Catalyst-Based Biomolecular Logic Gates
Abstract
1. Introduction
1.1. Fundamentals of Logic Gates
1.2. Early History of Biomolecular Logic Gates
2. Protein-Based Logic Gates
2.1. Allosteric Enzymes as Logic Gates
2.2. Engineered Protein Logic Gates
2.3. Biosensors Based on Enzymatic Logic Gates
2.4. Protease-Based Logic Gates
3. Nucleic Acid-Based Logic Gates
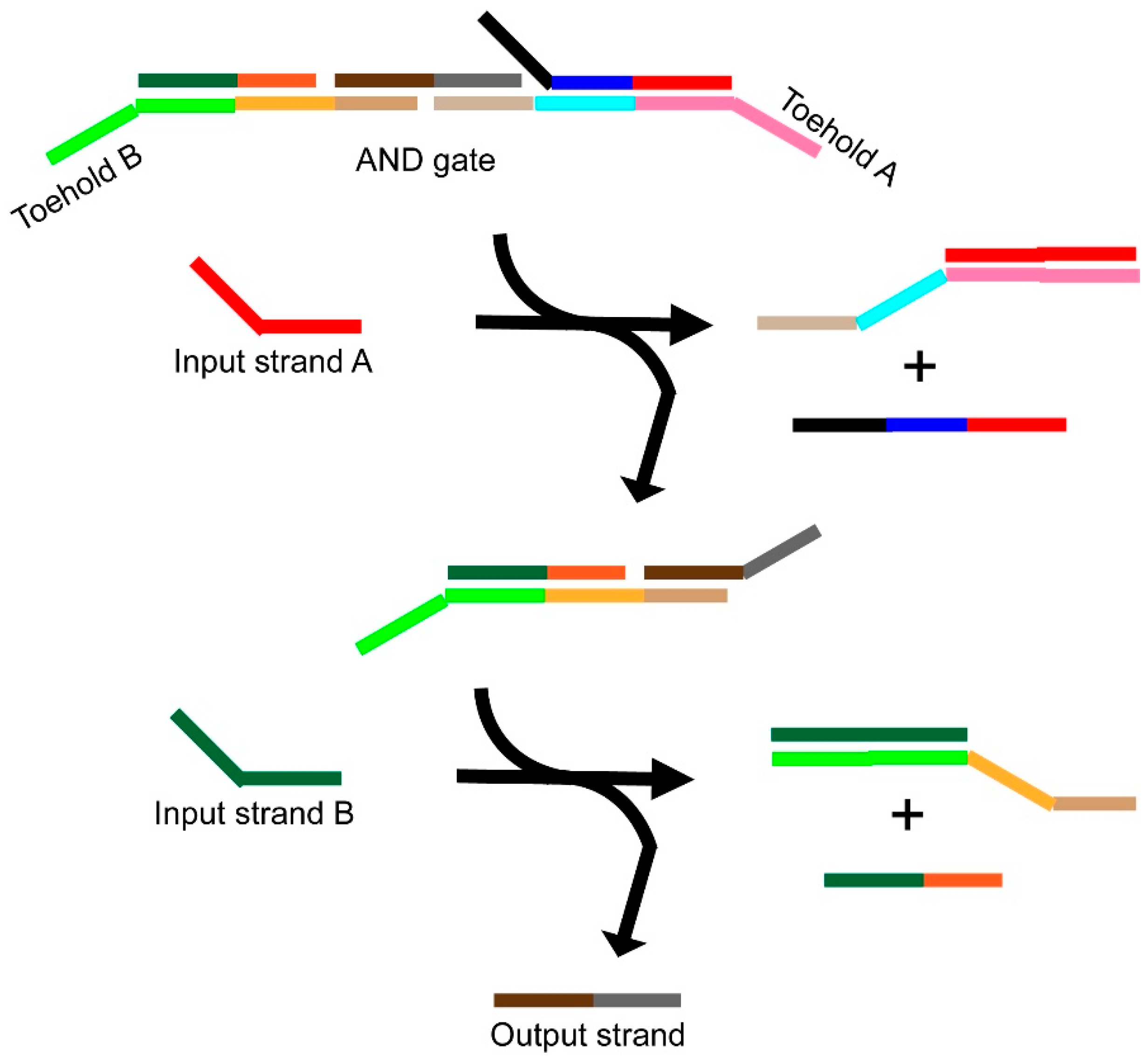
3.1. Catalytic (Entropy-Driven) Toehold-Mediated Strand Displacement
3.2. Using DNAzymes as Part of Logic Gates
3.3. Catalytic G-Quadruplexes for Visualizing the Output of DNA Logic Gates
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cui, S.; Lv, X.; Xu, X.; Chen, T.; Zhang, H.; Liu, Y.; Li, J.; Du, G.; Ledesma-Amaro, R.; Liu, L. Multilayer Genetic Circuits for Dynamic Regulation of Metabolic Pathways. ACS Synth. Biol. 2021, 10, 1587–1597. [Google Scholar] [CrossRef]
- Hoynes-O’Connor, A.; Moon, T.S. Programmable genetic circuits for pathway engineering. Curr. Opin. Biotechnol. 2015, 36, 115–121. [Google Scholar] [CrossRef]
- Katz, E.; Wang, J.; Privman, M.; Halámek, J. Multianalyte digital enzyme biosensors with built-in boolean logic. Anal. Chem. 2012, 84, 5463–5469. [Google Scholar] [CrossRef]
- Halámek, J.; Windmiller, J.R.; Zhou, J.; Chuang, M.C.; Santhosh, P.; Strack, G.; Arugula, M.A.; Chinnapareddy, S.; Bocharova, V.; Wang, J.; et al. Multiplexing of injury codes for the parallel operation of enzyme logic gates. Analyst 2010, 135, 2249–2259. [Google Scholar] [CrossRef]
- Privman, M.; Tam, T.K.; Bocharova, V.; Halámek, J.; Wang, J.; Katz, E. Responsive interface switchable by logically processed physiological signals: Toward “smart” actuators for signal amplification and drug delivery. ACS Appl. Mater. Interfaces 2011, 3, 1620–1623. [Google Scholar] [CrossRef]
- Zhou, J.; Halámek, J.; Bocharova, V.; Wang, J.; Katz, E. Bio-logic analysis of injury biomarker patterns in human serum samples. Talanta 2011, 83, 955–959. [Google Scholar] [CrossRef]
- Evans, A.C.; Thadani, N.N.; Suh, J. Biocomputing nanoplatforms as therapeutics and diagnostics. J. Control. Release 2016, 240, 387–393. [Google Scholar] [CrossRef]
- Privman, V.; Arugula, M.A.; Halámek, J.; Pita, M.; Katz, E. Network analysis of biochemical logic for noise reduction and stability: A system of three coupled enzymatic and gates. J. Phys. Chem. B 2009, 113, 5301–5310. [Google Scholar] [CrossRef][Green Version]
- Rafael, S.P.; Vallée-Bélisle, A.; Fabregas, E.; Plaxco, K.; Palleschi, G.; Ricci, F. Employing the metabolic “branch point effect” to generate an all-or-none, digital-like response in enzymatic outputs and enzyme-based sensors. Anal. Chem. 2012, 84, 1076–1082. [Google Scholar] [CrossRef]
- Halámek, J.; Zavalov, O.; Halámková, L.; Korkmaz, S.; Privman, V.; Katz, E. Enzyme-based logic analysis of biomarkers at physiological concentrations: And gate with double-sigmoid “filter” response. J. Phys. Chem. B 2012, 116, 4457–4464. [Google Scholar] [CrossRef]
- Zavalov, O.; Bocharova, V.; Halámek, J.; Halámková, L.; Korkmaz, S.; Arugula, M.A.; Chinnapareddy, S.; Katz, E.; Privman, V. Two-Input Enzymatic Logic Gates Made Sigmoid by Modifications of the Biocatalytic Reaction Cascades. Int. J. Unconv. Comput. 2012, 8, 347–365. [Google Scholar]
- Bakshi, S.; Zavalov, O.; Halámek, J.; Privman, V.; Katz, E. Modularity of biochemical filtering for inducing sigmoid response in both inputs in an enzymatic and gate. J. Phys. Chem. B 2013, 117, 9857–9865. [Google Scholar] [CrossRef]
- Benenson, Y. Biomolecular computing systems: Principles, progress and potential. Nat. Rev. Genet. 2012, 13, 455–468. [Google Scholar] [CrossRef]
- Bray, D. Protein molecules as computational elements in living cells. Nature 1995, 376, 307–312. [Google Scholar] [CrossRef]
- Magnasco, M.O. Chemical kinetics is turing universal. Phys. Rev. Lett. 1997, 78, 1190–1193. [Google Scholar] [CrossRef]
- Buisman, H.J.; ten Eikelder, H.M.M.; Hilbers, P.A.J.; Liekens, A.M.L. Computing algebraic functions with biochemical reaction networks. Artif. Life 2009, 15, 5–19. [Google Scholar] [CrossRef]
