Abstract
Halogenating enzymes have evolved in considerable mechanistic diversity. The apparent need for secondary metabolism coincides with the current need to introduce halogens in synthetic products. The potential of halogenating enzymes and, especially, vanadate-dependent haloperoxidases has been insufficiently exploited for synthetic purposes. In this work, we identified potential halogenase sequences by screening algal, fungal, and protobacterial sequence databases, structural modeling of putative halogenases, and mapping and comparing active sites. In a final step, individual haloperoxidases were expressed and kinetically characterized. A vanadate-dependent haloperoxidase from Rhodoplanes roseus was heterologously expressible by E. coli and could be purified to homogeneity. The kinetic data revealed a higher turnover number than the known VClPO-CI and no inhibitory effect from bromide, rendering this enzyme a promising biocatalyst. Other predicted haloperoxidases were not expressed successfully yet but these enzymes were predicted to be present in a wide taxonomic variety.
1. Introduction
Halogens can have a tremendous impact on the physicochemical properties of chemical compounds. The heavier halogens chlorine, bromine, and iodine are large and electron-withdrawing substituents and can form halogen bonds via their σ-hole [1,2,3,4,5,6]. On the one hand, introducing halogens is an important lever for chemical transformations such as substitutions, metal-halogen exchanges, or eliminations. On the other hand, these properties can have a considerable impact on the biological activity of an active compound. Therefore, halogens are a component of many active substances or their synthesis intermediates and taxonomically broad secondary metabolites [7,8,9]. Consequently, several halogenating enzymes have evolved in nature [9,10]. The most prominent halogenating enzymes are the flavin-dependent halogenases [11,12] and haloperoxidases [13,14,15]. Here, we focus on the enzyme family forming hypohalides from hydrogen peroxide and the respective halides. For catalytic activity, either a heme [13,16,17] or vanadate needs to be bound. It was hypothesized that the electrophilic hypohalide species reacts remotely from the active site and, thus, the regioselectivity depends exclusively on the reaction partners.
However, the number of halogenating enzymes studied is rather limited [9], although these enzymes can be synthetically valuable tools for synthesis, as precise doses of hypohalides can be used during the reaction to avoid over-halogenation. From a synthetic point of view, it is also problematic that, for example, in the case of the best-described haloperoxidase from Curvularia inaequalis, there is an evident substrate excess inhibition by chloride [18].
Here, we searched for novel haloperoxidases that are not catalytically inhibited at a substrate excess and do not release hypohalide from the active center such that the reaction takes place selectively in the active center. Initially, we analyzed sequence databases with algal, fungal, and proteobacterial sequences, thus focusing on a large taxonomic breadth. Representatives identified in this step as haloperoxidases with only moderate sequence similarity to known haloperoxidases were subjected to structure predictions and binding site comparisons to identify active sites with new structural characteristics. Selected candidates were subjected to expression studies and biochemical analyses (see Figure 1).
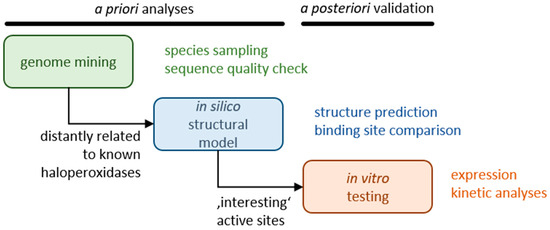
Figure 1.
A priori mining approach.
2. Results
2.1. Identification of Haloperoxidases from Sequence Databases
To obtain potential halogenase sequences, GenBank was screened for sequences from the Phaeophyceae (brown algae) using simple similarity searches on published sequences, e.g., from Ascophyllum nodosum [19]. The GenBank search yielded several sequences from Laminaria digitata, Saccharina japonica, and one from Fucus distichus (AAC35279.1). For further analysis, one sequence was chosen for Laminaria digitata (CAQ51446.1), which was found after stress elicitation and described as a vanadium-dependent haloperoxidase potentially involved in iodation. Additionally, one sequence was selected for Saccharina japonica (AYP65253.1) [20]. To extend the coverage, the transcriptome shotgun assembly database of NCBI was searched, which yielded multiple sequence candidates for Sagassum spp. (S. fusiforme GFKJ01014004.1, S. muticum GFKI01033812, and S. vulgare GEHA01064915) [21] and an additional sequence for Fucus ceranoides (HACY01003800.1). After comparison with the published sequences, the most likely reading frames were identified and potential frameshifts were corrected.
Similarly, to identify non-algal sequences, the sequence underlying the chloroperoxidase structure PDB ID 1IDQ_A from Curvularia inaequalis [22] was used as bait. The most similar fungal sequences not stemming from Curvularia (with a sequence similarity between 81 and 84%) were picked for further analysis and represented sequences from several Bipolaris species as well as one each from Stemphylium lycopersici and Exserohilum turcica. All species stem from the fungal Pleosporaceae family and mostly represent plant pathogens.
In addition, sequences were selected from proteobacteria (a sequence similarity between 43 and 49%) and from the cyanobacterium Nostoc minutum (a sequence similarity of 37%) for further analysis.
2.2. Clustering of Sequences of Selected Haloperoxidases
To visualize relative distances between sequences, a cladogram was constructed (Figure 2, left) based on a multiple sequence alignment (MSA, Figure S1). Brown algae sequences are clearly separate from those of bacteria and fungi, whereas the latter two are more similar to each other. This corresponds to a higher sequence identity (Figure S2) between bacteria and fungi sequences, mostly ~45%, with the exceptions of Luteitalea pratensis (~20–25%) and Deltaproteobacterium bacterium (~50%). By contrast, the sequence identity of algae sequences is in the range of 15–20%.
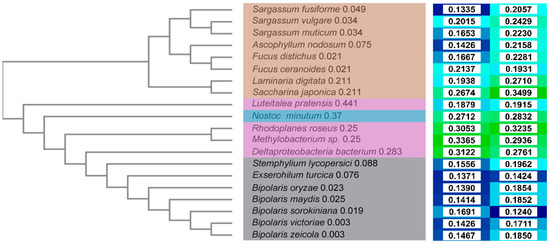
Figure 2.
