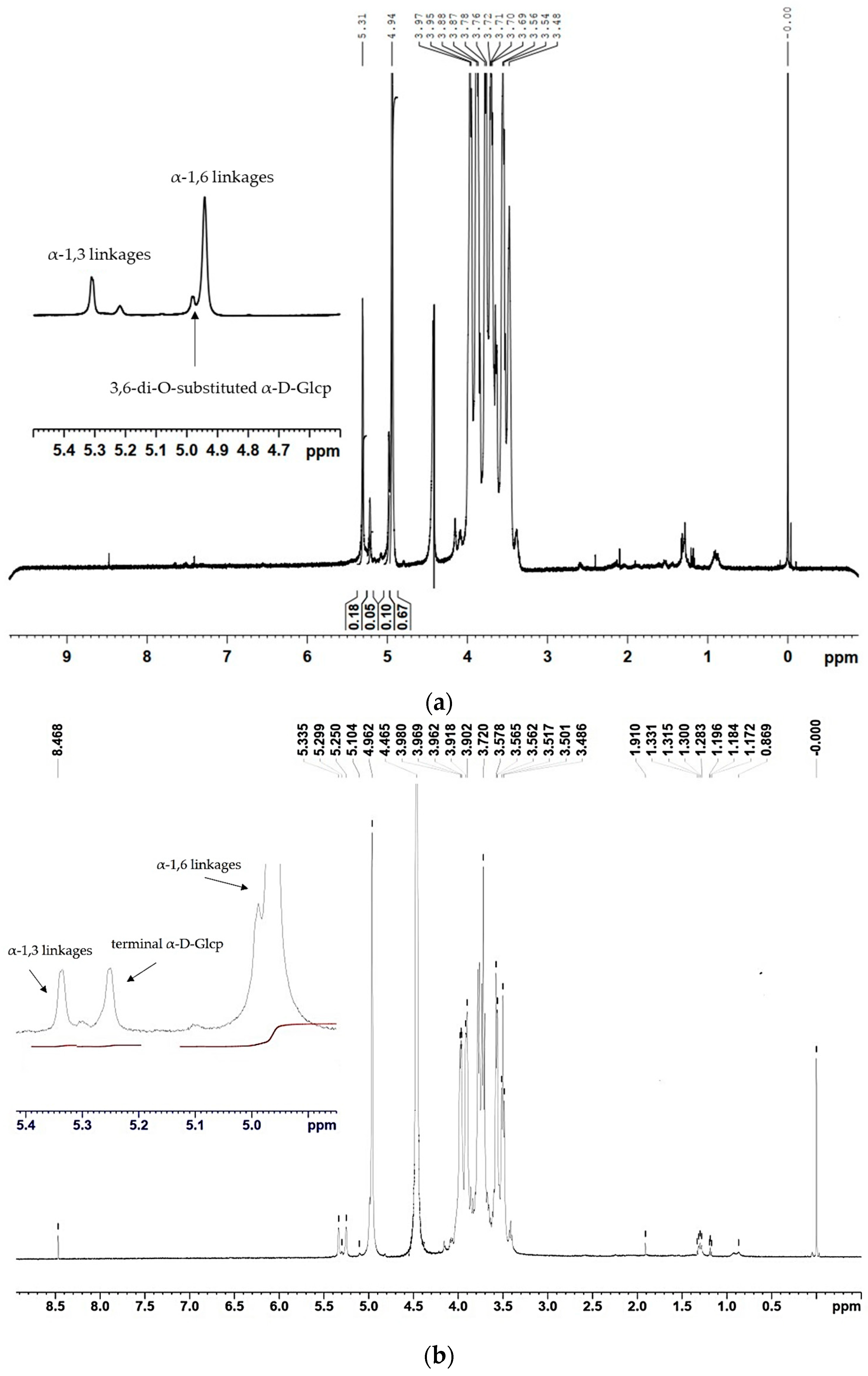
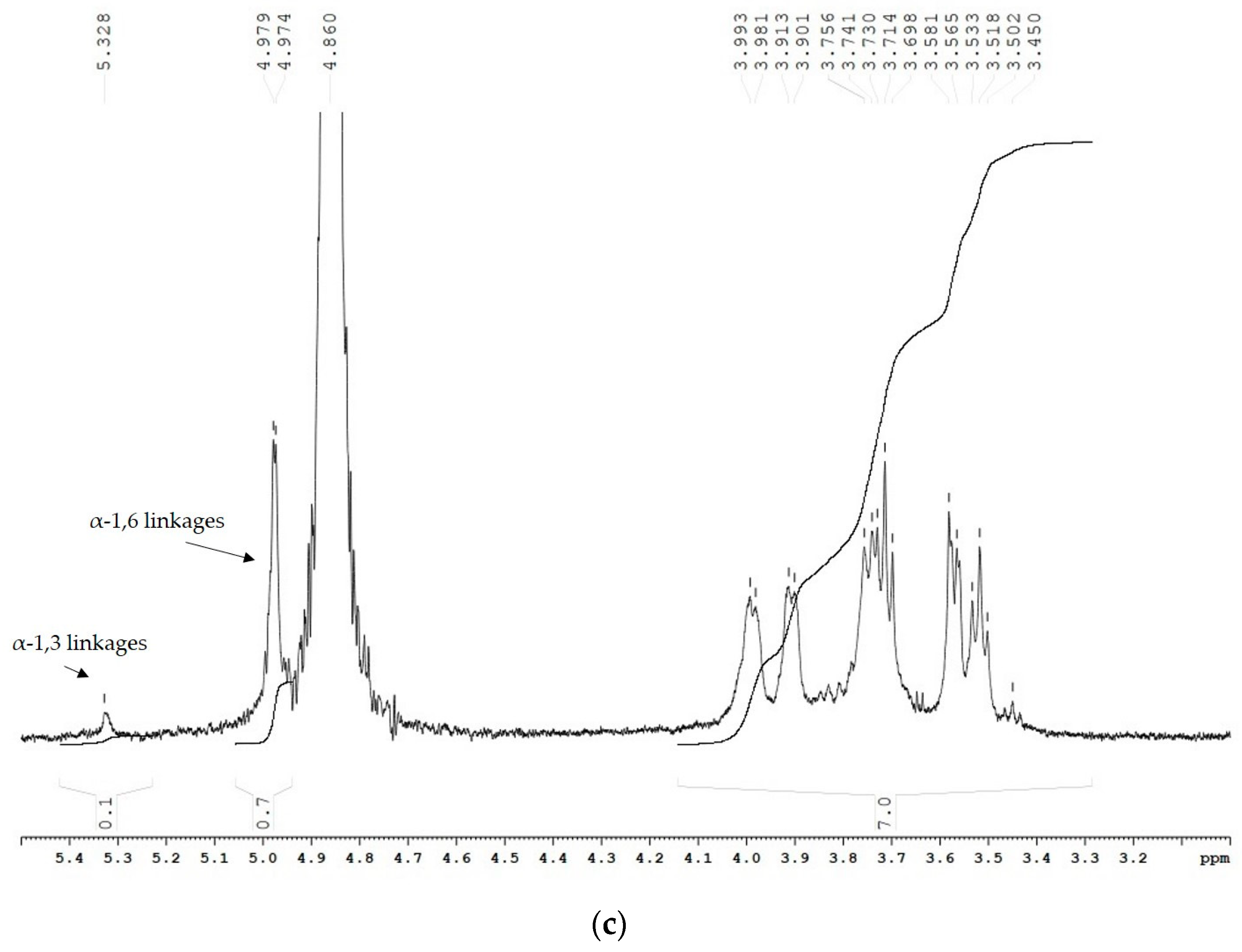
2.3. Structure and Linkage Specificity of URE 13-300 and U13M1 Polysaccharides
2.4. Physical Properties of URE 13-300 and U13M1 Glucans
3. Materials and Methods
3.1. Bacterial Strains, Plasmid Template and Media
3.2. Site-Directed Mutagenesis
3.3. Obtaining of Glucansucrase URE 13–300 Preparations
3.4. SDS-PAGE Analysis
3.5. Enzyme Activity Assay
3.6. Kinetic Studies
3.7. Effect of Metal Ions on Enzyme Activity
3.8. In Vitro Glucan Synthesis
3.9. NMR Analysis
3.10. Water Holding Capacity
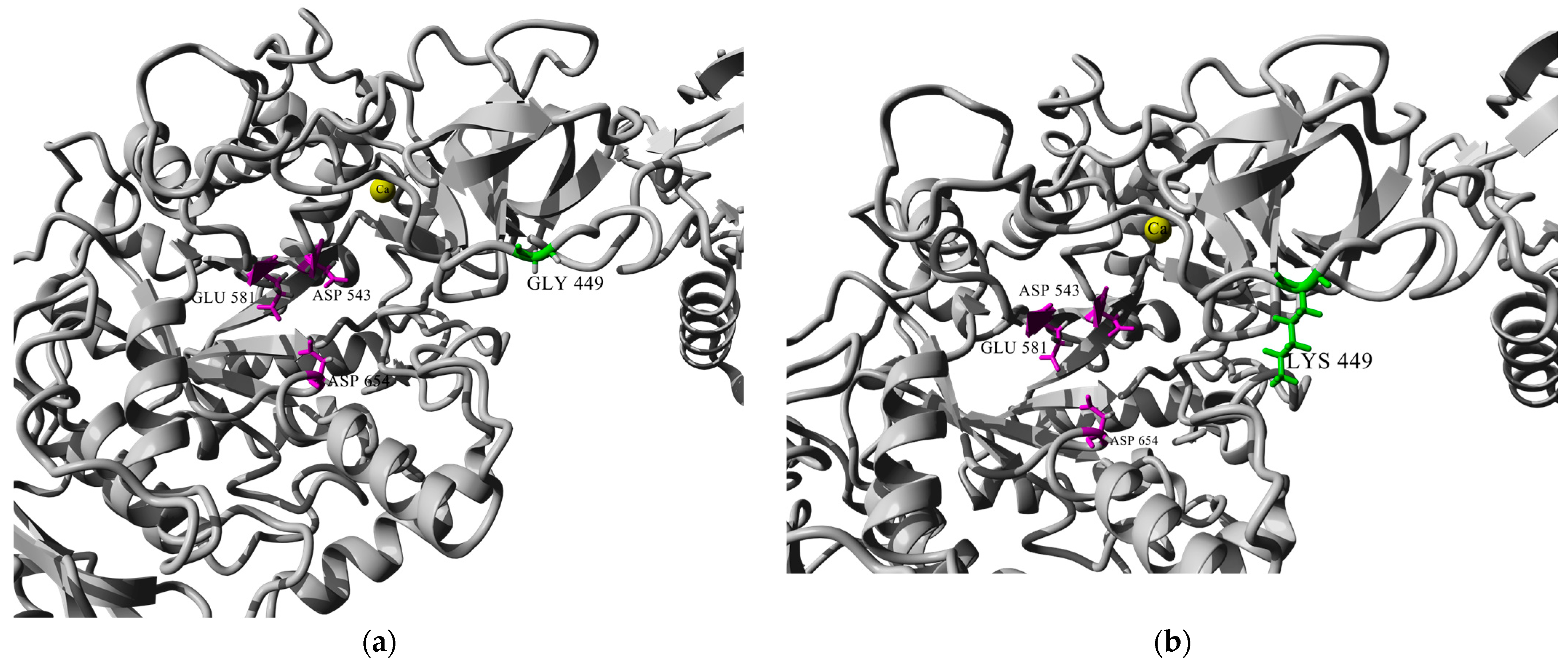
3.11. Sequence Analysis and Homology Modeling
3.12. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol. Adv. 2018, 36, 196–207. [Google Scholar] [CrossRef]
- Faucard, P.; Grimaud, F.; Lourdin, D.; Maigret, J.E.; Moulis, C.; Remaud-Siméon, M.; Putaux, J.L.; Potocki-Véronèse, G.; Rolland-Sabaté, A. Macromolecular structure and film properties of enzymatically-engineered high molar mass dextrans. Carbohydr. Polym. 2018, 181, 337–344. [Google Scholar] [CrossRef]
- van Hijum, S.A.F.T.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. R. 2006, 70, 157–176. [Google Scholar] [CrossRef]
- Moulis, C.; André, I.; Remaud-Simeon, M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell. Mol. Life Sci. 2016, 73, 2661–2679. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Meng, X.; Dijkhuizen, L.; Liu, W. Structures, physico-chemical properties, production and (potential) applications of sucrose-derived α-d-glucans synthesized by glucansucrases. Carbohydr. Polym. 2020, 249, 116818. [Google Scholar] [CrossRef]