- Privman, V.; Strack, G.; Solenov, D.; Pita, M.; Katz, E. Optimization of enzymatic biochemical logic for noise reduction and scalability: How many biocomputing gates can be interconnected in a circuit? J. Phys. Chem. B 2008, 112, 11777–11784. [Google Scholar] [CrossRef]
- Privman, V. Control of noise in chemical and biochemical information processing. Isr. J. Chem. 2011, 51, 118–131. [Google Scholar] [CrossRef]
- Teo, J.J.Y.; Woo, S.S.; Sarpeshkar, R. Synthetic Biology: A Unifying View and Review Using Analog Circuits. IEEE Trans. Biomed. Circuits Syst. 2015, 9, 453–474. [Google Scholar] [CrossRef]
- Grozinger, L.; Amos, M.; Gorochowski, T.E.; Carbonell, P.; Oyarzún, D.A.; Stoof, R.; Fellermann, H.; Zuliani, P.; Tas, H.; Goñi-Moreno, A. Pathways to cellular supremacy in biocomputing. Nat. Commun. 2019, 10, 5250. [Google Scholar] [CrossRef]
- Katz, E. Boolean Logic Gates Realized with Enzyme-catalyzed Reactions—Unusual Look at Usual Chemical Reactions. ChemPhysChem 2019, 20, 9–22. [Google Scholar] [CrossRef]
- Sugita, M. The Switching Circuit Logically or Functionally Equivalent to a System of Biochemical Reactions. J. Phys. Soc. Japan 1961, 16, 737–740. [Google Scholar] [CrossRef]
- Credi, A. Molecules that make decisions. Angew. Chem.-Int. Ed. 2007, 46, 5472–5475. [Google Scholar] [CrossRef]
- Szaciłowski, K. Digital information processing in molecular systems. Chem. Rev. 2008, 108, 3481–3548. [Google Scholar] [CrossRef]
- Ratner, M.A.; Jortner, J. Molecular Electronics: Some Directions. In Molecular Electronics; Blackwell Science Ltd.: Hoboken, NJ, USA, 1997; pp. 5–72. [Google Scholar]
- De Silva, P.A.; Gunaratne, N.H.Q.; McCoy, C.P. A molecular photoionic and gate based on fluorescent signalling. Nature 1993, 364, 42–44. [Google Scholar] [CrossRef]
- Sivan, S.; Tuchman, S.; Lotan, N. A biochemical logic gate using an enzyme and its inhibitor. Part II: The logic gate. BioSystems 2003, 70, 21–33. [Google Scholar] [CrossRef]
- Sugita, M. Functional analysis of chemical systems in vivo using a logical circuit equivalent. V. Molecular biological interpretation of the self-reproducing automata theory and chemico-physical interpretation of information in biological systems. J. Theor. Biol. 1975, 53, 223–237. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Okamoto, M.; Katsurayama, A.; Tsukiji, M.; Aso, Y.; Hayashi, K. Dynamic behavior of enzymatic system realizing two-factor model. J. Theor. Biol. 1980, 83, 1–16. [Google Scholar] [CrossRef]
- Okamoto, M.; Sakai, T.; Hayashi, K. Biochemical switching device: Monocyclic enzyme system. Biotechnol. Bioeng. 1988, 32, 527–537. [Google Scholar] [CrossRef]
- Rosen, R. Two-factor models, neural nets and biochemical automata. J. Theor. Biol. 1967, 15, 282–297. [Google Scholar] [CrossRef]
- Arkin, A.; Ross, J. Computational functions in biochemical reaction networks. Biophys. J. 1994, 67, 560–578. [Google Scholar] [CrossRef]
- Sivan, S.; Lotan, N. A biochemical logic gate using an enzyme and its inhibitor. 1. The inhibitor as switching element. Biotechnol. Prog. 1999, 15, 964–970. [Google Scholar] [CrossRef]
- Zauner, K.P.; Conrad, M. Enzymatic computing. Biotechnol. Prog. 2001, 17, 553–559. [Google Scholar] [CrossRef]
- Baron, R.; Lioubashevski, O.; Katz, E.; Niazov, T.; Willner, I. Logic gates and elementary computing by enzymes. J. Phys. Chem. A 2006, 110, 8548–8553. [Google Scholar] [CrossRef]
- Baron, R.; Lioubashevski, O.; Katz, E.; Niazov, T.; Willner, I. Elementary arithmetic operations by enzymes: A model for metabolic pathway based computing. Angew. Chem.-Int. Ed. 2006, 45, 1572–1576. [Google Scholar] [CrossRef]
- Niazov, T.; Baron, R.; Katz, E.; Lioubashevski, O.; Willner, I. Concatenated logic gates using four coupled biocatalysts operating in series. Proc. Natl. Acad. Sci. USA 2006, 103, 17160–17163. [Google Scholar] [CrossRef]
- Vlasov, Y.G.; Tarantov, Y.A.; Bobrov, P.V. Analytical characteristics and sensitivity mechanisms of electrolyte-insulator-semiconductor system-based chemical sensors-a critical review. Anal. Bioanal. Chem. 2003, 376, 788–796. [Google Scholar] [CrossRef]