Comparison of sequences and model quality of haloperoxidases. Cladogram of the selected haloperoxidases (left). The sequences are color-coded according to the taxonomy (center): algae in brown, bacteria in magenta, cyanobacteria in cyan, and fungi in grey. The tree is calculated based on the distances generated from the pairwise scores (numbers in the middle column). The quality of the structural models is expressed by TopScore (second right column) and TopScoreSingle values (right column) [23], where lower values and blue color indicate better quality.
Among algae, the separation corresponds to the species tree, separating the order Laminariales (Laminaria digitata and Saccharina japonica) from the Fucales order. Within the Fucales order, the Fucaceae family sequences (Fucus spp. and Ascophyllum nodosum) are separated from the Sargassaceae family (Sargassum spp.). Consequently, L. digitata and S. japonica are the most diverse in the group and form a separate cluster (identity range of 51.6–58.8% to all other algae sequences, and 63.50% between the two). The other two main clusters are either formed by the sequences from the genus Sargassum (identity ~90% within the cluster) and the order Fucales (identity of 95.8% between F. distichus and F. ceranoides) together with A. nodosum (identity range of 62.3–66.9% in the group, and 86.2–95.8% in the cluster). Among bacteria, L. pratensis and N. minutum are the most divergent in the group (identity range of 21.5–22.9% and 43.0–45.4%, respectively, in the group, and 28.0% between the two). R. roseus and Methylobacter sp. form another cluster (identity between the two of 55.3%), whereas the sequence from Deltaproteobacteria bacterium is separated and has a higher similarity to the group of fungi (identity range of 22.2–43.5% and 50.6–52.1%, respectively, for the bacteria and fungi groups). Among fungi, S. lycopersici and E. turcica are branching out and are separated from the sequences of the genus Bipolaris (identity range of 82.4–86.1% in the group, and 83.9% between the two). In the Bipolaris cluster, B. victoriae and B. zeicola are the most similar (identity of 99.3%), while B. sorokiniana, B. maydis, and B. orizae show, respectively, increasing diversity.
2.3. Structural Modeling of Selected Haloperoxidases
As no experimentally derived structures were available for the 20 selected haloperoxidase enzymes, a structural model for each sequence was constructed. In general, the models from fungi and some algae sequences are of very good quality (TopScore [23] values ~0.15; TopScore values are bounded between 0 (very good) and 1 (bad)), and for other algal and bacteria sequences, the models are of lower, albeit still good quality (TopScore values ~0.25–0.3) [24] (Figure 2, Figure 3, Figure S3, Figure S4 and Figure S5). This is due to different template availabilities and differences in the sequence identities to the templates used for modeling (see Table S1 for details). Interestingly, the Ascophyllum sequence, which is very similar to the known structure from A. crysall (PDB ID 1QI9_B), had a similar TopScore value as the more distant sequence from S. fusiforme (Table S1), indicating a good modeling performance of TopModel [25]. The high-quality structural models (Figure 2 and Figure S2) allow for the inference of the location of the catalytic residues after alignment as well as the probable functional assembly.
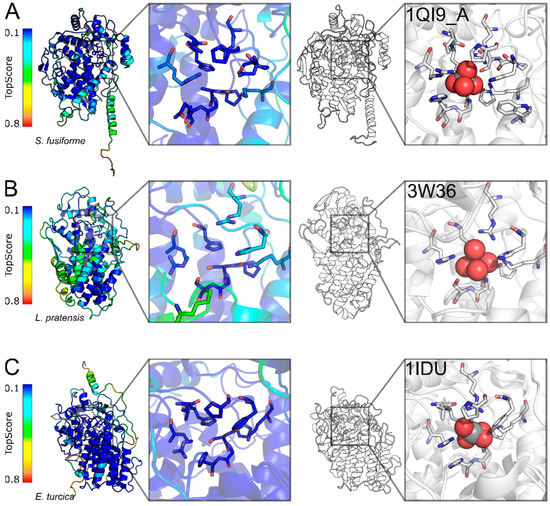
Figure 3.
Examples of high-quality structural models of haloperoxidases. One example for each taxonomic group is reported with Sargassum fusiforme for algae (A), Luteitalea pratensis for bacteria (B), and Exserohilum turcica for fungi (C). The local quality of the models is assessed with TopScore where darker blue color indicates better quality (left). The best template selected for each example is also reported for comparison as white colored structure (right). The vanadium cofactor is shown as spheres and residues located ≤5 Å from it are reported as sticks.
2.3.1. Structural Models of Haloperoxidases from Algae
For models from algae, the best templates generally have a sequence identity of ~55–65% to the target sequences (Table S1). Exceptions include sequences of the genus Fucus, where the best templates have an identity of ~85% and A. nodosum, for which a structure with an identical sequence was deposited (PDB ID: 1QI9, 100% identity and 99.6% coverage). Most algae sequences were modeled based on templates in multimeric functional assembly. Since the interface is close to the catalytic pocket, generating at least a dimeric structural model is required. The regions modeled with lower quality (Table S4) include unstructured N- and C-termini and some short loop regions located mostly at a large distance (>20 Å) from the catalytic pocket. Notably, the N-terminus is placed right before a helix involved in dimerization (~23 residues long, Table S4), which is pointing out and oriented towards the putative dimerization interface. To make constructing a dimeric model easier, a truncated model without the low-quality N-terminus, thus, starting from the dimerization helix, was considered instead for the next steps.
2.3.2. Structural Models of Haloperoxidases from Bacteria
For models from bacteria, the best templates available have an identity of ~45–50% to the target sequences (Table S2), indicating that less well-suited templates are available for the selected sequences. Compared to models from algae, the catalytic pocket is less exposed due to a more compact helical bundle, which is generally modeled with high quality. Additionally, in most models, a β-hairpin located on top of the pocket can be found, likely acting as a lid. Exceptions include the cases of L. pratensis and N. minutum, where this region is mostly unfolded but still of good quality (TopScore < 0.5). Other than unstructured N- and C-termini, low-quality regions (Table S5) include some loop regions located mostly at a large distance (>25–30 Å) from the catalytic pocket. In the case of Methylobacterium sp., while the folding of the β-hairpin lid is conserved, its quality is low (TopScore > 0.5), especially in the first β-strand.