- Côté, G.L.; Skory, C.D. Effects of mutations at threonine-654 on the insoluble glucan synthesized by Leuconostoc mesenteroides NRRL B-1118 glucansucrase. Appl. Microbiol. Biotechnol. 2014, 98, 6651–6658. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Waiyaseesang, N.; Panpetch, P.; Charoenwongpaiboon, T.; Nepogodiev, S.A.; Ekgasit, S.; Field, R.A.; Pichayangkura, R. Characterisation of insoluble α-1,3-/α-1,6 mixed linkage glucan produced in addition to soluble α-1,6-linked dextran by glucansucrase (DEX-N) from Leuconostoc citreum ABK-1. Int. J. Biol. Macromol. 2020, 152, 473–482. [Google Scholar] [CrossRef]
- Ogawa, K.; Yui, T.; Okamura, K.; Misaki, A. Crystalline features of streptococcal (1→3)-α-D-glucans of human saliva. Biosci. Biotech. Biochem. 1994, 58, 1326–1327. [Google Scholar] [CrossRef]
- Yui, T.; Goto, K.; Kawano, Y.; Ogawa, K. Molecular modeling study of highly branching (1→3)-α-D-glucan, a model polysaccharide for cariogenic glucan, using the N-H mapping method. Biosci. Biotechnol. Biochem. 2000, 64, 52–60. [Google Scholar] [CrossRef]
- Côté, G.L. Alternan. In Polysaccharides, I. Polysaccharides from Prokaryotes, 5th ed.; Vandamme, E.J., DeBaets, S., Steinbüchel, A., Eds.; Wiley: Weinheim, Germany, 2002; Volume 5, pp. 323–350. [Google Scholar]
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Misaka, T.; Abe, K.; Ko-bayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186. [Google Scholar] [CrossRef]
- Vujičić-Žagar, A.; Pijning, T.; Kralj, S.; López, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406–21411. [Google Scholar] [CrossRef]
- Pijning, T.; Vujičić-Žagar, A.; Kralj, S.; Dijkhuizen, L.; Dijkstra, B.W. Flexibility of truncated and full-length glucansucrase GTF180 enzymes from Lactobacillus reuteri 180. FEBS J. 2014, 281, 2159–2171. [Google Scholar] [CrossRef]
- Molina, M.; Cioci, G.; Moulis, C.; Séverac, E.; Remaud-Siméon, M. Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure-Function Relationship Studies and Engineering. Microorganisms 2021, 9, 1607. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.B.; Li, M.Q.; Hu, X.Q.; Li, Y. Functional analysis of truncated and site-directed mutagenesis dextransucrases to produce different type dextrans. Enzyme Microb. Technol. 2017, 102, 26–34. [Google Scholar] [CrossRef]