- Privman, V.; Pedrosa, V.; Melnikov, D.; Pita, M.; Simonian, A.; Katz, E. Enzymatic AND-gate based on electrode-immobilized glucose-6-phosphate dehydrogenase: Towards digital biosensors and biochemical logic systems with low noise. Biosens. Bioelectron. 2009, 25, 695–701. [Google Scholar] [CrossRef]
- Mailloux, S.; Gerasimova, Y.V.; Guz, N.; Kolpashchikov, D.M.; Katz, E. Bridging the Two Worlds: A Universal Interface between Enzymatic and DNA Computing Systems. Angew. Chem.-Int. Ed. 2015, 54, 6562–6566. [Google Scholar] [CrossRef]
- Katz, E. Transduction of Signals Generated by Enzyme Logic Gates. In Enzyme-Based Computing Systems; Wiley-VCH: Weinheim, Germany, 2019; pp. 113–149. ISBN 9783527819997. [Google Scholar]
- Chen, Z.; Kibler, R.D.; Hunt, A.; Busch, F.; Pearl, J.; Jia, M.; VanAernum, Z.L.; Wicky, B.I.M.M.; Dods, G.; Liao, H.; et al. De novo design of protein logic gates. Science 2020, 368, 78–84. [Google Scholar] [CrossRef]
- Daringer, N.M.; Dudek, R.M.; Schwarz, K.A.; Leonard, J.N. Modular Extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol. 2014, 3, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.A.K.; Der, B.S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychalski, E.A.; Ross, D.; Densmore, D.; Voigt, C.A. Genetic circuit design automation. Science 2016, 352. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Zhou, S.; Deng, Y. Transcription-Factor-based Biosensor Engineering for Applications in Synthetic Biology. ACS Synth. Biol. 2021, 10, 911–922. [Google Scholar] [CrossRef]
- Moškon, M.; Komac, R.; Zimic, N.; Mraz, M. Distributed biological computation: From oscillators, logic gates and switches to a multicellular processor and neural computing applications. Neural Comput. Appl. 2021, 33, 8923–8938. [Google Scholar] [CrossRef]
- Guiziou, S.; Ulliana, F.; Moreau, V.; Leclere, M.; Bonnet, J. An Automated Design Framework for Multicellular Recombinase Logic. ACS Synth. Biol. 2018, 7, 1406–1412. [Google Scholar] [CrossRef]
- Gorman, S.D.; Boehr, D.D. Energy and Enzyme Activity Landscapes of Yeast Chorismate Mutase at Cellular Concentrations of Allosteric Effectors. Biochemistry 2019, 58, 4058–4069. [Google Scholar] [CrossRef]
- O’Rourke, K.F.; Sahu, D.; Bosken, Y.K.; D’Amico, R.N.; Chang, C.-e.A.; Boehr, D.D. Coordinated Network Changes across the Catalytic Cycle of Alpha Tryptophan Synthase. Structure 2019, 27, 1405–1415.e5. [Google Scholar] [CrossRef]
- Boehr, D.D.; D’Amico, R.N.; O’Rourke, K.F. Engineered control of enzyme structural dynamics and function. Protein Sci. 2018, 27, 825–838. [Google Scholar] [CrossRef]
- Gorman, S.D.; D’Amico, R.N.; Winston, D.S.; Boehr, D.D. Engineering allostery into proteins. Adv. Exp. Med. Biol. 2019, 1163, 359–384. [Google Scholar] [CrossRef]
- Zhong, W.; Cui, L.; Goh, B.C.; Cai, Q.; Ho, P.; Chionh, Y.H.; Yuan, M.; Sahili, A.E.; Fothergill-Gilmore, L.A.; Walkinshaw, M.D.; et al. Allosteric pyruvate kinase-based “logic gate” synergistically senses energy and sugar levels in Mycobacterium tuberculosis. Nat. Commun. 2017, 8, 1986. [Google Scholar] [CrossRef] [PubMed]
- Katz, E. Conclusions and Perspectives: Where Are We Going? In Enzyme-Based Computing Systems; Wiley-VCH: Weinheim, Germany, 2019; pp. 371–382. ISBN 9783527819997. [Google Scholar]
- Agliari, E.; Altavilla, M.; Barra, A.; Dello Schiavo, L.; Katz, E. Notes on stochastic (bio)-logic gates: Computing with allosteric cooperativity. Sci. Rep. 2015, 5, 9415. [Google Scholar] [CrossRef] [PubMed]
- Galstyan, V.; Funk, L.; Einav, T.; Phillips, R. Combinatorial Control through Allostery. J. Phys. Chem. B 2019, 123, 2792–2800. [Google Scholar] [CrossRef] [PubMed]
- Guntas, G.; Mitchell, S.F.; Ostermeier, M. A Molecular Switch Created by In Vitro Recombination of Nonhomologous Genes. Chem. Biol. 2004, 11, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Guntas, G.; Mansell, T.J.; Kim, J.R.; Ostermeier, M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 11224–11229. [Google Scholar] [CrossRef]
