Abstract
In this study, we overcame the limitations of single-enzyme system catalysis by codisplaying Candida rugosa lipase 1 (CRL1) and Rhizopus oryzae lipase (ROL) on the cell surfaces of the whole-cell catalyst Pichia pastoris to produce biodiesel from tallow seed oil. We screened double antibiotic-resistant strains on tributyrin plates, performed second electroporation based on single-displayed ROL on GS115/KpRS recombinants and single-displayed CRL1 on GS115/ZCS recombinants and obtained an ROL/CRL1 codisplay on P. pastoris GS115 surfaces. The maximum activity of the codisplaying GS115/pRCS recombinant was 470.59 U/g dried cells, which was 3.9-fold and 1.3-fold higher than that of single-displayed ROL and CRL1, respectively. When self-immobilized lipases were used as whole-cell catalysts, the rate of methyl ester production from GS115/pRCS harboring ROL and CRL1 was 1.4-fold higher than that obtained with single-displayed ROL. Therefore, biodiesel catalysis by synergetic codisplayed enzymes is an alternative biodiesel production strategy.
1. Introduction
Lipases (EC 3.1.1.3) are enzymes that hydrolyze triacylglycerols to single fatty acids and exhibit ester bond hydrolase activity with good substrate specificity and stereoselectivity [1]. Lipases facilitate catalysis at the oil-water interface and in the organic phase, and those from Candida spp., Rhizopus spp., and Pseudomonas spp. are widely used as catalysts in the production of medicines, biodiesel, cosmetics, and food. Rhizopus oryzae lipases (ROLs) are indispensable in oil processing and the food industry. Candida rugosa lipases (CRLs) are widely used in industrial production [2].
Previous studies confirmed that there is synergetic catalytic efficacy between CRL1 and ROL. The biodiesel conversion rate of this combination is superior to that of ROL or CRL alone [3,4,5,6]. Lee et al. reported biodiesel production using a combination of C. rugosa and R. oryzae lipases and supercritical carbon dioxide [5]. The ROL-CRL mixture eliminated the acyl migration step. The CRL:ROL ratio required for efficient biodiesel production was also investigated [4]. Continuous biodiesel scale-up was developed by using immobilized lipases to reduce costs and conserve energy. Biodiesel production from jatropha and waste oil was tested in a packed-bed reactor containing coimmobilized ROL and CRL lipases and the biodiesel conversion rate was ~99%. In contrast, the biodiesel conversion rates for separate ROL and CRL were 70% and 20%, respectively [6].
The alternative immobilization method [7] or microbial surface display (MSF) expresses targeted genes as fusion proteins on the surfaces of ribosomes [8], viruses [9], or cells [10,11]. The peptides/proteins have relatively independent spatial structures and biological activity [12,13,14]. MSF facilitates the study of peptide properties, mutual recognition, and action [15] and can be used to screen specific peptide structures, immobilize proteins, and direct protein evolution [16,17,18]. MSF is used in biological research, industrial production, whole-cell catalysis [19], vaccine production [20], peptide drug screening [21], and biosensor development [22]. It requires neither purification nor fixation before use. In recent years, MSF lipases have attracted attention as whole-cell biocatalysts with good operational stability and reusability [22,23]. A current research trend is the use of MSF lipases to synthesize renewable, nontoxic alternative fuels for biodiesel engines [23,24,25,26].
Enzyme cell surface display has not yet been commercialized for biorefinery. In yeast, targeted protein production is inefficient and catalytic bioactivity is inadequate. In our previous biodiesel production study, we codisplayed synergetic Candida antarctica lipase B and Thermomyces lanuginosus lipases with the GPI-anchor cell wall protein Sed1 [27]. To the best of our knowledge, however, few reports have addressed C. rugosa and R. oryzae lipase codisplay for synergetic biodiesel preparation.
Based on the advantages of ROL-CRL combinations and the high performance of the anchoring protein Sed1, we fused R. oryzae and C. rugosa lipase genes with the Sed1 anchor gene. The Sed1-ROL and Sed1-CRL1 fusion proteins were expressed under the control of the AOX1 promoter. We used a second electroporation to codisplay ROL plus CRL1 on the surface of Pichia pastoris GS115. We compared the optimal substrate, temperature, and pH for the codisplayed lipases against those for the single-displayed ROL and CRL1. We further evaluated the potential industrial applications of the codisplayed lipases on a whole-cell catalyst by using them to generate biodiesel through the transesterification of tallow seed oil with short-chain alcohols.
