Novel Biotransformation of Maslinic Acid to MA-2-O-β-D-Glucoside by UDP-Glycosyltransferases from Bacillus subtilis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression and Purification of YojK, YjiC, and UGT109A3
2.2. Enzymatic Activities of UTGs for MA Glycosylation
2.3. Identification of MA Glycosylation Products
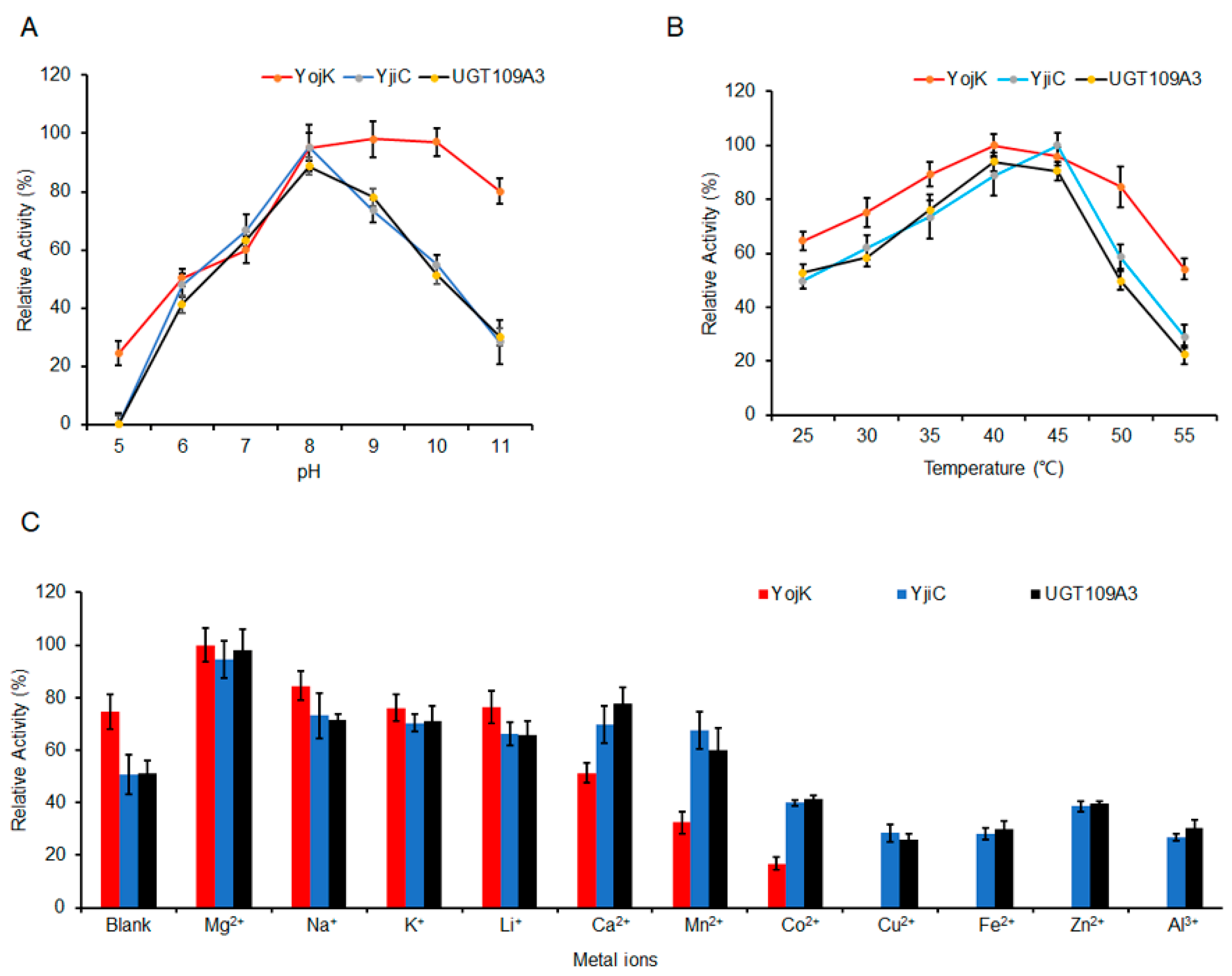
2.4. The Optimization of MA Glycosylation by UGTs
2.5. Exploring the Essential Residues of YojK, YjiC, and UGT109A3
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plasmid Construction and Heterologous Protein Expression
3.3. Protein Purification
3.4. Enzymatic Assays for MA Glycosylation In Vitro
3.5. HPLC Analysis
3.6. LC-MS and NMR Identification of the MA-Glycosides
3.7. Determination of Solubility
3.8. The Optimization of Enzymatic Glycosylation Reaction
3.9. Molecular Docking
3.10. Site-Directed Mutagenesis
3.11. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sheng, H.; Sun, H. Synthesis, biology and clinical significance of pentacyclic triterpenes: A multi-target approach to prevention and treatment of metabolic and vascular diseases. Nat. Prod. Rep. 2011, 28, 543–593. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, S.; Kim, J.; Mijakovic, I.; Jung, K.H.; Choi, G.; Kim, S.C.; Kim, Y.J. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 2019, 37, 107394. [Google Scholar] [CrossRef]
- Lu, K.W.; Yang, M.D.; Peng, S.F.; Chen, J.C.; Chen, P.Y.; Chen, H.Y.; Lu, T.J.; Chueh, F.S.; Lien, J.C.; Lai, K.C.; et al. Maslinic Acid Induces DNA Damage and Impairs DNA Repair in Human Cervical Cancer HeLa Cells. Anticancer Res. 2020, 40, 6869–6877. [Google Scholar] [CrossRef]
- Yap, W.H.; Ooi, B.K.; Ahmed, N.; Lim, Y.M. Maslinic acid modulates secreted phospholipase A2-IIA (sPLA2-IIA)-mediated inflammatory effects in macrophage foam cells formation. J. Biosci. 2018, 43, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Cabra, N.; Vega-Granados, K.; Moya-Andérico, L.; Vukomanovic, M.; Parra, A.; Álvarez de Cienfuegos, L.; Torrents, E. Novel Oleanolic and Maslinic Acid Derivatives as a Promising Treatment against Bacterial Biofilm in Nosocomial Infections: An in Vitro and in Vivo Study. ACS Infect. Dis. 2019, 5, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.; Rivas, F.; Lopez, P.E.; Garcia-Granados, A.; Martinez, A.; Albericio, F.; Marquez, N.; Muñoz, E. Solution- and solid-phase synthesis and anti-HIV activity of maslinic acid derivatives containing amino acids and peptides. Bioorg. Med. Chem. 2009, 17, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, P.; Zhao, D.; Ye, L.; Dai, L.; Zhang, X.; Sun, Y.; Zheng, J.; Bi, C. Construction of Escherichia coli cell factories for crocin biosynthesis. Microb. Cell Fact. 2019, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, J.; Yao, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Yang, J.; Sun, Y. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 2017, 248, 69–76. [Google Scholar] [CrossRef]