- Wright, C.M.; Majumdar, A.; Tolman, J.R.; Ostermeier, M. NMR chracterization of an engineered domain fusion between maltose binding protein and TEM β-lactamase provides Insight into Its structure and allosteric mechanism. Proteins Struct. Funct. Bioinform. 2010, 78, 1423–1430. [Google Scholar] [CrossRef]
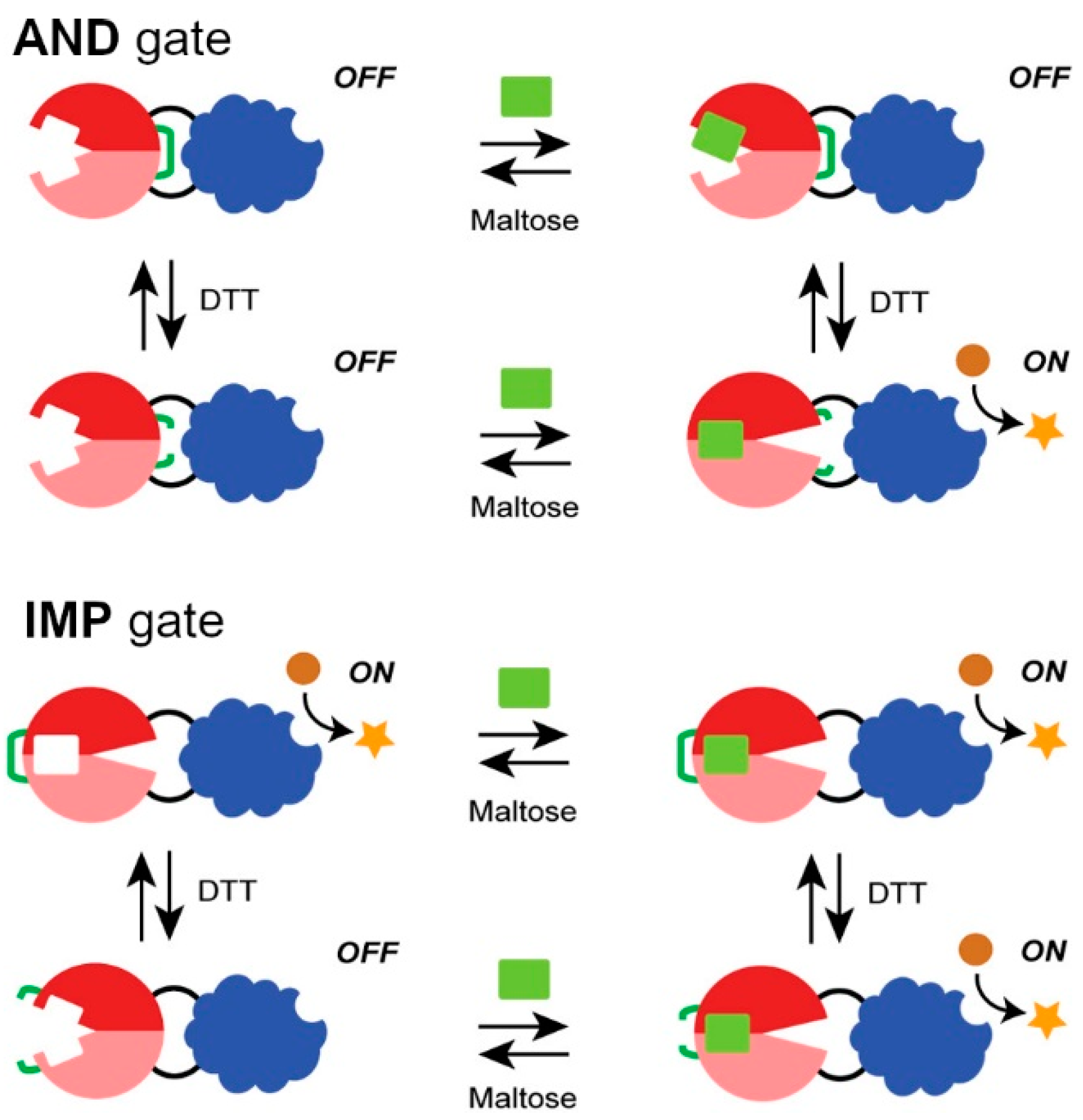
- Choi, J.H.; Ostermeier, M. Rational design of a fusion protein to exhibit disulfide-mediated logic gate behavior. ACS Synth. Biol. 2015, 4, 400–406. [Google Scholar] [CrossRef]
- Guo, Z.; Johnston, W.A.; Whitfield, J.; Walden, P.; Cui, Z.; Wijker, E.; Edwardraja, S.; Lantadilla, I.R.; Ely, F.; Vickers, C.; et al. Generalizable Protein Biosensors Based on Synthetic Switch Modules. J. Am. Chem. Soc. 2019, 141, 8128–8135. [Google Scholar] [CrossRef]
- Bollella, P.; Edwardraja, S.; Guo, Z.; Alexandrov, K.; Katz, E. Control of Allosteric Protein Electrochemical Switches with Biomolecular and Electronic Signals. J. Phys. Chem. Lett. 2020, 11, 5549–5554. [Google Scholar] [CrossRef]
- Bollella, P.; Bellare, M.; Kadambar, V.K.; Guo, Z.; Alexandrov, K.; Melman, A.; Katz, E. Boolean Logic Networks Mimicked with Chimeric Enzymes Activated/Inhibited by Several Input Signals. ChemPhysChem 2020, 21, 589–593. [Google Scholar] [CrossRef]
- Bollella, P.; Guo, Z.; Edwardraja, S.; Krishna Kadambar, V.; Alexandrov, K.; Melman, A.; Katz, E. Self-powered molecule release systems activated with chemical signals processed through reconfigurable Implication or Inhibition Boolean logic gates. Bioelectrochemistry 2021, 138, 107735. [Google Scholar] [CrossRef] [PubMed]
- Lopez, R.J.; Babanova, S.; Artyushkova, K.; Atanassov, P. Surface modifications for enhanced enzyme immobilization and improved electron transfer of PQQ-dependent glucose dehydrogenase anodes. Bioelectrochemistry 2015, 105, 78–87. [Google Scholar] [CrossRef] [PubMed]
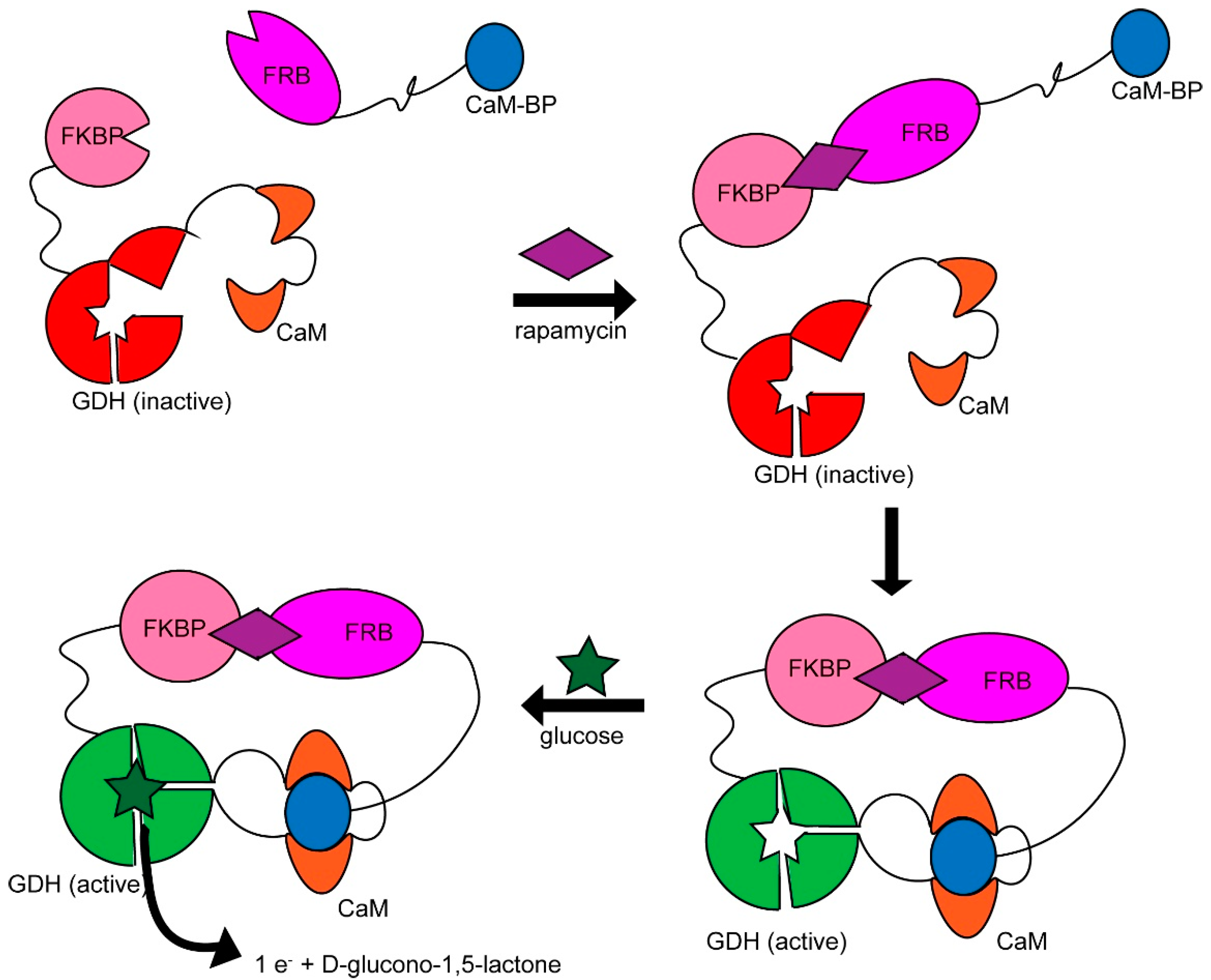
- Vishweshwaraiah, Y.L.; Chen, J.; Chirasani, V.R.; Tabdanov, E.D.; Dokholyan, N.V. Two-input protein logic gate for computation in living cells. Nat. Commun. 2021, 12, 6615. [Google Scholar] [CrossRef] [PubMed]
- Karginov, A.V.; Ding, F.; Kota, P.; Dokholyan, N.V.; Hahn, K.M. Engineered allosteric activation of kinases in living cells. Nat. Biotechnol. 2010, 28, 743–747. [Google Scholar] [CrossRef]
- Wang, J.; Jain, A.; McDonald, L.R.; Gambogi, C.; Lee, A.L.; Dokholyan, N.V. Mapping allosteric communications within individual proteins. Nat. Commun. 2020, 11, 3862. [Google Scholar] [CrossRef]
- Omersa, N.; Aden, S.; Kisovec, M.; Podobnik, M.; Anderluh, G. Design of Protein Logic Gate System Operating on Lipid Membranes. ACS Synth. Biol. 2020, 9, 316–328. [Google Scholar] [CrossRef]
- Okhokhonin, A.V.; Domanskyi, S.; Filipov, Y.; Gamella, M.; Kozitsina, A.N.; Privman, V.; Katz, E. Biomolecular Release from Alginate-modified Electrode Triggered by Chemical Inputs Processed through a Biocatalytic Cascade—Integration of Biomolecular Computing and Actuation. Electroanalysis 2018, 30, 426–435. [Google Scholar] [CrossRef]
- Gamella, M.; Privman, M.; Bakshi, S.; Melman, A.; Katz, E. DNA Release from Fe3+-Cross-Linked Alginate Films Triggered by Logically Processed Biomolecular Signals: Integration of Biomolecular Computing and Actuation. ChemPhysChem 2017, 18, 1811–1821. [Google Scholar] [CrossRef]
- Honarvarfard, E.; Gamella, M.; Poghossian, A.; Schöning, M.J.; Katz, E. An enzyme-based reversible Controlled NOT (CNOT) logic gate operating on a semiconductor transducer. Appl. Mater. Today 2017, 9, 266–270. [Google Scholar] [CrossRef]
- Jablonski, M.; Poghossian, A.; Molinnus, D.; Keusgen, M.; Katz, E.; Schöning, M.; Notice, C. Enzyme-based XOR logic gate with electronic transduction of the output signal. Int. J. Unconv. Comput. 2019, 14, 375–383. [Google Scholar]
- Karschuck, T.L.; Filipov, Y.; Bollella, P.; Schöning, M.; Katz, E. Not-XOR (NXOR) logic gate based on an enzyme-catalyzed reaction. Int. J. Unconv. Comput. 2019, 14, 235–242. [Google Scholar]
- Filipov, Y.; Domanskyi, S.; Wood, M.L.; Gamella, M.; Privman, V.; Katz, E. Experimental Realization of a High-Quality Biochemical XOR Gate. ChemPhysChem 2017, 18, 2908–2915. [Google Scholar] [CrossRef] [PubMed]