2. Results and Discussion
2.1. ROL and CRL1 Localization on P. pastoris GS115
Figure 1 shows that GS115/KpRS displaying ROL emitted strong green fluorescence at 545 nm excitation, whereas GS115/ZCS displaying CRL1 emitted strong red fluorescence at 495 nm excitation. The negative control cells did not fluoresce. Hence, ROL and CRL1 were displayed on P. pastoris cell surfaces.
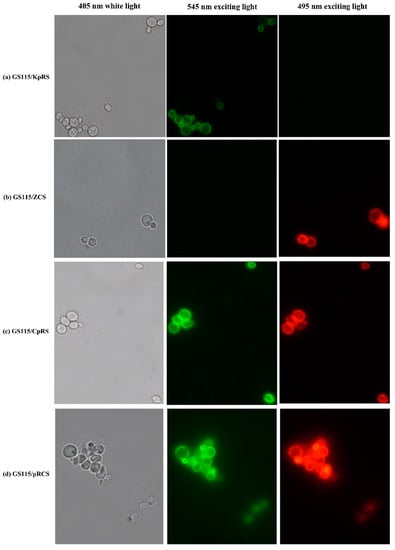
Figure 1.
Fluorescence microscopy of recombinant strains displaying Rhizopus oryzae lipase (ROL) and Candida rugosa lipase 1 (CRL1). (a,c,d) Pichia pastoris GS115 recombinant cells displayed ROL localized to the cell surfaces with clear green fluorescence at 545 nm excitation. All cells were immunologically incubated with anti-Flag primary antibody and FITC-conjugated anti-IgG secondary antibody. (b,c,d) GS115 recombinants displayed CRL1 localized to the cell surfaces with clear red fluorescence at 495 nm excitation. All cells were incubated with primary anti-HA antibody and Alexa Fluor 555-labeled donkey antimouse immunoglobulin G IgG (H+L) secondary antibody. No fluorescence was detected in the control GS115 cells harboring pPICZα or pPIC9K.
Flow cytometric analysis of the single-displayed and codisplayed lipases on the recombinants revealed that >95.95% of the P. pastoris GS115 cells had coimmobilized the lipases via the SED1 cell surface anchor protein. Compared with the control cells, the GS115/pPIC9K and GS115/pPICZαA, FL1 (green fluorescence), and FL3 (red fluorescence) channels had strong positive signals. The floating rates were 97.48% for ROL (Figure 2a) on GS115/KpRS and 96.50% for CRL1 on GS115/ZCS (Figure 2b). For the double-displaying lipases, the rates of the displayed ROL and CRL1 on GS115/CpRS were 97.75% and 98.91%, respectively (Figure 2c). The simultaneous display rate for ROL plus CRL1 on GS115/CpRS was 96.66%. The rates of single-displayed ROL and CRL1 on GS115/pRCS were 98.81% and 97.14%, respectively (Figure 2d), and the floating rate of the codisplayed ROL plus CRL1 on GS115/pRCS was 95.95%.
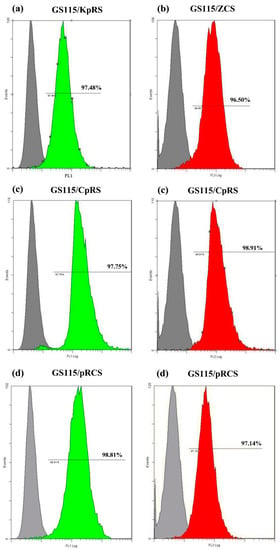
Figure 2.
Determination of expression levels of single-displayed Rhizopus oryzae lipase (ROL) and Candida rugosa lipase 1 (CRL1) and codisplayed ROL plus CRL1 proteins on the Pichia pastoris GS115 surfaces and comparison with those on the control GS115 cells through FACS. (a) The ROL display rate was ~97.48% for the GS115/KpRS recombinants based on the green fluorescence peaks. (b) The CRL1 display rate was ~96.50% for the GS115/ZCS recombinants based on the red fluorescence peaks. (c) The ROL and CRL1 expression rates were 97.75% and 98.91%, respectively, on the P. pastoris GS115/CpRS surfaces. (d) The ROL and CRL1 expression rates were 98.81% and 97.14%, respectively, on the P. pastoris GS115/pRCS surfaces. The gray histogram represents the negative control.