- Luo, S.L.; Dang, L.Z.; Zhang, K.Q.; Liang, L.M.; Li, G.H. Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett. Appl. Microbiol. 2015, 60, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Gurung, R.B.; Kim, E.H.; Oh, T.J.; Sohng, J.K. Enzymatic synthesis of apigenin glucosides by glucosyltransferase (YjiC) from Bacillus licheniformis DSM 13. Mol. Cells 2013, 36, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, J.; Yang, J.; Men, Y.; Zeng, Y.; Cai, Y.; Sun, Y. Enzymatic Synthesis of Novel Glycyrrhizic Acid Glucosides Using a Promiscuous Bacillus Glycosyltransferase. Catalysts 2018, 8, 615. [Google Scholar] [CrossRef]
- Nadeem, A.; Xu, K.; Wang, J.N.; Li, C. Novel catalytic glycosylation of Glycyrrhetinic acid by UDP-Glycosyltransferases from Bacillus subtilis. Biochem. Eng. J. 2020, 162 (Suppl. 16), 107723. [Google Scholar]
- Ali, M.Y.; Chang, Q.; Yan, Q.; Qian, Z.; Guo, X.; Thow, K.; Wu, J.; Zhang, Y.; Feng, Y. Highly Efficient Biosynthesis of Glycyrrhetinic Acid Glucosides by Coupling of Microbial Glycosyltransferase to Plant Sucrose Synthase. Front. Bioeng. Biotechnol. 2021, 9, 645079. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, H.; Qu, Y.; Zhang, X.; Zhang, J.; Dai, L. Biocatalytic Synthesis of a Novel Bioactive Ginsenoside Using UDP-Glycosyltransferase from Bacillus subtilis 168. Catalysts 2020, 10, 289. [Google Scholar] [CrossRef]
- Xu, S.H.; Chen, H.L.; Fan, Y.; Xu, W.; Zhang, J. Application of tandem biotransformation for biosynthesis of new pentacyclic triterpenoid derivatives with neuroprotective effect. Bioorg. Med. Chem. Lett. 2020, 30, 126947. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Chen, R.; Li, J.; Wang, R.; Chen, D.; Dou, X.; Dai, J. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org. Lett. 2014, 16, 4874–4877. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Qin, L.; Hu, Y.; Huang, J.W.; Hu, Z.; Min, J.; Sun, Y.; Guo, R.T. Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity. Biochem. Biophys. Res. Commun. 2021, 534, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, C.; Xiang, Q.; Zhao, N.; Luo, Y.; Bao, R. Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis. Int. J. Biol. Macromol. 2021, 166, 806–817. [Google Scholar] [CrossRef]
- Bahadur, G.R.; Youn, G.S.; Dipesh, D.; Thi, L.T.; Rae, J.N.; Jin, J.H.; Jin, O.T.; Kyung, S.J. Synthesis of Curcumin Glycosides with Enhanced Anticancer Properties Using One-Pot Multienzyme Glycosylation Technique. J. Microbiol. Biotechnol. 2017, 27, 1639–1648. [Google Scholar]