- Filipov, Y.; Gamella, M.; Katz, E. Nano-species Release System Activated by Enzyme-based XOR Logic Gate. Electroanalysis 2018, 30, 1281–1286. [Google Scholar] [CrossRef]
- Chung, H.K.; Lin, M.Z. On the cutting edge: Protease-based methods for sensing and controlling cell biology. Nat. Methods 2020, 17, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Gräwe, A.; Ranglack, J.; Weber, W.; Stein, V. Engineering artificial signalling functions with proteases. Curr. Opin. Biotechnol. 2020, 63, 1–7. [Google Scholar] [CrossRef]
- Chen, Z.; Elowitz, M.B. Programmable protein circuit design. Cell 2021, 184, 2284–2301. [Google Scholar] [CrossRef]
- Gao, X.J.; Chong, L.S.; Kim, M.S.; Elowitz, M.B. Programmable protein circuits in living cells. Science 2018, 361, 1252–1258. [Google Scholar] [CrossRef]
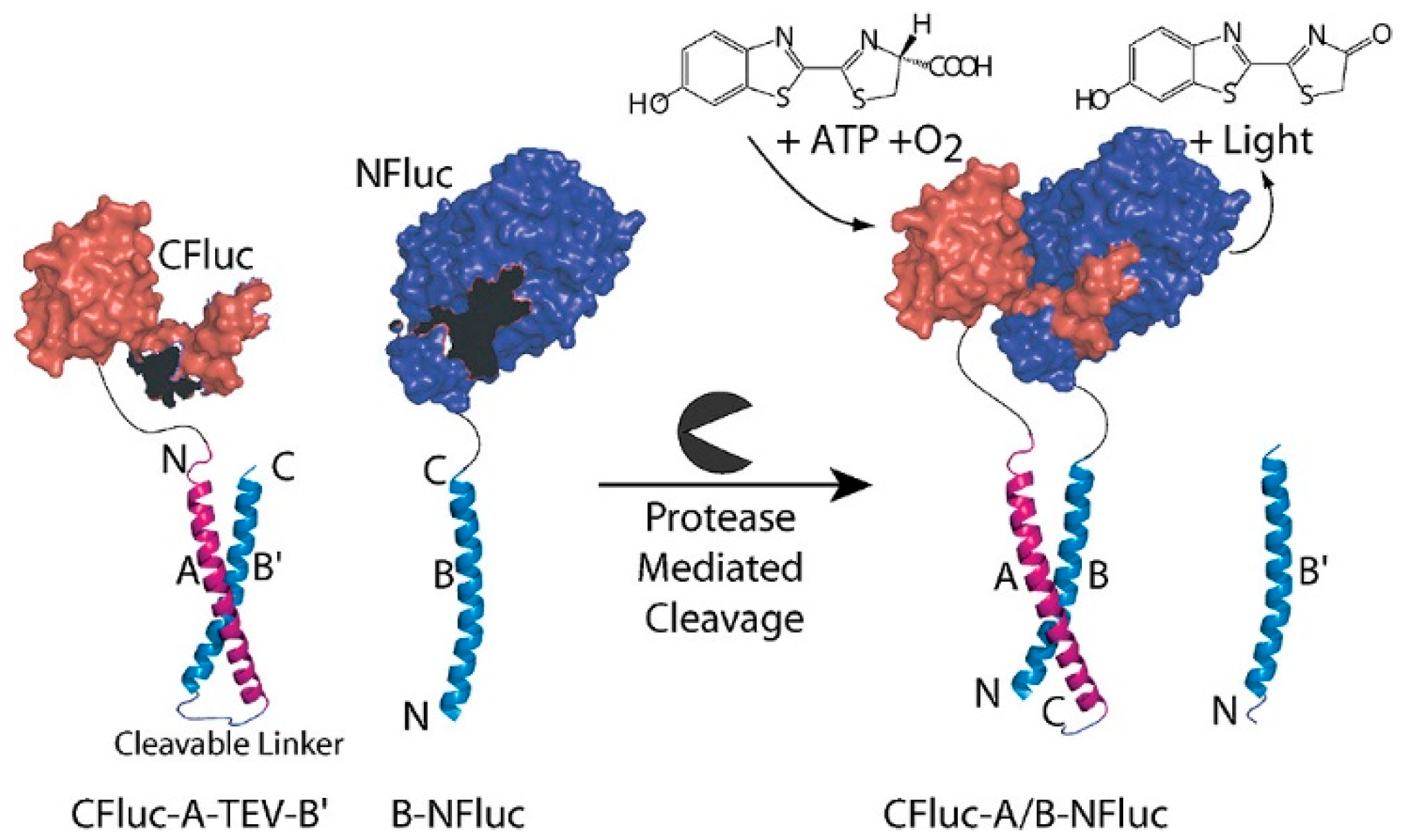
- Shekhawat, S.S.; Porter, J.R.; Sriprasad, A.; Ghosh, I. An autoinhibited coiled-coil design strategy for split-protein protease sensors. J. Am. Chem. Soc. 2009, 131, 15284–15290. [Google Scholar] [CrossRef][Green Version]
- Fernandez-Rodriguez, J.; Voigt, C.A. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 2016, 44, 6493–6502. [Google Scholar] [CrossRef]
- Seelig, G.; Soloveichik, D.; Zhang, D.Y.; Winfree, E. Enzyme-Free Nucleic Acid Logic Circuits. Science 2006, 314, 1585–1589. [Google Scholar] [CrossRef]
- Yang, J.; Wu, R.; Li, Y.; Wang, Z.; Pan, L.; Zhang, Q.; Lu, Z.; Zhang, C. Entropy-driven DNA logic circuits regulated by DNAzyme. Nucleic Acids Res. 2018, 46, 8532–8541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Turberfield, A.J.; Yurke, B.; Winfree, E. Engineering Entropy-Driven Reactions and Networks Catalyzed by DNA. Science 2009, 318, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009, 131, 17303–17314. [Google Scholar] [CrossRef]
- Thubagere, A.J.; Thachuk, C.; Berleant, J.; Johnson, R.F.; Ardelean, D.A.; Cherry, K.M.; Qian, L. Compiler-aided systematic construction of large-scale DNA strand displacement circuits using unpurified components. Nat. Commun. 2017, 8, 14373. [Google Scholar] [CrossRef]
- Qian, L.; Winfree, E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 99, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, F.; Yan, H.; Liu, Y. DNA based arithmetic function: A half adder based on DNA strand displacement. Nanoscale 2016, 8, 3775–3784. [Google Scholar] [CrossRef]
- Frezza, B.M.; Cockroft, S.L.; Ghadiri, M.R. Modular multi-level circuits from immobilized DNA-based logic gates. J. Am. Chem. Soc. 2007, 129, 14875–14879. [Google Scholar] [CrossRef]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Towards a DNA Nanoprocessor: Reusable Tile-Integrated DNA Circuits. Angew. Chem.-Int. Ed. 2016, 55, 10244–10247. [Google Scholar] [CrossRef]
- Qian, L.; Winfree, E. Parallel and scalable computation and spatial dynamics with DNA-based chemical reaction networks on a surface. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; Volume 8727, pp. 114–131. [Google Scholar]
- Woods, D.; Doty, D.; Myhrvold, C.; Hui, J.; Zhou, F.; Yin, P.; Winfree, E. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature 2019, 567, 366–372. [Google Scholar] [CrossRef]
- Srinivas, N.; Parkin, J.; Seelig, G.; Winfree, E.; Soloveichik, D. Enzyme-free nucleic acid dynamical systems. Science 2017, 358. [Google Scholar] [CrossRef]
- Fern, J.; Scalise, D.; Cangialosi, A.; Howie, D.; Potters, L.; Schulman, R. DNA Strand-Displacement Timer Circuits. ACS Synth. Biol. 2017, 6, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Han, H.; Guo, C.; Lin, X.; Chen, D.; Yang, S.; Yang, Q.; Li, F. Information processing using an integrated DNA reaction network. Nanoscale 2021, 13, 5706–5713. [Google Scholar] [CrossRef] [PubMed]
- Orbach, R.; Willner, B.; Willner, I. Catalytic nucleic acids (DNAzymes) as functional units for logic gates and computing circuits: From basic principles to practical applications. Chem. Commun. 2015, 51, 4144–4160. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Stefanovic, D.; Rudchenko, S. Exercises in molecular computing. Acc. Chem. Res. 2014, 47, 1845–1852. [Google Scholar] [CrossRef] [PubMed]