2.2. Functional Activity of Surface-Displayed Recombinant Strains
We screened almost 30 recombinant strains on tributyrin plates (0.5% v/v) containing double antibiotics and compared the hydrolysis zones formed. We evaluated the functional activity of the recombinant P. pastoris GS115 strains displaying lipases and predicted their applications. The enzyme activity of GS115/KpRS was in the range of 40–120 U/g dried cells. The highest enzyme activity was 121.99 U/g dried cells (Figure 3a). Compared with that in GS115/KpRS, GS115/ZCS showed an enzyme activity of >100 U/g dried cells, and its highest enzyme activity was 359.62 U/g dried cells (Figure 3b).
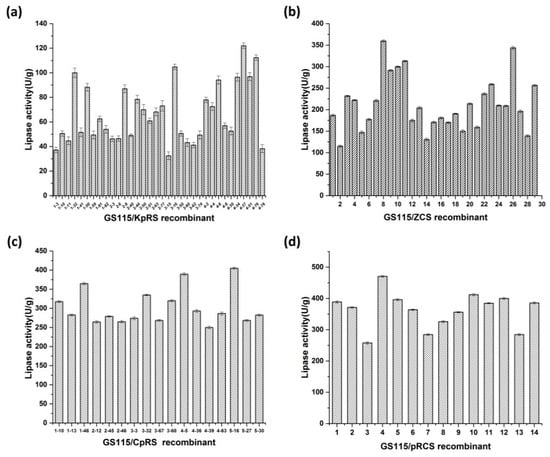
Figure 3.
Screening of olive oil hydrolysis by Pichia pastoris GS115/KpRS, GS115/ZCS, GS115/CpRS, and GS115/pRCS in a shake-flask fermentation assay. (a) Lipase activity of recombinant GS115/KpRS strains. (b) Lipase activity of recombinant GS115/ZCS strains. (c) Lipase activity of recombinant GS115/CpRS strains. (d) Lipase activity of recombinant GS115/pRCS strains.
Figure 3c shows that the maximum enzyme activity of the codisplayed GS115/CpRS was 405.00 U/g dried cells. The codisplayed GS115/pRCS had a maximum enzyme activity of 470.59 U/g dried cells (Figure 3d), which was 3.9-fold and 1.3-fold higher than that of single-displayed ROL and CRL1, respectively. The enzyme activity of the codisplayed lipases was higher than that of the single-displayed ROL and CRL1 because the enzymes were synergetic. GS115/pRCS had higher bioactivity on olive oil than GS115/CpRS. GS115/pRCS was generated through a second electroporation into host GS115 cells harboring single ROL with a linearized ZCS vector. GS115/CpRS was generated through a second electroporation into host GS115 cells harboring single CRL1 with a linearized KpRS vector.
2.3. Surface-Displayed GS115/KpRS, GS115/ZCS, and GS115/pRCS Characterization
We investigated substrate chain length specificity at the optimal temperature and pH for cell surface display using C2, C4, C8, C10, C12, C14, and C16 p-nitrophenolates as substrates. Figure 4a shows that the recombinant strains of single-displayed ROL and CRL1 had preferential enzyme activity on pNPC (C8). The optimal substrate for codisplayed ROL and CRL1 on GS115/pRCS was also pNPC (C8). The single-displayed lipase strains ROL and CRL1 also preferred pNPC (C8).
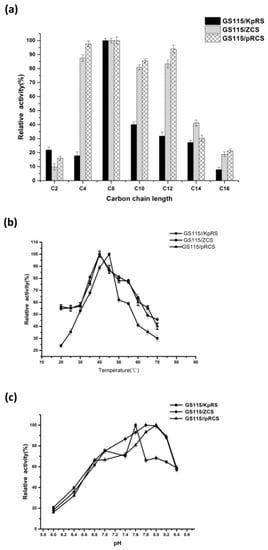
Figure 4.