2.3.3. Structural Models of Haloperoxidases from Fungi
For models from fungi, the best templates available have a very high sequence identity of ~85% to the target sequence (Table S3). Structurally, these resemble the models from bacteria due to a very compact helical bundle supporting the catalytic pocket and the presence of a β-hairpin lid. The sequence conservation of this lid is very high, with only one position not fully conserved (M, K, and R, respectively, for S. lycopersici, E. turcica, and models from the genus Bipolaris) and three positions with “strongly similar properties” (indicated with a colon in the MSA; Table S6 and Figure S1). In these models, low-quality regions (Table S6) are located in unstructured N- and C-termini and in extremely short loop regions located mostly at a large distance (>25–30 Å) from the catalytic pocket.
2.4. Mapping and Comparison of the Catalytic Sites
For the structural models, including dimeric models for algae, molecular interaction fields (MIF) were calculated with DrugScore2018 [26,27] and represented with Zernike descriptors [28]. In doing so, the description of the molecular recognition properties of the binding sites was compressed, allowing an alignment-independent pairwise comparison using the Manhattan distance as a measure. That way, proteins with similar molecular recognition properties can be identified, likely interacting and reacting with similar substrates.
L. pratensis and B. sorokiniana are branching out (Figure 4 and Figure S6) and have the most diverse binding sites in the set. For the latter, this result is surprising given the high identity (82.4–86.1%) with the other members of the Bipolaris group. These are mainly part of one cluster, with subclusters by E. turcica and R. roseus (normalized distance of 0.57), and B. oryzae (normalized distance within the cluster of 0.50–0.55), as well as B. victoriae and B. zeicola (normalized distance of 0.43). Therefore, interestingly, the model from the bacterium R. roseus displays molecular recognition similarities to some other models from fungi. Instead, a bacterial model that differs from other examples is the one of Deltaproteobacteria bacterium. This result is expected, given the low to average sequence identity of 22.2–43.5% and 50.6–52.1% for the bacteria and fungi models, respectively.
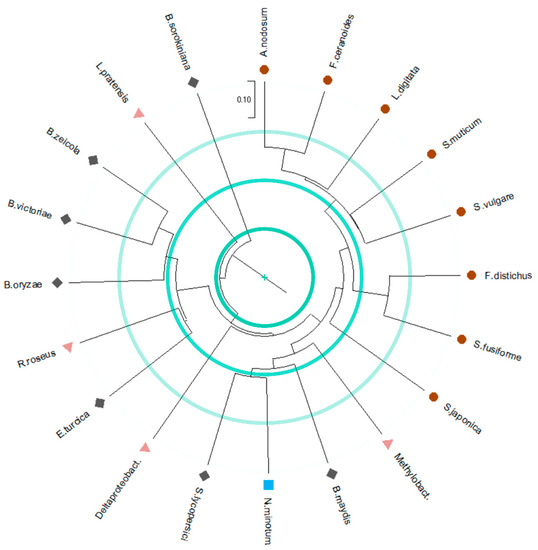
Figure 4.
Clustering of the molecular recognition properties of the selected haloperoxidase structural models. Hierarchical clustering was performed for 20 haloperoxidase binding sites. The binding sites were encoded as DrugScore2018 MIFs and compressed using Zernike descriptors. Manhattan distances between the Zernike descriptors were used for similarity assessment. The sequences are color-coded according to the taxonomy: algae in brown circles, bacteria in magenta triangles, cyanobacteria in cyan squares, and fungi in grey diamond shapes. The tree is drawn to scale with branch lengths in the same units as those of the distances used to infer the tree. Cyan concentric lines represent 25%, 50%, and 75% from the center of the maximum distance, respectively.
All the other examples form two main clusters, one formed by models from fungi and bacteria and another one entirely formed by models from algae. In the first, apparently unrelated systems are clustered together, including Methylobacter sp. (normalized distance within the cluster of 0.54–0.66), B. maydis (normalized distance within the cluster of 0.55–0.66), N. minutum and S. lycopersici (normalized distance of 0.52). In the second, S. japonica is branching out and has the most different binding site (normalized distance within the cluster of 0.58–0.68). Interestingly, S. fusiforme is separated from the other models from the Sargassum genus and is instead part of a subcluster with F. distichus (normalized distance of 0.39), which are separated from the other examples (normalized distance within the cluster of 0.44–0.63 and 0.59–0.70, respectively). Then, S. vulgare and S. muticum form a subcluster (normalized distance of 0.49) and are separated from the other examples (normalized distance within the cluster of 0.46–0.54 and 0.46–0.51, respectively). Finally, L. digitata, F. ceranoides, and A. nodosum form a subcluster, with the first being the most different (normalized distance within the cluster of 0.43–0.52) of the subcluster and the last two the most similar (normalized distance of 0.36) of the entire set. Other highly similar binding sites include S. fusiforme and F. distichus (normalized distance of 0.39), and B. victoriae and B. zeicola (normalized distance of 0.43).
2.5. Expression of Four Putative Haloperoxidase Sequences
To confirm the bioinformatics results achieved for this group of genes and putative enzymes, four sequences were chosen from A. nodosum, F. ceranoides, S. fusiforme, and R. roseus to test for haloperoxidase activity. These cases include algae sequences with similar binding sites from A. nodosum and F. ceranoides, as shown in the previous analysis of the predicted structural models, but also algae sequences with a more diverse binding site from S. fusiforme. Finally, a more divergent example from the bacteria R. roseus was also included. The expression system was based on a previously established expression platform using E. coli BL21 Arctic Express (DE3) as the heterologous expression host [16].
SDS-PAGE analysis following expression of all four constructs in 50 mL cultures using the optimized haloperoxidase expression system showed a high amount of inclusion body formation visible for all constructs except that from R. roseus (see Figure S7). However, all four proteins seemed to be produced in E. coli to a certain extent. Following these first small-scale attempts, large-scale expression of all genes was tested using 500 mL cultures. After cell disruption via ultrasonication, the integrated N-terminal His-Tag was used to perform an affinity chromatography-based isolation on an Ni-column (see Section 4 for details). Due to scale-up problems for the constructs of A. nodosum, S. fusiforme and F. ceranoides, all three proteins did not show the desired purity required for biochemical characterization and no haloperoxidase activity was found in initial activity tests. Only the construct of R. roseus showed sufficient purity (Figure 5) and detectable haloperoxidase activity. Therefore, all further experiments focused on this enzyme, which will be referred to as VPO-RR from now on.