- Xu, K.; Zhao, Y.J.; Ahmad, N.; Wang, J.N.; Lv, B.; Wang, Y.; Ge, J.; Li, C. O-glycosyltransferases from Homo sapiens contributes to the biosynthesis of Glycyrrhetic Acid 3-O-mono-β-D-glucuronide and Glycyrrhizin in Saccharomyces cerevisiae. Synth. Syst. Biotechnol. 2021, 6, 173–179. [Google Scholar] [CrossRef] [PubMed]


| C | 1H NMR (ppm) | 13C NMR (ppm) |
|---|---|---|
| 1 | 1.93, 1H, dd, J = 4.4, 13.0 Hz 0.88, 1H, overlapped | 44.71 |
| 2 | 3.55, 1H, m | 79.44 |
| 3 | 2.91, 1H, d, J = 9.2 Hz | 80.41 |
| 4 | - | 39.52 |
| 5 | 0.79, 1H, br d, J = 11.3 Hz | 54.73 |
| 6 | 1.49, 1H, m 1.33, 1H, m | 18.37 |
| 7 | 1.61, 1H, m 1.43, 1H, m | 32.75 |
| 8 | - | 39.36 |
| 9 | 1.56, 1H, m | 47.50 |
| 10 | - | 38.11 |
| 11 | 1.85, 1H, m 1.48, 1H, m | 23.12 |
| 12 | 5.17, 1H, br s | 121.67 |
| 13 | - | 144.58 |
| 14 | - | 41.85 |
| 15 | 1.68, 1H, m 0.98, 1H, m | 27.67 |
| 16 | 1.85, 2H, m | 23.56 |
| 17 | - | 45.93 |
| 18 | 2.76, 1H, J = 3.6, 15.6 Hz | 41.32 |
| 19 | 1.60, 1H, m 1.05, 1H, m | 46.25 |
| 20 | - | 30.91 |
| 21 | 1.32, 1H, m 1.14, 1H, m | 33.89 |
| 22 | 1.43, 1H, m 1.25, 1H, m | 32.64 |
| 23 | 0.74, 3H, s | 17.75 |
| 24 | 0.96, 3H, s | 29.26 |
| 25 | 0.90, 3H, s | 16.62 |
| 26 | 0.72, 3H, s | 17.32 |
| 27 | 1.10, 3H, s | 26.15 |
| 28 | - | 179.29 |
| 29 | 0.88, 3H, s | 33.36 |
| 30 | 0.88, 3H, s | 23.87 |
| 1′ | 4.23, 1H, d, J = 7.8 Hz | 102.82 |
| 2′ | 2.94, 1H, t, J = 8.2 Hz | 73.80 |
| 3′ | 3.14, 1H, m | 77.00 |
| 4′ | 3.06, 1H, t, J = 9.0 Hz | 70.41 |
| 5′ | 3.15, 1H, m | 77.18 |
| 6′ | 3.65, 1H, brd, J = 13.6 Hz 3.43, 1H, dd, J = 5.8, 13.6 Hz | 61.42 |
| Enzymes | Km (mM) | Kcat (s−1) |
|---|---|---|
| YojK | 0.23 ± 0.02 | 4.61 ± 0.35 |
| YjiC | 0.48 ± 0.05 | 3.31 ± 0.12 |
| UGT109A3 | 0.52 ± 0.03 | 3.78 ± 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, F.; Chen, J.; Zhang, Y.; Sun, Y.; Liu, Y.; Yu, Y.; Xu, K.; Cai, H. Novel Biotransformation of Maslinic Acid to MA-2-O-β-D-Glucoside by UDP-Glycosyltransferases from Bacillus subtilis. Catalysts 2022, 12, 884. https://doi.org/10.3390/catal12080884
Hu F, Chen J, Zhang Y, Sun Y, Liu Y, Yu Y, Xu K, Cai H. Novel Biotransformation of Maslinic Acid to MA-2-O-β-D-Glucoside by UDP-Glycosyltransferases from Bacillus subtilis. Catalysts. 2022; 12(8):884. https://doi.org/10.3390/catal12080884
Chicago/Turabian StyleHu, Fen, Jiaxin Chen, Yunfeng Zhang, Yuxi Sun, Yan Liu, Yuan Yu, Ke Xu, and Haifeng Cai. 2022. "Novel Biotransformation of Maslinic Acid to MA-2-O-β-D-Glucoside by UDP-Glycosyltransferases from Bacillus subtilis" Catalysts 12, no. 8: 884. https://doi.org/10.3390/catal12080884
APA StyleHu, F., Chen, J., Zhang, Y., Sun, Y., Liu, Y., Yu, Y., Xu, K., & Cai, H. (2022). Novel Biotransformation of Maslinic Acid to MA-2-O-β-D-Glucoside by UDP-Glycosyltransferases from Bacillus subtilis. Catalysts, 12(8), 884. https://doi.org/10.3390/catal12080884