Characteristics of the displayed targeted lipases Rhizopus oryzae lipase (ROL) and Candida rugosa lipase 1 (CRL1). (a) Effects of substrate specificity on single-displayed ROL and CRL1 and codisplayed lipases on Pichia pastoris strains. Bioactivity was measured by spectrophotometry at 40 °C in 10 mM acetonitrile containing C2, C4, C8, C10, C12, C14, and C16 p-nitrophenol esters. The optimal substrate for single-displayed ROL and CRL1 and codisplayed lipases was pNP-octanoate (C8). (b) Effects of temperature on single-displayed ROL and CRL1 and codisplayed lipase activity on P. pastoris strains. Surface-displayed lipase activity at 20 °C–70 °C was measured at pH 8.0 for 10 min. Relative activity was calculated by comparing bioactivity on pNP-octanoate (C8) substrate at 40 °C and pH 8.0. The optimal temperature was 45 °C for ROL on GS115/KpRS. The optimal temperature was 40 °C for CRL1 on GS115/ZCS and codisplayed lipases on GS115/pRCS. Data are means ± standard deviations (SDs) of three independent tests. (c) Effects of pH on single-displayed ROL and CRL1 and codisplayed lipase activity on P. pastoris strains. Effects of pH on single-displayed and codisplayed lipase activity were determined by comparing the original bioactivity on pNP-octanoate (C8) at 40 °C and pH 6.0–8.5 for 10 min. The optimal pH values were 7.6, 7.8–8.0, and 8.0 for single-displayed ROL and CRL1 and codisplayed ROL plus CRL1 on GS115, respectively. Data are means ± SDs of three independent tests.
Figure 4b shows that single-displayed GS115/KpRS had the highest enzyme activity on pNPC (C8) at 45 °C. In contrast, the optimal temperature for free ROL lipase was 30 °C when pNPC was the substrate [27]. Previous reports suggested that cell wall protection stabilizes self-immobilized CRL1 [12,19]. The optimal temperature for single-displayed CRL1 in GS115/ZCS and free-form CRL1 was 40 °C [2,27]. Lipases codisplayed on GS115/pRCS presented with maximum enzyme activity at 40 °C.
The optimal pH for the displayed lipases on recombinant yeast were determined for the optimal substrate and temperature. The pH was in the range of 6.0–8.4. Figure 4c shows that the optimal pH was 7.6 for single-displayed ROL on GS115/KpRS. For free-form ROL, it was 8.0 [28]. The optimal pH range was 7.8–8.0 for free-form CRL1 and single-displayed CRL1 in GS115/ZCS [2,29]. The optimal pH was 8.0 for the codisplayed lipases on GS115/pRCS.
2.4. Application of Surface-Displayed Lipases as Whole-Cell Biocatalysts
The rates of biodiesel conversion from tallow tree seed oil and methanol were 22.35% and 30.98%, respectively, for single-displayed ROL on GS115/KpRS and codisplayed lipases on GS115/pRCS. No fatty acid methyl ester was generated using single-displayed CRL1 on GS115/ZCS alone. The fatty acid methyl ester production rate was 1.4-fold higher for codisplayed lipases on GS115/pRCS harboring ROL and CRL1than that obtained using single-displayed ROL. Hence, the combination of ROL plus CRL1 synergistically catalyzes biodiesel production when the enzymes are coexpressed and self-immobilized on yeast cells.
The reaction rate varies with enzyme concentration [5,30]. The low biodiesel yield obtained here might be the consequence of the low lipase activity in 0.5 g dried cells. High cell density fermentation on GS115/pRCS will be performed to produce numerous lipases in 10 L fermenters. Future research will determine the optimal concentration of dual-displayed lipases required for biodiesel production. To accelerate biodiesel conversion catalyzed by self-immobilized dual lipases, the oil source, methanol, and water content must be optimized for transesterification.