- Swistowska, A.M.; Gronert, S.; Wittrock, S.; Collisi, W.; Hecht, H.-J.; Hofer, B. Identification of structural determinants for sub-strate binding and turnover by glucosyltransferase R supports the permutation hypothesis. FEBS Lett. 2007, 581, 4036–4042. [Google Scholar] [CrossRef]
- Tsumori, H.; Minami, T.; Kuramitsu, H.K. Identification of essential amino acids in the Streptococcus mutans glucosyltransfer-ases. J. Bacteriol. 1997, 179, 3391–3396. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Guionnet, M.; Schörghuber, J.; Lichtenecker, R.; Moulis, C.; Remaud-Siméon, M.; Lippens, G. Futile Encounter Engineering of the DSR-M Dextransucrase Modifies the Resulting Polymer Length. Biochemistry 2019, 58, 2853–2859. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90. [Google Scholar] [CrossRef]
- Hellmuth, H.; Wittrock, S.; Kralj, S.; Dijkhuizen, L.; Hofer, B.; Seibel, J. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: Toward tailor-made polymer and oligosaccharide products. Biochemistry 2008, 47, 6678–6684. [Google Scholar] [CrossRef]
- Moulis, C.; Joucla, G.; Harrison, D.; Fabre, E.; Potocki-Veronese, G.; Monsan, P.; Remaud-Simeon, M. Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 2006, 281, 31254–31267. [Google Scholar] [CrossRef]
- Bozonnet, S.; Dols-Laffargue, M.; Fabre, E.; Pizzut, S.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 2002, 184, 5753–5761. [Google Scholar] [CrossRef]
- Brison, Y.; Fabre, E.; Moulis, C.; Portais, J.C.; Monsan, P.; Remaud-Siméon, M. Synthesis of dextrans with controlled amounts of alpha-1,2 linkages using the transglucosidase GBD-CD2. Appl. Microbiol. Biotechnol. 2010, 86, 545–554. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Wang, X.; Grijpstra, P.; van Leeuwen, S.S.; Pijning, T.; Dijkhuizen, L. Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase activity. Appl. Microbiol. Biotechnol. 2018, 102, 7935–7950. [Google Scholar] [CrossRef]