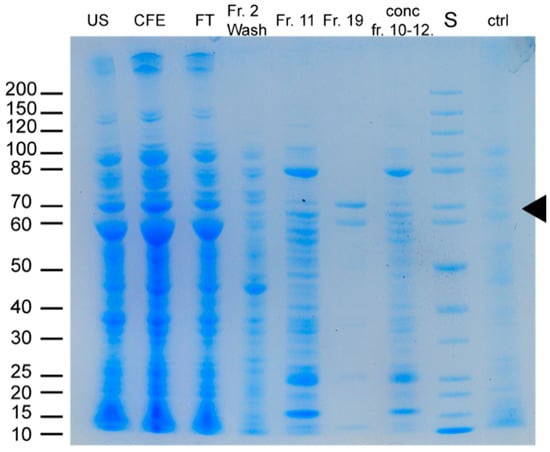
Figure 5.
SDS-PAGE analysis of the chromatographic isolation of VPO-RR using an Ni-column. Chaperone (implemented in E. coli ArcticExpress (DE3) (Agilent Technologies)) size of Cpn60: ~60 kDa; Cpn10: ~10 kDa. US: cell debris after ultrasonication; CFE: cell-free extract; FT: flow-through of Ni-NTA column; Fr.: fractions collected during Ni-NTA isolation; conc.: concentrated fractions after pooling; S: Standard; ctrl: empty-vector control of E. coli ArcticExpress (DE3).
2.6. Biochemical Characterization of VPO-RR
In the first step, the kinetics of the enzyme was determined using a two-dimensional Michaelis–Menten function as previously reported [16]. For this experiment, potassium bromide was chosen as the halide source (Figure 6).
where v0 is the initial activity; vmax the maximal activity; KM a Michaelis constant; c a molar concentration; and Ki an inhibitory constant.
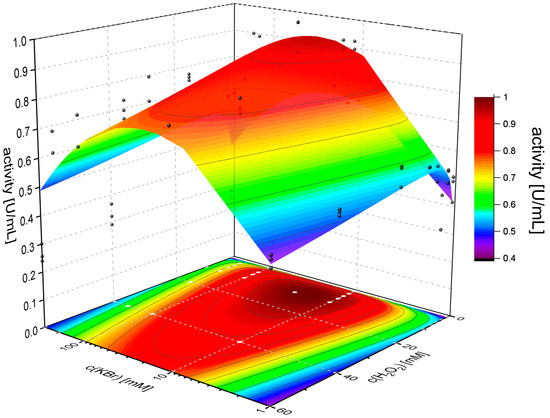
Figure 6.
Two-dimensional kinetics of VPO-RR with hydrogen peroxide and potassium bromide as substrates. The activity was determined using the MeDAC assay with triplicates for each concentration pair. The volumetric activities were plotted against concentrations (black and white dots) and fitted using the substrate-excess inhibition term by Murray [29] in Origin (surface and contour plot).
The turnover number kcat was determined using the given values of vmax and each affinity constant (KM), respectively, and compared to the parameters of a literature-known enzyme, the vanadium-dependent chloroperoxidase (VClPO) from Curvularia inaequalis, short VClPO-CI (Table 1).

Table 1.
Kinetic parameters of VPO-RR compared to parameters of VClPO-CI from previous work [16] [a].
The kinetic profile of VPO-RR shows some remarkable differences compared to known homologs. The so-far generally assumed substrate-excess inhibition by halides is not as pronounced as with VClPO-CI (405 mM compared to 56.8 mM) [30,31]. However, hydrogen peroxide inhibits the enzyme with Ki = 178 mM, in contrast to VClPO-CI, which is not inhibited. The overall turnover number of VPO-RR exceeds the one of VClPO-CI by a hundred-fold (1304 s−1 compared to 9.8 s−1). In conclusion, VPO-RR shows several advantages in regard to kinetic properties compared to literature-known haloperoxidases. Alternately, the apparent inhibition by hydrogen peroxide may be a lack of stability against the strongly oxidizing effect of the peroxide rather than a molecular excess inhibition, which are virtually indistinguishable in this experiment. The overall higher turnover number and lower inhibitory effect of bromide compared to VClPO-CI may allow for biotransformations with higher efficiency.
As reaction temperatures are a frequent optimization parameter in synthetic applications, the overall temperature stability of VPO-RR was investigated. Over a period of three hours, the enzyme was incubated at different temperatures ranging from 25 to 85 °C. After the incubation, the samples were centrifuged at maximum speed and the supernatant was measured using the MeDAC assay. All activities determined were normalized to the activity at room temperature (here 25 °C) and compared (Figure 7).
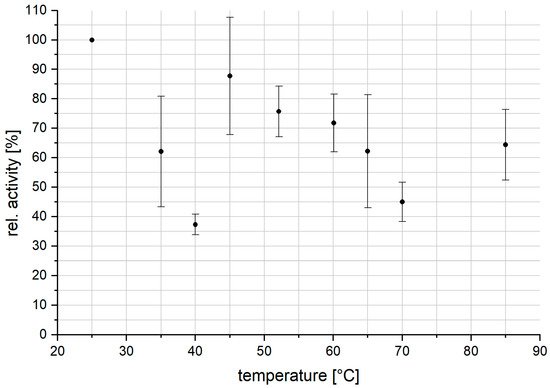
Figure 7.
Relative activity of VPO-RR after incubation at different temperatures for three hours. The activity of each sample was normalized to the activity of the sample incubated at 25 °C. The uncertainties indicate the standard deviation of triplicate measurements.
Although the temperature stability measurements suffer from large variations, principal information can be drawn from this series. Similar to VClPO-CI, VPO-RR exhibits good stability against higher temperatures, e.g., with a loss in the relative activity of 28.2% after incubation at 60.1 °C. Comparing slightly elevated temperatures such as 40 °C to high temperatures such as 70 °C showed a behavioral change. In several attempts, the activity of VPO-RR dropped drastically after incubation at 40 °C (60% decrease), whereas no such significant decrease in activity was observed for samples incubated at 45 °C (13% decrease) to 60 °C (28% decrease). It might be possible that an energetically favored structure is reached at elevated temperatures compared to the structure of VPO-RR at 25 °C. Failure to reach this state due to insufficiently high temperatures, such as 35 or 40 °C, might lead to an undesirable or partially undesirable state where enzymatic activity is severely affected. Whether the nature of this phenomenon is generally attributable to the tertiary structure of the enzyme or the ortho-vanadate cofactor itself has to be elucidated further.