3. Materials and Methods
3.1. Strains, Vectors, and Culture Media
Escherichia coli DH5α (TaKaRa Biotechnology Co. Ltd., Dalian, China) was the host for the DNA manipulations. P. pastoris strain GS115 (Mut+, his4−) for heterologous target lipase expression, Saccharomyces cerevisiae strain for cloning the sed1 anchoring gene, and pPIC9K and pPICZαA expression vectors were purchased from Invitrogen (Carlsbad, CA, USA). Escherichia coli/ROL-9K and E. coli/ZXFα–CRL1 were constructed to amplify ROL and CRL1. The E. coli was cultivated in Luria-Bertani (LB) medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) sodium chloride supplemented with 100 μg/mL ampicillin or 50 μg/mL kanamycin). The pMD18-T (simple) with Amp+ resistance was purchased from TaKaRa Biotechnology Co. Ltd. (Dalian, China) and used as the cloning vector. Yeast extract-peptone-dextrose (YPD) medium (2% (w/v) tryptone, 1% (w/v) yeast extract, 2% (w/v) dextrose), buffered glycerol-complex (BMGY) medium (2% (w/v) tryptone, 1% (w/v) yeast extract, 1% glycerol, 1 M potassium phosphate, pH 7.0, 1.34% yeast nitrogen base (YNB), and 4 × 10−5% (w/v) biotin), or BMMY medium (2% (w/v) tryptone, 1% (w/v) yeast extract, 1 M potassium phosphate (pH 7.0), 1.34% (w/v) YNB, 4 × 10−5% (w/v) biotin, and 0.25% (v/v) methanol) was used to cultivate P. pastoris GS115 and the recombinants. Minimal dextrose medium (1.34% (w/v YNB, 4 × 10−5% (w/v) biotin, 2% (w/v) dextrose, and 2% (w/v) agar) was used to select the P. pastoris transformants. Yeast YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose, and 2% (w/v) agar) containing G418 (Gibco BRL, Grand Island, NY, USA) at final concentrations of 0.25 mg/mL, 0.5 mg/mL, 0.75 mg/mL, 1.0 mg/mL, 1.5 mg/mL, 1.75 mg/mL, 2.0 mg/mL, 3.0 mg/mL, or 4.0 mg/mL was used to screen for transformants with multiple copies. Tributyrin medium (2% (w/v) tryptone, 1% (w/v) yeast extract, 2% (w/v) agar, 10% (w/v) 1 M potassium phosphate (pH 6.0), 10% of 10 × YNB, and 4 × 10−5% (w/v) biotin) was used to assay extracellular lipase activity. PrimeSTARTM DNA polymerase, restriction enzymes, and a DNA ligation kit (v. 2.0) were purchased from TaKaRa Biotechnology Co. Ltd. (Dalian, China). The gel extraction kit and the plasmid mini kit I were purchased from Omega Bio-Tek (Norcross, GA, USA).
3.2. Plasmid Construction
The ROL gene was amplified with ROL-9K/E. coli total DNA as the template and ROL-F/ROL-R as the primer pair. The amplified ROL product was digested with EcoRI and MluI. The Sed1 anchoring gene was amplified with S. cerevisiae total DNA as the template and Sed1-F/Sed1-R as the primer pair. The amplified Sed1 product was digested with MluI and NotI. Figure 5a shows that the digested products were ligated into the pPIC9K vector between the EcoRI and NotI sites. The recombinant plasmid product was designated pPIC9K-ROL-Sed1 (KpRS).
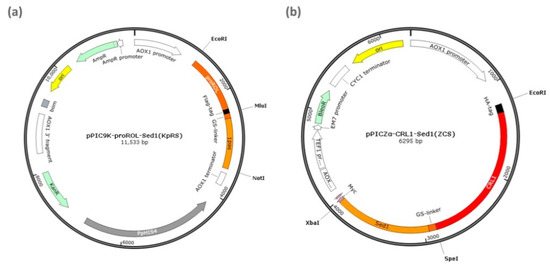
Figure 5.
Display plasmids encoding sed-targeted genes. (a) pPIC9K-ROL-sed1 (KpRS) encoding Rhizopus oryzae lipase (ROL) and sed1 fusion protein. Flag-tag between proROl and sed1 was used for immunofluorescence detection. (b) pPICZα-CRL1-sed1 (ZCS) encoding Candida rugosa lipase 1 (CRL1) and sed1 fusion protein. The HA peptide at the N-terminal of CRL1 was used in the immunofluorescence analysis.