- Bivolarski, V.; Vasileva, T.; Valerie, G.; Iliev, I. Synthesis of glucooligosaccharides with prebiotic potential by glucansucrase URE 13-300 acceptor reactions with maltose, raffinose and lactose. Eng. Life Sci. 2018, 18, 904–913. [Google Scholar] [CrossRef]
- Iliev, I.; Vasileva, T.; Bivolarski, V.; Yovcheva, T.; Marudova, M.; Viraneva, A.; Bodurov, I. Obtaining of Water-Insoluble Glucan Through Transferase Enzyme Reaction. BG Patent 67404 B1, 21 November 2023. [Google Scholar]
- Meng, X.; Dobruchowska, J.M.; Pijning, T.; López, C.A.; Kamerling, J.P.; Dijkhuizen, L. Residue Leu940 has a crucial role in the linkage and reaction specificity of the glucansucrase GTF180 of the probiotic bacterium Lactobacillus reuteri 180. J. Biol. Chem. 2014, 289, 32773–32782. [Google Scholar] [CrossRef]
- Funane, K.; Ishii, T.; Ono, H.; Kobayashi, M. Changes in linkage pattern of glucan products induced by substitution of Lys residues in the dextransucrase. FEBS Lett. 2005, 579, 4739–4745. [Google Scholar] [CrossRef]
- Molina, M.; Moulis, C.; Monties, N.; Pizzut-Serin, S.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud-Simeon, M. Deciphering an Undecided Enzyme: Investigations of the Structural Determinants Involved in the Linkage Specificity of Alternansucrase. ACS Catal. 2019, 9, 2222–2237. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Miao, M.; Ma, Y.; Jiang, B.; Cui, S.W.; Jin, Z.; Zhang, T. Characterisations of Lactobacillus reuteri SK24. 003 glucansucrase implications for a-gluco-poly-and oligosaccharides biosynthesis. Food Chem. 2017, 222, 105–112. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Eeuwema, W.; Gerwig, G.; Dijkhuizen, L.; Kamerling, J.P. Structural characterization of bioengineered alpha-D-glucans produced by mutant glucansucrase GTF180 enzymes of Lactobacillus reuteri strain 180. Biomacromolecules 2009, 10, 580–588. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural Analysis of Bioengineered α-d-Glucan Produced by a Triple Mutant of the Glucansucrase GTF180 Enzyme from Lactobacillus reuteri Strain 180: Generation of (α1→4) Linkages in a Native (1→3) (1→6)-α-d-Glucan. Biomacromolecules 2008, 9, 2251–2258. [Google Scholar] [CrossRef][Green Version]
- Vuillemin, M.; Grimaud, F.; Claverie, M.; Rolland-Sabaté, A.; Garnier, C.; Lucas, P.; Monsan, P.; Dols-Lafargue, M.; Remaud-Siméon, M.; Moulis, C. A dextran with unique rheological properties produced by the dextransucrase from Oenococcus kitaharae DSM 17330. Carbohydr Polym. 2018, 179, 10–18. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Miller, A.W.; Robyt, J.F. Detection of dextransucrase and levansucrase on polyacrylamide gels by the periodic acid-Schiff stain: Staining artifacts and their prevention. Anal. Biochem. 1986, 156, 357–363. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ahmed, Z.; Wang, Y.; Anjum, N.; Ahmad, A.; Khan, S.T. Characterization of exopolysaccharide produced by Lactobacillus kefiranofaciens ZW3 isolated from Tibet kefir—Part II. Food Hydrocoll. 2013, 30, 343–350. [Google Scholar] [CrossRef]
- Schormann, N.; Patel, M.; Thannickal, L.; Purushotham, S.; Wu, R.; Mieher, J.L.; Wu, H.; Deivanayagam, C. The catalytic domains of Streptococcus mutans glucosyltransferases: A structural analysis. Acta Cryst. 2023, F79, 119–127. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Dobruchowska, J.M.; Gerwig, G.J.; Dijkhuizen, L. Characterization of the Functional Roles of Amino Acid Residues in Acceptor-binding Subsite +1 in the Active Site of the Glucansucrase GTF180 from Lactobacillus reuteri 180. J. Biol. Chem. 2015, 290, 30131–30141. [Google Scholar] [CrossRef]