Another important aspect of haloperoxidases is the selectivity for the corresponding halide. Here, one needs to differentiate between a preferred halide, i.e., one that leads to a higher turnover number, and the enzyme’s general acceptance of a halide. Using the MeDAC assay, bromination and chlorination experiments were performed under the same conditions. As a reference, the bromination and chlorination using the MeDAC assay with VClPO-CI, similar to recent work, were included in the SI to illustrate the applicability of the assay for this purpose (see Table S7) [16]. In direct comparison, no chlorination activity could be measured for VPO-RR, while bromination occurred with a volumetric activity of 1.34 U ml−1 (Table 2). Hence, VPO-RR does not accept chloride as a halide source under these conditions.

Table 2.
Halide acceptance of VPO-RR [a].
3. Discussion
Halogenating enzymes do not yet have anywhere near the potential to overtake mega-ton processes such as chlor-alkali electrolysis, on which the entire production of halogenating reagents is based. Here, we identified potential halogenase sequences by screening algal, fungal, and protobacterial sequence databases, structural modeling of putative halogenases, and mapping and comparing active sites. After test expressions of four putative halogenase sequences, the vanadium-dependent haloperoxidase VPO-RR was biochemically characterized, which showed beneficial kinetic properties, including an overall higher turnover number and lower inhibitory effect of bromide compared to VClPO-CI.
At present, there is a gap in the scope of known halogenating enzymes. On the one hand, some enzymes are involved in the biosynthesis of a particular natural compound, such as many flavin-dependent halogenases (e.g., Chl from Streptomyces aureofaciens [32] or CmdE from Chondromyces crocatus Cm c5 [33]), but their high substrate selectivity renders these enzymes unsuitable as general synthetic tools. On the other hand, the vanadate-dependent haloperoxidases only produce hypohalous acid rather than provide an enzyme-substrate complex and, thus, lack an enzyme-driven substrate selectivity [14,34,35]. Furthermore, vanadate-dependent haloperoxidases are still underrepresented in the set of halogenases used for synthetic purposes. Accordingly, predominantly VClPO-CI from Curvularia inaequalis [14] and VBrPO-CO from Corallina officinalis [36] were extensively described. Identifying and characterizing VPO-RR here thus provides a valuable expansion of the known set of vanadate-dependent haloperoxidases.
The pipeline for halogenase identification applied here, including structural characterization and comparison of molecular recognition properties of active sites, is unique because it benefited from screening taxonomically wide sequence databases at the beginning. The structural modeling resulted in overall very good to good models as predicted by TopScore [25,37] based on good available templates, such that the structural quality of the active site should also be high. Recently, we compared the quality of 10,336 enzyme structural models generated with TopModel [25,37] used here and AlphaFold2 [38] and found that the average scores differed only by ~5% [24]. Particularly, if good templates are available, as was the case here, the comparative modeling approach by TopModel resulted in very good models [24]. For binding site comparison, MIFs based on the established knowledge-based potentials for scoring protein–ligand interactions, DrugScore2018 [27,39] were used. This provides a balanced description of enthalpic and entropic interaction contributions and also allows for the consideration of metal ions in the recognition process. Finally, compressing this information as Zernike descriptors [28] yields an alignment-independent way of comparing active sites, which is advantageous in view of the identities of the selected halogenase sequences down to ~20% and, hence, the resulting variability of the overall enzyme structure.
Our analyses show that this group of haloperoxidases is taxonomically widespread, although they must be assigned to secondary metabolism. The sequences chosen here for further analysis are representatives of the Phaeophyta and thus still closely related to the described Rhodophyta C. officinalis, Ascomycota as well as Curvularia inaequalis, but also cyanobacteria and bacteria. Both the phylogenetic tree and cladogram build upon the haloperoxidase sequences and the clustering of the active site geometry (see Figure 4) represents the approximate taxonomic relation. They also underline the variety in the assumed structures indicating different physicochemical properties.
Four sequences (from A. nodosum, F. ceranoides, S. fusiforme, and R. roseus,) were selected for test expressions. These organisms are representatives of the Phaeophyta and thus still closely related to the described Rhodophyta C. officinalis, Ascomycota, and Curvularia inaequalis, but also cyanobacteria and bacteria. However, only the bacterial sequence from R. roseus was expressible within E. coli. The kinetic data revealed that VPO-RR is not strongly inhibited by halides, in contrast to what was described for VClPO-CI [18,31,40]. Furthermore, its 100-fold higher catalytic turnover number renders this enzyme a potential advancement over literature-known haloperoxidases for biocatalytic processes. The temperature stability of VClPO-CI could be observed for VPO-RR; for temperatures up to 85 °C, a remaining activity of well over 60% was observed. Regarding its halide selectivity, only bromide was found to be an accepted halide source, whereas chloride showed no measurable activity. Whether the enzyme is capable of selective halogenations or possesses substrate binding properties is the focus of current investigations.
In summary, we present a structure-based pipeline to identify new halogenases and characterize VPO-RR as a new member of vanadium-dependent haloperoxidases. Its beneficial kinetic properties might allow for biotransformations with higher efficiency. Therefore, a more detailed biochemical characterization will be essential.
4. Materials and Methods
4.1. Structural Modeling of Selected Haloperoxidases
As no experimentally derived structures were available for the 20 selected haloperoxidase sequences, a homology model was constructed for each. To do so, the template-based structure prediction program TopModel [25,37] was applied using the “normal run” mode. In order to build models from algae arranged in a dimeric assembly, a monomer was first built, taking into account the best dimeric template selected by TopModel (Table S1, PDB ID: 1QI9). Next, the low-quality N-terminus was truncated as no dimeric template information is available for this region, meaning a clash might occur at the interface. Since for algae the functional assembly is consistent and involves the interaction between dimerization helices, the model was duplicated, superimposed to the reference template structure, and minimized. Both monomer and dimer structures were preprocessed with the Protein Preparation Wizard [41] of Schrödinger’s Maestro Suite. The truncated dimeric models were energy-minimized using the OPLS 2005 force field [42] with standard cutoff values for van der Waals, electrostatic, and H-bond interactions until the average RMSD of non-hydrogen atoms reached 0.30 Å. Bond orders and missing hydrogen atoms were assigned, and the H-bond network was optimized.