The CRL1 gene was amplified with the E. coli/ZXFα–CRL1 total DNA template. The amplified CRL1 product was digested with EcoRI and SpeI. The Sed1 anchoring gene was amplified from S. cerevisiae total DNA with the Sed1-F2/Sed1-R2 primer pair and ligated into pPICZαA at the EcoRI and XbaI sites after SpeI and XbaI treatment. The recombinant plasmid product was designated pPICZα-CRL1-sed1 (ZCS) (Figure 5b). All constructed recombinant plasmids were transformed into E. coli DH5α and the transformants were screened in LB medium containing 50 μg/mL kanamycin. The GS115/CpRS and GS115/pRCS recombinants were generated with the linearized expression vectors KpRS and ZCS and by second electroporation into the GS115/ZCS and GS115/KpRS host cells. The PCR program was 94 °C for 5 min, 30 cycles of 94 °C for 1 min, 58 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min. All plasmid sequence constructs were confirmed by DNA sequencing at Tianyi Huiyuan (Wuhan, China). The primers used are listed in Table 1.

Table 1.
Primers used in the present study.
3.3. Yeast Transformation and Inducible Expression
The linearized expression vector plasmids were verified by sequencing and introduced into P. pastoris GS115 competent cells by electroporation with a Gene Pulser (Bio-Rad Laboratories, Hercules, CA, USA; 1500 V; 25 μF; 200 Ω; 0.2 cm cuvette) according to the manufacturer’s instructions. Transformants were screened on G418 YPD plates containing 300 μg/mL zeocin and incubated at 28 °C for 2–3 d. Colonies surrounded by large hydrolysis zones were selected, inoculated into YPD medium, and incubated at 200 rpm and 30 °C overnight. Correct recombinant cultures were inoculated into BMGY medium at 200 rpm and 28 °C. At OD600 = 4.0–6.0, the cultures were centrifuged at 5,000 rpm and 4 °C for 3 min, and the recombinant-bearing sediments were resuspended in 50 mL sterile BMMY induction medium for the activity assay. Fresh methanol was added to a final 1% (v/v) concentration every 24 h.
3.4. Immunofluorescence and Flow Cytometric Assays
An immunofluorescence assay was performed on the displayed ROL using mouse monoclonal Flag-tag and fluorescein isothiocyanate-conjugated (FITC) goat antimouse immunoglobulin G (IgG) detection. For the displayed CRL1, mouse monoclonal anti-HA primary antibody and Alexa Fluor 555-labeled donkey antimouse IgG (H+L) secondary antibody were used. Recombinants with the strongest enzyme activity on olive oil were incubated in a shaking flask for 120 h, centrifuged at 12,000 rpm, harvested, resuspended in phosphate-buffered saline (PBS; pH 7.4) at a final 1 mg/mL bovine serum albumin concentration, combined with the rabbit monoclonal Flag-tag primary antibody, and incubated at 4 °C for 2 h. The cells were washed three times with PBS, incubated with FITC-labeled goat antimouse IgG or Alexa Fluor 555-labeled donkey antimouse IgG (H+L) on ice in the dark for 1–2 h, resuspended in PBS, and observed by fluorescence microscopy (Nikon Corp., Chiyoda, Japan) and flow cytometry (Beckman Coulter, Fullerton, CA, USA). P. pastoris GS115 harboring empty pPIC9K or pPICZαA plasmids was subjected to the same treatment and served as negative controls.
3.5. Plate Assay
Normally growing recombinants were inoculated on tributyrin agar (0.5% v/v), incubated at 28 °C for 5 d, and exposed to 200 μL anhydrous methanol every 24 h to induce ROL and CRL1 lipases. Lipase activity was evaluated based on the hydrolysis zones around the colonies.
3.6. Displayed Recombinant Activity Assay
The activity of the displayed recombinant was measured by the acid–base titration method [31]. One milliliter of resuspended cells was added to 4 mL substrate (25% (v/v) olive oil emulsified with 2% (w/v) polyvinyl alcohol) plus 5 mL of 50 mM phosphate buffer (pH 8.0), and the mixture was incubated at 150 rpm and 40 °C for 10 min. The reaction was immediately stopped with 15 mL ethanol. Two drops of 1% (w/v) phenolphthalein reagent were added, the mixture was titrated with 50 mM NaOH, and the volume of NaOH solution consumed was measured and recorded. The assay was repeated in triplicate.