- Kang, H.K.; Oh, J.S.; Kim, D. Molecular characterization and expression analysis of the glucansucrase DSRWC from Weissella cibaria synthesizing α-(1→6) glucan. FEMS Microbiol. Lett. 2009, 292, 33–41. [Google Scholar] [CrossRef]
- İspirli, H.; Yüzer, M.O.; Skory, C.; Colquhoun, I.J.; Sağdıç, O.; Dertli, E. Characterization of a glucansucrase from Lactobacillus reuteri E81 and production of malto-oligosaccharides. Biocatal. Biotransformation 2019, 37, 421–430. [Google Scholar] [CrossRef]
- Yu, L.; Qian, Z.; Ge, J.; Du, R. Glucansucrase Produced by Lactic Acid Bacteria: Structure, Properties, and Applications. Fermentation 2022, 8, 629. [Google Scholar] [CrossRef]
- Li, M.Q.; Zhang, H.B.; Li, Y.; Hu, X.-Q.; Yang, J.W. The thermoduric effects of site-directed mutagenesis of proline and lysine on dextransucrase from Leuconostoc mesenteroides 0326. Int. J. Biol. Macromol. 2018, 107, 1641–1649. [Google Scholar] [CrossRef]
- Robyt, J.F.; Walseth, T.F. Production, purification, and properties of dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 1979, 68, 95–111. [Google Scholar] [CrossRef]
- Yi, A.-R.; Lee, S.-R.; Jang, M.-U.; Park, J.-M.; Eom, H.-J.; Han, N.S.; Kim, T.-J. Cloning of dextransucrase gene from Leuconostoc citreum HJ-P4 and its high-level expression in E. coli by low temperature induction. J. Microbiol. Biotechnol. 2009, 19, 829–835. [Google Scholar]
- van Leeuwen, S.S.; Kralj, S.; van Geel-Schutten, I.H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural analysis of the α-D-glucan (EPS180) produced by the Lactobacillus reuteri strain 180 glucansucrase GTF180 enzyme. Carbohydr. Res. 2008, 343, 1237–1250. [Google Scholar] [CrossRef]
- Patel, S.; Majumder, A.; Goyal, A. Potentials of exopolysaccharides from lactic acid bacteria. Indian J. Microbiol. 2012, 52, 3–12. [Google Scholar] [CrossRef]
- Naessens, M.; Cerdobbel, A.; Soetaert, W.; Vandamme, E.J. Leuconostoc dextransucrase and dextran: Production, properties and applications. J. Chem. Technol. Biotechnol. 2005, 80, 8445–8860. [Google Scholar] [CrossRef]