4.2. Calculation of DrugScore2018 Potential Fields
Binding sites were characterized and represented with descriptors based on DrugScore2018 [27,39] potential fields. Potential fields of six different probe atoms were considered: C_ar (aromatic interactions), C_3 (aliphatic interactions), O_2 (H-bond acceptor), N_3 (H-bond donor), O_3 (H-bond donor and acceptor), Cl (representative for halogens). For all computed DrugScore2018 potential fields, a grid spacing of 0.375 Å was used. Since the vanadate cofactor was a constant in all models for the classification, it was not considered for potential field generation.
4.3. Calculation of Zernike Descriptors and Clustering
The DrugScore2018 potential interaction fields were transformed to a set of descriptors using a series expansion in 3D Zernike polynomials as described by Nisius et al. [28]. First, different descriptors for one individual protein and different probe atoms were merged. Second, the pairwise distances between all proteins (Manhattan distance) of the stacked Zernike descriptors were calculated. A matrix of normalized distance values is reported in Figure S6. These distances were then used for clustering. The UPGMA (unweighted pair group method with arithmetic mean) agglomerative bottom-up hierarchical clustering method as implemented in the MEGA (Molecular Evolutionary Genetics Analysis) software [43] was applied to generate the tree reported in Figure 4 from the distance matrix in Figure S6.
4.4. Expression and Isolation of Haloperoxidases
The expression and isolation procedure was established according to recent work [16]. The platform for expression of the haloperoxidase genes consisted of E. coli ArcticExpress (DE3) as the heterologous host and a pET21-vector backbone, using N-terminal His-Tags for later isolation.
For small-scale test expressions, 50 mL cultures in TB-media were inoculated with a cell amount that corresponded to a starting OD600 of 0.05 in 250 mL flasks. Then, 0.05% glucose and 0.2% lactose were supplemented to enable autoinduction later on, with 100 µg mL−1 ampicillin as the antibiotic. After an incubation period of 3 h at 37 °C, 2% ethanol (v/v) was added and the temperature decreased to 13 °C. The cultivation was continued for 72 h before the cells were centrifuged and the dry pellets stored at −20 °C.
The isolation was performed on ice with resuspended pellets in lysis buffer (20%, Bis-TRIS acetate, 100 mM, pH 7 with 12.5 mM imidazole) using ultrasonication for 5 min, cooling for 5 min, and an additional ultrasonication step. The lysate was centrifuged for 1 h at 10.000× g and the supernatant separated from the cell debris. SDS samples were prepared according to the final OD600 values of the culture, aiming for a final OD600 in the SDS sample of 1 for SDS-GEL analysis using the NuPage system. For cell debris samples, the pellet was resuspended with the same amount of lysis buffer (20%) as before.
For large-scale expression, 500 mL cultures were used with the exact same concentrations of supplements. For isolation, an EDTA-free protease inhibitor tablet (cOmplete Tablet, Mini, EASYpack, Roche) was added to the resuspended pellet in lysis buffer and stirred for 20 min. For cell disruption, the ultrasonication steps were increased to 10 min each. After centrifugation, the cleared lysate was loaded onto a 5 mL Ni-NTA Superflow Cartridge (Qiagen). Initial experiments were carried out on an ÄKTA prime system using a linear elution gradient with Bis-TRIS acetate (BTA) buffer 100 mM at pH 7 and BTA with 250 mM imidazole included. Peak fractions were collected and analyzed via SDS-PAGE, and fractions containing the target protein were pooled and dialyzed overnight in assay buffer (BTA 100 mM pH 6) in the presence of 100 µM sodium ortho-vanadate. The dialysate was concentrated using PALL concentrators with a molecular cutoff of 10 kDa. Final protein concentrations were measured using molecular extinction coefficients for absorption at 280 nm for each individual protein calculated by ExpPASy assuming oxidative conditions (εox (VPO-RR) = 100,730 mM−1 cm−1).
4.5. Activity Measurement and Kinetics of VPO-RR
Activity of haloperoxidases was measured using the MeDAC assay [18]. A 1 µL of enzyme solution was added to 249 µL of assay buffer, containing 5 mM of potassium bromide, 3 mM of hydrogen peroxide, 30 µM MeDAC (in DMSO) with 0.4% v/v DMSO (as the final concentration) in BTA 100 mM pH 6. The fluorescence was measured over 10 min at an excitation wavelength of 425 nm and emission wavelengths of 450 and 550 nm. Using a calibration function included in a calculation sheet, the activity was determined by plotting the concentration of BrMeDAC against time and determining the slope of the linear fit. Multiplying with corresponding dilution factors yielded the final volumetric activities in U mL−1.
For the two-dimensional Michaelis–Menten kinetics, the substrate-inhibition term by Murray [29] was used as a fit function in OriginPro 2018. The enzyme was mixed with MeDAC and added to the wells filled with predefined concentrations of hydrogen peroxide and potassium bromide. Measurement and evaluation were performed according to the general activity measurements. The resulting volumetric activities were plotted in Origin and fitted with the custom Michaelis–Menten function. The complete function is accessible in the Supplementary Materials (see Equation (S4)).
4.6. Temperature Stability
To test the thermal stability of VPO-RR, 20 µL of enzyme solution was placed in a PCR tube and incubated for 3 h at the given temperatures. The samples were centrifuged for 5 min at maximum speed, and 1 µL of the solution was used according to the activity assay mentioned before (vide supra). The experiments were performed in biological triplicates. The averaged 25 °C sample was used as a reference and set to 100% of relative activity.