- Lakin, M.R.; Stojanovic, M.N.; Stefanovic, D. Implementing Molecular Logic Gates, Circuits Stefanovic, and Cascades Using DNAzymes. Adv. Unconv. Comput. 2017, 1–28. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Stefanovic, D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003, 21, 1069–1074. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Stefanovic, D. Deoxyribozyme-Based Half-Adder. J. Am. Chem. Soc. 2003, 125, 6673–6676. [Google Scholar] [CrossRef]
- Lederman, H.; Macdonald, J.; Stefanovic, D.; Stojanovic, M.N. Deoxyribozyme-based three-input logic gates and construction of a molecular full adder. Biochemistry 2006, 45, 1194–1199. [Google Scholar] [CrossRef]
- Macdonald, J.; Li, Y.; Sutovic, M.; Lederman, H.; Pendri, K.; Lu, W.; Andrews, B.L.; Stefanovic, D.; Stojanovic, M.N. Medium scale integration of molecular logic gates in an automaton. Nano Lett. 2006, 6, 2598–2603. [Google Scholar] [CrossRef]
- Pei, R.; Matamoros, E.; Liu, M.; Stefanovic, D.; Stojanovic, M.N. Training a molecular automaton to play a game. Nat. Nanotechnol. 2010, 5, 773–777. [Google Scholar] [CrossRef]
- Elbaz, J.; Wang, F.; Remacle, F.; Willner, I. PH-programmable DNA logic arrays powered by modular DNAzyme libraries. Nano Lett. 2012, 12, 6049–6054. [Google Scholar] [CrossRef] [PubMed]
- Prokup, A.; Hemphill, J.; Deiters, A. DNA computation: A photochemically controlled and gate. J. Am. Chem. Soc. 2012, 134, 3810–3815. [Google Scholar] [CrossRef] [PubMed]
- Orbach, R.; Remacle, F.; Levine, R.D.; Willner, I. DNAzyme-based 2:1 and 4:1 multiplexers and 1:2 demultiplexer. Chem. Sci. 2014, 5, 1074–1081. [Google Scholar] [CrossRef]
- Orbach, R.; Wang, F.; Lioubashevski, O.; Levine, R.D.; Remacle, F.; Willner, I. A full-adder based on reconfigurable DNA-hairpin inputs and DNAzyme computing modules. Chem. Sci. 2014, 5, 3381–3387. [Google Scholar] [CrossRef]
- Li, W.; Yang, Y.; Yan, H.; Liu, Y. Three-input majority logic gate and multiple input logic circuit based on DNA strand displacement. Nano Lett. 2013, 13, 2980–2988. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Li, T.; Dong, S.; Wang, E. Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mater. 2013, 25, 2440–2444. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Semova, S.; Kolpashchikov, D.; Macdonald, J.; Morgan, C.; Stefanovic, D. Deoxyribosyme-based ligase logic gates and their initial circuits. J. Am. Chem. Soc. 2005, 127, 6914–6915. [Google Scholar] [CrossRef]
- Penchovsky, R.; Breaker, R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005, 23, 1424–1433. [Google Scholar] [CrossRef]
- Penchovsky, R. Engineering integrated digital circuits with allosteric ribozymes for scaling up molecular computation and diagnostics. ACS Synth. Biol. 2012, 1, 471–482. [Google Scholar] [CrossRef]
- Kahan-Hanum, M.; Douek, Y.; Adar, R.; Shapiro, E. A library of programmable DNAzymes that operate in a cellular environment. Sci. Rep. 2013, 3, 1535. [Google Scholar] [CrossRef]
- Brown, C.W.; Lakin, M.R.; Stefanovic, D.; Graves, S.W. Catalytic molecular logic devices by DNAzyme displacement. ChemBioChem 2014, 15, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Lakin, M.R.; Brown, C.W.; Horwitz, E.K.; Fanning, M.L.; West, H.E.; Stefanovic, D.; Graves, S.W. Biophysically inspired rational design of structured chimeric substrates for DNAzyme Cascade Engineering. PLoS ONE 2014, 9, e110986. [Google Scholar] [CrossRef] [PubMed]
- Morihiro, K.; Ankenbruck, N.; Lukasak, B.; Deiters, A. Small molecule release and activation through DNA computing. J. Am. Chem. Soc. 2017, 139, 13909–13915. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, T.; Zhang, L.; Dong, S.; Wang, E. G-quadruplex DNAzyme based molecular catalytic beacon for label-free colorimetric logic gates. Biomaterials 2011, 32, 7318–7324. [Google Scholar] [CrossRef] [PubMed]
- Orbach, R.; Remacle, F.; Levine, R.D.; Willner, I. Logic reversibility and thermodynamic irreversibility demonstrated by DNAzyme-based Toffoli and Fredkin logic gates. Proc. Natl. Acad. Sci. USA 2012, 109, 21228–21233. [Google Scholar] [CrossRef]