- Saravanan, C.; Kumar, H.; Shetty, P.K.H. Isolation and characterization of exopolysaccharide from Leuconostoc lactis KC117496 isolated from idli batter. Int. J. Biol. Macromol. 2016, 90, 100–106. [Google Scholar] [CrossRef]
| CaCl2, mM | URE 13-300 | U13M1 |
|---|---|---|
| Relative Activity, % | Relative Activity, % | |
| Control | 100 | 100 |
| 5 | 152 ± 2.3 | 117 ± 2.0 |
| 10 | 142 ± 1.5 | 123 ± 1.6 |
| 15 | 156 ± 1.5 | 127 ± 3.2 |
| 20 | 176 ± 4.1 | 133 ± 1.2 |
| 25 | 178 ± 3.0 | 110 ± 2.4 |
| Metal Ions, 5 mM | URE 13-300 | U13M1 |
|---|---|---|
| Relative Activity, % | Relative Activity, % | |
| Control | 100 | 100 |
| MgCl2 | 110 ± 3.1 | 54 ± 1.2 |
| FeSO4 | 154 ± 0.5 | 44 ± 1.0 |
| BaCl2 | 128 ± 1.6 | 32 ± 0.6 |
| MnSO4 | 120 ± 0.8 | 22 ± 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelova, S.; Vasileva, T.; Bivolarski, V.; Iliev, I. Impact of G449 Single-Point Mutation on Glucansucrase URE 13-300 Enzyme Properties and Polysaccharide Structure. Catalysts 2023, 13, 1455. https://doi.org/10.3390/catal13121455
Angelova S, Vasileva T, Bivolarski V, Iliev I. Impact of G449 Single-Point Mutation on Glucansucrase URE 13-300 Enzyme Properties and Polysaccharide Structure. Catalysts. 2023; 13(12):1455. https://doi.org/10.3390/catal13121455
Chicago/Turabian StyleAngelova, Stanimira, Tonka Vasileva, Veselin Bivolarski, and Ilia Iliev. 2023. "Impact of G449 Single-Point Mutation on Glucansucrase URE 13-300 Enzyme Properties and Polysaccharide Structure" Catalysts 13, no. 12: 1455. https://doi.org/10.3390/catal13121455
APA StyleAngelova, S., Vasileva, T., Bivolarski, V., & Iliev, I. (2023). Impact of G449 Single-Point Mutation on Glucansucrase URE 13-300 Enzyme Properties and Polysaccharide Structure. Catalysts, 13(12), 1455. https://doi.org/10.3390/catal13121455