4.7. Halide Selectivity
For the halide selectivity test, the assay was performed under normal conditions using potassium bromide and potassium chloride, respectively. After the determination of activity, the values were directly compared to each other. For samples where no linear area could be determined, the values were set to zero.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal12101195/s1; Supplemental Methods; Figure S1: Multiple sequence alignment for the selected haloperoxidases; Figure S2: Percent identity matrix for the selected haloperoxidases; Figure S3: Structural models of haloperoxidases from algae sequences; Figure S4: Structural models of haloperoxidases from bacteria sequences; Figure S5: Structural models of haloperoxidases from fungi sequences; Figure S6: Matrix of normalized pairwise Manhattan distances between all proteins of the stacked Zernike descriptors; Figure S7: SDS-PAGE analysis of the test expression of all four putative VPO constructs; Figure S8: SDS-PAGE of samples obtained during isolation procedures for the enzymes VPO-SF, VPO-FC, and VPO-AN; Figure S9: Example measurement of chlorinating activity for VClPO-CI using the MeDAC assay; Table S1: Threading information; Table S2: Threading information; Table S3: Threading information; Table S4: Critical evaluation of algae structural models; Table S5: Critical evaluation of bacteria structural models; Table S6: Critical evaluation of fungi structural models; Table S7: Comparison between bromination and chlorination of MeDAC using VClPO-CI; Supplemental References.
Author Contributions
Conceptualization, B.U., J.P., T.C. and H.G.; Formal analysis, N.P., A.V.F., K.D., B.U., J.P., T.C. and H.G.; Investigation, N.P., A.V.F., K.D. and B.P.; Supervision, B.U., J.P., T.C. and H.G.; Visualization, N.P., A.V.F., K.D., B.U., J.P., T.C. and H.G.; Writing—original draft, N.P., A.V.F. and K.D.; Writing—review and editing, B.U., J.P., T.C. and H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the Ministry of Innovation, Science, and Research within the framework of the NRW Strategieprojekt BioSC (No. 313/323-400-002 13) by the seed fund ‘HaloEnz’ granted to H.G., B.U., J.P., and T.C. and, in part, by Deutsche Forschungsgemeinschaft (DFG) through GRK 2158/2 (project number 270650915) to H.G. and J.P.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
We are grateful for computational support from the “Zentrum für Informations und Medientechnologie” at the Heinrich-Heine-Universität Düsseldorf and the computing time provided by the John von Neumann Institute for Computing (NIC) to HG on the supercomputer JUWELS at Jülich Supercomputing Centre (JSC) (user IDs: HKF7, VSK33).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, Y.; Wang, Y.; Zhu, W. Nonbonding interactions of organic halogens in biological systems: Implications for drug discovery and biomolecular design. Phys. Chem. Chem. Phys. 2010, 12, 4543–4551. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, Y.; Xu, Z.; Li, H.; Liu, H.; Zhu, W. Halogen bonding for rational drug design and new drug discovery. Expert Opin. Drug Discov. 2012, 7, 375–383. [Google Scholar] [CrossRef]
- Sirimulla, S.; Bailey, J.B.; Vegesna, R.; Narayan, M. Halogen interactions in protein–ligand complexes: Implications of halogen bonding for rational drug design. J. Chem. Inf. Model. 2013, 53, 2781–2791. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yang, Z.; Liu, Y.; Lu, Y.; Chen, K.; Zhu, W. Halogen bond: Its role beyond drug–target binding affinity for drug discovery and development. J. Chem. Inf. Model. 2014, 54, 69–78. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, L.; Cui, D.; Yao, Z.; Gao, B.; Lin, J.; Wei, D. The Important Role of Halogen Bond in Substrate Selectivity of Enzymatic Catalysis. Sci. Rep. 2016, 6, 34750. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, Z.; Chen, T.; Chen, T.; Wang, Z.; Tian, G.; Shi, J.; Wang, X.; Lu, Y.; Yan, X.; et al. Utilization of halogen bond in lead optimization: A case study of rational design of potent phosphodiesterase type 5 (PDE5) inhibitors. J. Med. Chem. 2011, 54, 5607–5611. [Google Scholar] [CrossRef] [PubMed]
- Gribble, G.W. The diversity of naturally produced organohalogens. Chemosphere 2003, 52, 289–297. [Google Scholar] [CrossRef]
- Kesner, M. Bromine and Bromine Compounds from the Dead Sea, Israel Products in the Service of People; The Weizmann Institute of Science: Rehovoth, Israel, 1999. [Google Scholar]
- Fejzagić, A.V.; Gebauer, J.; Huwa, N.; Classen, T. Halogenating Enzymes for Active Agent Synthesis: First Steps Are Done and Many Have to Follow. Molecules 2019, 24, 4008–4041. [Google Scholar] [CrossRef]
- Wagner, C.; El Omari, M.; König, G.M. Biohalogenation: Nature’s way to synthesize halogenated metabolites. J. Nat. Prod. 2009, 72, 540–553. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Ayub, M.J.; Furnham, N.; Thornton, J.M.; Laskowski, R.A. Chopping and changing: The evolution of the flavin-dependent monooxygenases. J. Mol. Biol. 2016, 428, 3131–3146. [Google Scholar] [CrossRef]
- Dong, C.; Flecks, S.; Unversucht, S.; Haupt, C.; Van Pee, K.-H.; Naismith, J.H. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science 2005, 309, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Kuhnel, K.; Blankenfeldt, W.; Terner, J.; Schlichting, I. Crystal structures of chloroperoxidase with its bound substrates and complexed with formate, acetate, and nitrate. J. Biol. Chem. 2006, 281, 23990–23998. [Google Scholar] [CrossRef] [PubMed]
- van Schijndel, J.W.; Vollenbroek, E.G.; Wever, R. The chloroperoxidase from the fungus Curvularia inaequalis; a novel vanadium enzyme. Biochim. Biophys. Acta Biomembr. 1993, 1161, 249–256. [Google Scholar] [CrossRef]