- Li, H.; Guo, S.; Liu, Q.; Qin, L.; Dong, S.; Liu, Y.; Wang, E. Implementation of Arithmetic Functions on a Simple and Universal Molecular Beacon Platform. Adv. Sci. 2015, 2, 1500054. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Zhou, Z.; Dong, S.; Wang, E. Molecular aptamer beacon tuned DNA strand displacement to transform small molecules into DNA logic outputs. Chem. Commun. 2014, 50, 3321–3323. [Google Scholar] [CrossRef]
- Miyoshi, D.; Inoue, M.; Sugimoto, N. DNA logic gates based on structural polymorphism of telomere DNA molecules responding to chemical input signals. Angew. Chem.-Int. Ed. 2006, 45, 7716–7719. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Potassium-lead-switched G-quadruplexes: A new class of DNA logic gates. J. Am. Chem. Soc. 2009, 131, 15082–15083. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Moshe, M.; Elbaz, J.; Willner, I. Sensing of UO22+ and design of logic gates by the application of supramolecular constructs of ion-dependent DNAzymes. Nano Lett. 2009, 9, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, H.; Wang, D.; Zhao, Y.; Li, N.; Liu, F. DNA tetraplexes-based toehold activation for controllable DNA strand displacement reactions. J. Am. Chem. Soc. 2013, 135, 13628–13631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.M.; Liang, R.P.; Qiu, J.D. Colorimetric logic gates based on ion-dependent DNAzymes. J. Phys. Chem. C 2013, 117, 12352–12357. [Google Scholar] [CrossRef]
- Shlyahovsky, B.; Li, Y.; Lioubashevski, O.; Elbaz, J.; Willner, I. Logic gates and antisense DNA devices operating on a translator nucleic acid scaffold. ACS Nano 2009, 3, 1831–1843. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, L.; Zhang, Y.M.; Liang, R.P.; Qiu, J.D. DNA colorimetric logic gates based on triplex-helix molecular switch. J. Phys. Chem. C 2014, 118, 14410–14417. [Google Scholar] [CrossRef]
- Fan, D.; Wang, K.; Zhu, J.; Xia, Y.; Han, Y.; Liu, Y.; Wang, E. DNA-based visual majority logic gate with one-vote veto function. Chem. Sci. 2015, 6, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
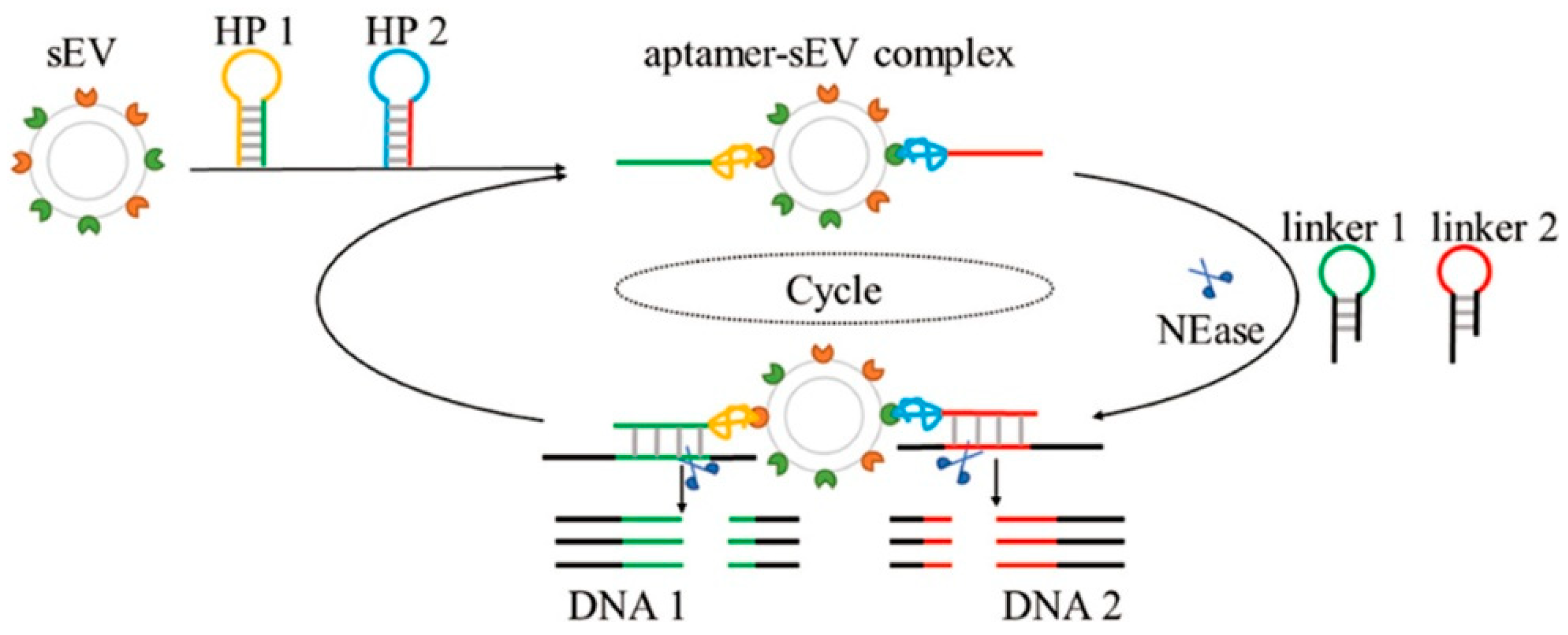
- Yu, Y.; Guo, Q.; Jiang, W.; Zhang, H.; Cai, C. Dual-Aptamer-Assisted and Logic Gate for Cyclic Enzymatic Signal Amplification Electrochemical Detection of Tumor-Derived Small Extracellular Vesicles. Anal. Chem. 2021, 93, 11298–11304. [Google Scholar] [CrossRef]
- Fink, T.; Lonzarić, J.; Praznik, A.; Plaper, T.; Merljak, E.; Leben, K.; Jerala, N.; Lebar, T.; Strmšek, Ž.; Lapenta, F.; et al. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat. Chem. Biol. 2019, 15, 115–122. [Google Scholar] [CrossRef]
- Barnea, G.; Strapps, W.; Herrada, G.; Berman, Y.; Ong, J.; Kloss, B.; Axel, R.; Lee, K.J. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. USA 2008, 105, 64–69. [Google Scholar] [CrossRef]
- Lee, Y.; Son, J.Y.; Kang, J.I.; Park, K.M.; Park, K.D. Hydrogen Peroxide-Releasing Hydrogels for Enhanced Endothelial Cell Activities and Neovascularization. ACS Appl. Mater. Interfaces 2018, 10, 18372–18379. [Google Scholar] [CrossRef]
- Kang, J.I.; Park, K.M. Advances in gelatin-based hydrogels for wound management. J. Mater. Chem. B 2021, 9, 1503–1520. [Google Scholar] [CrossRef]
- Badelt, S.; Grun, C.; Sarma, K.V.; Wolfe, B.; Shin, S.W.; Winfree, E. A domain-level DNA strand displacement reaction enumerator allowing arbitrary non-pseudoknotted secondary structures. J. R. Soc. Interface 2020, 17, 20190866. [Google Scholar] [CrossRef] [PubMed]
- Cherry, K.M.; Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 2018, 559, 370–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. RNA-cleaving DNAzymes: Old catalysts with new tricks for intracellular and in vivo applications. Catalysts 2018, 8, 550. [Google Scholar] [CrossRef]
- Li, X.; Yang, F.; Zhou, W.; Yuan, R.; Xiang, Y. Targeted and direct intracellular delivery of native DNAzymes enables highly specific gene silencing. Chem. Sci. 2020, 11, 8966–8972. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Zhu, G.; Chen, T.; Donovan, M.J.; Tan, W. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J. Am. Chem. Soc. 2015, 137, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhao, Y.; Xu, K.; Zhang, C.; Ma, Q.; Qi, L.; Chao, D.; Zheng, T.; Yang, L.; Miao, Y.; et al. Highly Specific, Single-step Cancer Cell Isolation with Multi-Aptamer-Mediated Proximity Ligation on Live Cell Membranes. Angew. Chem.-Int. Ed. 2020, 59, 23564–23568. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Fan, Z.; Yu, M.M.; Zhao, J.; Li, L. A Redox-Activatable DNA Nanodevice for Spatially-Selective, AND-Gated Imaging of ATP and Glutathione in Mitochondria. Nano Lett. 2021, 21, 10047–10053. [Google Scholar] [CrossRef]
- Wang, D.; Li, S.; Zhao, Z.; Zhang, X.; Tan, W. Engineering a Second-Order DNA Logic-Gated Nanorobot to Sense and Release on Live Cell Membranes for Multiplexed Diagnosis and Synergistic Therapy. Angew. Chem.-Int. Ed. 2021, 60, 15816–15820. [Google Scholar] [CrossRef]
- Li, Y.; Deng, J.; Han, Z.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.; Sun, J. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef]
- Kamar, O.; Sun, S.C.; Lin, C.H.; Chung, W.Y.; Lee, M.S.; Liao, Y.C.; Kolpashchikov, D.M.; Chuang, M.C. A mutation-resistant deoxyribozyme or gate for highly selective detection of viral nucleic acids. Chem. Commun. 2017, 53, 10592–10595. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, J.; Deiters, A. DNA computation in mammalian cells: MicroRNA logic operations. J. Am. Chem. Soc. 2013, 135, 10512–10518. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Zhang, C.; Lv, C.; Sun, Y.; Zhang, M.; Zhao, Y.; Yang, L.; Han, D.; Tan, W. Construction of a Multiple-Aptamer-Based DNA Logic Device on Live Cell Membranes via Associative Toehold Activation for Accurate Cancer Cell Identification. J. Am. Chem. Soc. 2019, 141, 12738–12743. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Stojanovic, M.N. Is there a future for DNA-based molecular devices in diabetes management? J. Diabetes Sci. Technol. 2007, 1, 440–444. [Google Scholar] [CrossRef]
- Gomes de Oliveira, A.G.; Dubovichenko, M.V.; ElDeeb, A.A.; Wanjohi, J.; Zablotskaya, S.; Kolpashchikov, D.M. RNA-Cleaving DNA Thresholder Controlled by Concentrations of miRNA Cancer Marker. ChemBioChem 2021, 22, 1750–1754. [Google Scholar] [CrossRef]
- Singh, A. Designing protein logic gates. Nat. Methods 2020, 17, 565. [Google Scholar] [CrossRef]
- Katz, E. Bioelectronic Interface between Enzyme-Based and DNA-Based Computing Systems. In Enzyme-Based Computing Systems; Wiley-VCH: Weinheim, Germany, 2019; pp. 335–355. ISBN 9783527819997. [Google Scholar]
- Groseclose, T.M.; Rondon, R.E.; Herde, Z.D.; Aldrete, C.A.; Wilson, C.J. Engineered systems of inducible anti-repressors for the next generation of biological programming. Nat. Commun. 2020, 11, 4440. [Google Scholar] [CrossRef]
- Groseclose, T.M.; Rondon, R.E.; Hersey, A.N.; Milner, P.T.; Kim, D.; Zhang, F.; Realff, M.J.; Wilson, C.J. Biomolecular Systems Engineering: Unlocking the Potential of Engineered Allostery via the Lactose Repressor Topology. Annu. Rev. Biophys. 2021, 50, 303–321. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, Y. Biocomputing for Portable, Resettable, and Quantitative Point-of-Care Diagnostics: Making the Glucose Meter a Logic-Gate Responsive Device for Measuring Many Clinically Relevant Targets. Angew. Chem.-Int. Ed. 2018, 57, 9702–9706. [Google Scholar] [CrossRef]
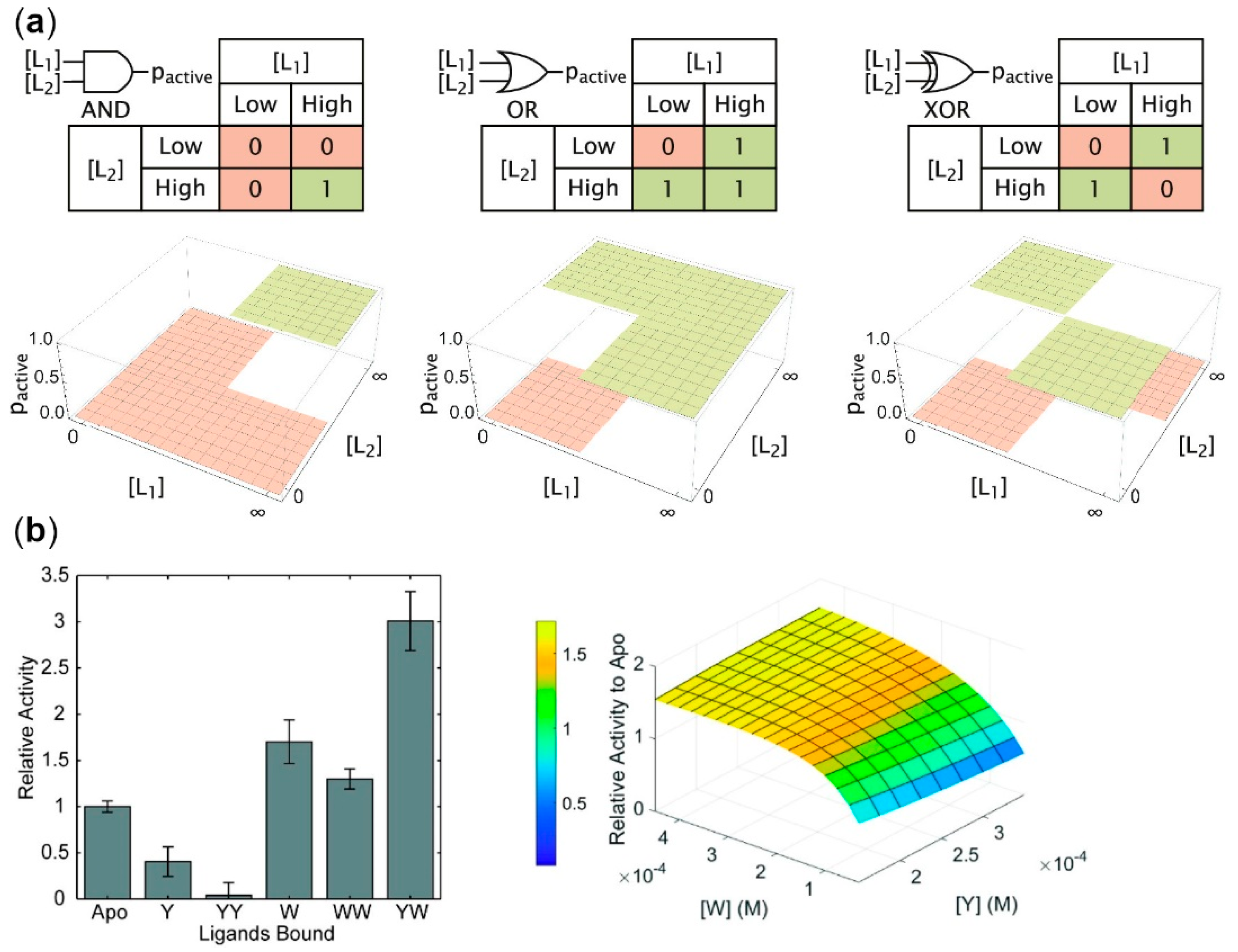



| Input Values | Output Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | AND | OR | INH | XOR | NAND | NOR | IMP | XNOR |
| 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winston, D.S.; Boehr, D.D. Catalyst-Based Biomolecular Logic Gates. Catalysts 2022, 12, 712. https://doi.org/10.3390/catal12070712
Winston DS, Boehr DD. Catalyst-Based Biomolecular Logic Gates. Catalysts. 2022; 12(7):712. https://doi.org/10.3390/catal12070712
Chicago/Turabian StyleWinston, Dennis S., and David D. Boehr. 2022. "Catalyst-Based Biomolecular Logic Gates" Catalysts 12, no. 7: 712. https://doi.org/10.3390/catal12070712
APA StyleWinston, D. S., & Boehr, D. D. (2022). Catalyst-Based Biomolecular Logic Gates. Catalysts, 12(7), 712. https://doi.org/10.3390/catal12070712