- Van Schijndel, J.W.P.M.; Barnett, P.; Roelse, J.; Vollenbroek, E.G.M.; Wever, R. The stability and steady-state kinetics of vanadium chloroperoxidase from the fungus Curvularia inaequalis. Eur. J. Biochem. 1994, 225, 151–157. [Google Scholar] [CrossRef]
- Getrey, L.; Krieg, T.; Hollmann, F.; Schrader, J.; Holtmann, D. Enzymatic halogenation of the phenolic monoterpenes thymol and carvacrol with chloroperoxidase. Green Chem. 2014, 16, 1104–1108. [Google Scholar] [CrossRef]
- Weichold, V.; Milbredt, D.; van Pée, K.-H. Specific Enzymatic Halogenation—From the Discovery of Halogenated Enzymes to Their Applications In Vitro and In Vivo. Angew. Chem. Int. Ed. 2016, 55, 6374–6389. [Google Scholar] [CrossRef]
- Fejzagić, A.V.; Myllek, S.; Hogenkamp, F.; Greb, J.; Pietruszka, J.; Classen, T. A Fluorescence-Based Assay System for the Determination of Haloperoxidase-Activity Using a Two-Dimensional Calibration Approach. ChemistryOpen 2020, 9, 959–966. [Google Scholar] [CrossRef]
- Weyand, M.; Hecht, H.; Kiess, M.; Liaud, M.; Vilter, H.; Schomburg, D. X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 A resolution. J. Mol. Biol. 1999, 293, 595–611. [Google Scholar] [CrossRef]
- Cosse, A.; Potin, P.; Leblanc, C. Patterns of gene expression induced by oligoguluronates reveal conserved and environment-specific molecular defense responses in the brown alga Laminaria digitata. N. Phytol. 2009, 182, 239–250. [Google Scholar] [CrossRef]
- Kumar, A.; Castellano, I.; Patti, F.P.; Delledonne, M.; Abdelgawad, H.; Beemster, G.T.S.; Asard, H.; Palumbo, A.; Buia, M.C. Molecular response of Sargassum vulgare to acidification at volcanic CO2 vents: Insights from de novo transcriptomic analysis. Mol. Ecol. 2017, 26, 2276–2290. [Google Scholar] [CrossRef]
- Messerschmidt, A.; Prade, L.; Wever, R. Implications for the catalytic mechanism of the vanadium-containing enzyme chloroperoxidase from the fungus Curvularia inaequalis by X-ray structures of the native and peroxide form. Biol. Chem. 1997, 378, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Mulnaes, D.; Gohlke, H. TopScore: Using Deep Neural Networks and Large Diverse Data Sets for Accurate Protein Model Quality Assessment. J. Chem. Theory Comput. 2018, 14, 6117–6126. [Google Scholar] [CrossRef] [PubMed]
- van der Weg, K.; Gohlke, H. TopEnzyme: A framework and database for structural coverage of the functional enzyme space. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mulnaes, D.; Porta, N.; Clemens, R.; Apanasenko, I.; Reiners, J.; Gremer, L.; Neudecker, P.; Smits, S.H.J.; Gohlke, H. TopModel: Template-Based Protein Structure Prediction at Low Sequence Identity Using Top-Down Consensus and Deep Neural Networks. J. Chem. Theory Comput. 2020, 16, 1953–1967. [Google Scholar] [CrossRef]
- Gohlke, H.; Hendlich, M.; Klebe, G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000, 295, 337–356. [Google Scholar] [CrossRef]
- Dittrich, J.; Schmidt, D.; Pfleger, C.; Gohlke, H. Converging a Knowledge-Based Scoring Function: Drug Score (2018). J. Chem. Inf. Model. 2019, 59, 509–521. [Google Scholar] [CrossRef]
- Nisius, B.; Gohlke, H. Alignment-independent comparison of binding sites based on DrugScore potential fields encoded by 3D Zernike descriptors. J. Chem. Inf. Model. 2012, 52, 2339–2347. [Google Scholar] [CrossRef]
- Murray, D.R. The inhibition of esterases by excess substrate. Biochem. J. 1930, 24, 1890–1896. [Google Scholar] [CrossRef]
- de Boer, E.; Wever, R. The reaction mechanism of the novel vanadium-bromoperoxidase. A steady-state kinetic analysis. J. Biol. Chem. 1988, 263, 12326–12332. [Google Scholar] [CrossRef]
- Wever, R.; Krenn, B.E.; Renirie, R. Marine Vanadium-Dependent Haloperoxidases, Their Isolation, Characterization, and Application. Methods Enzym. 2018, 605, 141–201. [Google Scholar] [CrossRef]
- Dairi, T.; Nakano, T.; Aisaka, K.; Katsumata, R.; Hasegawa, M. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci. Biotechnol. Biochem. 1995, 59, 1099–1106. [Google Scholar] [CrossRef]
- Rachid, S.; Krug, D.; Kunze, B.; Kochems, I.; Scharfe, M.; Zabriskie, T.M.; Blöcker, H.; Müller, R. Molecular and biochemical studies of chondramide formation—Highly cytotoxic natural products from Chondromyces crocatus Cm c5. Chem. Biol. 2006, 13, 667–681. [Google Scholar] [CrossRef]
- Fernandez-Fueyo, E.; van Wingerden, M.; Renirie, R.; Wever, R.; Ni, Y.; Holtmann, D.; Hollmann, F. Chemoenzymatic Halogenation of Phenols by using the Haloperoxidase from Curvularia inaequalis. ChemCatChem 2015, 7, 4035–4038. [Google Scholar] [CrossRef]
- Hemrika, W.; Renirie, R.; Macedo-Ribeiro, S.; Messerschmidt, A.; Wever, R. Heterologous Expression of the Vanadium-containing Chloroperoxidase from Curvularia inaequalis in Saccharomyces cerevisiae and Site-directed Mutagenesis of the Active Site Residues His496, Lys353, Arg360, and Arg490. J. Biol. Chem. 1999, 274, 23820–23827. [Google Scholar] [CrossRef]
- Carter, J.N.; Beatty, K.E.; Simpson, M.T.; Butler, A. Reactivity of recombinant and mutant vanadium bromoperoxidase from the red alga Corallina officinalis. J. Inorg. Biochem. 2002, 91, 59–69. [Google Scholar] [CrossRef]
- Mulnaes, D.; Koenig, F.; Gohlke, H. TopSuite Web Server: A Meta-Suite for Deep-Learning-Based Protein Structure and Quality Prediction. J. Chem. Inf. Model. 2021, 61, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Gohlke, H.; Klebe, G. Statistical potentials and scoring functions applied to protein–ligand binding. Curr. Opin. Chem. Biol. 2001, 11, 231–235. [Google Scholar] [CrossRef]
- Everett, R.R.; Soedjak, H.S.; Butler, A. Mechanism of dioxygen formation catalyzed by vanadium bromoperoxidase. Steady state kinetic analysis and comparison to the mechanism of bromination. J. Biol. Chem. 1990, 265, 15671–15679. [Google Scholar] [CrossRef]
- Schrödinger Software Suite 2017-1, Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2017.
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).