The assay determined the amount of dry cells and lipase liberating the equivalent of 1 μmol free fatty acid (FA) min−1 via olive oil hydrolysis. This amount of lipase was defined as one enzyme activity unit (U). The cell dry weight was measured by harvesting the cells in a preweighed centrifuge tube, centrifuging them at 8000 × g for 15 min, washing them in distilled water, and dividing them into two equal portions. One part was dried to a constant weight at 105 °C, whereas the other was used to estimate enzyme activity.
3.7. Characterization of the Displayed ROL and CRL1
Lipase activity toward p-nitrophenolate was measured according to a previously reported method. One enzyme activity unit (U) was defined as the amount of enzyme required to liberate 1 μmol p-nitrophenol per minute from the substrate p-nitrophenolate [32]. The enzyme activity was calculated using the calibration curve prepared by plotting OD410 against enzyme activity under the assay conditions.
To establish recombinant substrate chain length specificity, 10 mM acetonitrile solutions of C2, C4, C8, C10, C12, C14, and C16 p-nitrophenol esters were prepared. The system was preheated in a 40 °C water bath for 5 min. Then, a 50 μL cell suspension was added and incubated at 150 rpm and 40 °C for 10 min. The reaction was terminated with 4 mL of 95% (v/v) methanol. The suspension was centrifuged at 5000 rpm and 4 °C for 3 min, and the supernatant OD410 was measured using a spectrophotometer. The enzyme activity was calculated and the calibration curve, plotting OD410 against enzyme activity, indicated the optimal substrate chain length. For the blank control, the cell suspension was replaced with 50 μL Tris-HCl buffer. Each treatment was measured in triplicate.
The optimal temperature was determined at 20 °C–70 °C for 10 min. The optimal p-Nitrophenol Ester (pNP) was the substrate. The enzyme activity was calculated and the calibration curve, plotting OD410 against enzyme activity, identified the optimal reaction temperature. Each treatment was measured in triplicate.
The optimal pH was determined in a reaction system containing the optimal pNP substrate concentration and reaction temperature. The buffer was replaced with potassium phosphate and tris-HCl buffer in the 6.0–8.5 pH range. The enzyme activity was calculated and the calibration curve, plotting OD410 against enzyme activity, revealed the optimal pH. Each treatment was measured in triplicate. The effects of the p-nitrophenolate substrate on the enzymes were analyzed by pNP colorimetry [31]. A standard pNP curve was plotted based on the correlation between pNP concentration and OD410.
3.8. Surface-Displayed Recombinants as Whole-Cell Catalysts for Biodiesel Production
Single-displayed and codisplayed recombinants were collected, infiltrated with 5% (w/v) PEG2000 as a cryoprotectant, and freeze-dried. Whole cell-catalyzed transesterification was conducted in a tert-butanol reaction system contained in a 50 mL screw-capped flask. The reaction proceeded at 150 rpm and 40 °C for 96 h. The reagents included 4:1 molar methanol: oil, 2.19 g Chinese tallow tree seed oil, 1.1107 mL tert-butanol, 0.404 mL methanol, and 0.5 g whole-cell catalyst. At the end, 100 μL of each reaction mixture was collected and centrifuged at 12,000 rpm for 2 min. Then, 10 µL of each supernatant was mixed with 290 μL hexane and 300 μL heptadecanoic acid methyl ester-hexane solution as the internal standard. Thereafter, 1 μL mixture was used in cryogenic gas chromatography as described in a previous study [33]. The biodiesel methyl ester content was measured using gas chromatography (Fuli, Wenlin, China). The device was fitted with an Agilent HP-INNOWax capillary column (0.25 mm × 30 m; Agilent Technologies, Waldbronn, Germany). The column temperature was maintained at 180 °C for 2 min, raised to 230 °C at 3 °C/min, and maintained at 230 °C for 1 min. The injector and detector temperatures were 230 °C and 280 °C, respectively. The methyl ester yield (wt/wt%) was calculated as the amount of methyl ester produced by the whole-cell catalysts divided by the amount of oil used.
4. Conclusions
When codisplayed and self-immobilized on yeast cell surfaces with the SED1 anchor, ROL and CRL1 synergistically catalyzed biodiesel production. The maximum activity of the recombinant strain GS115/pRCS codisplaying ROL and CRL1 was 470.59 U/g dry cells and was 3.9- and 1.3-fold higher than that for single-displayed ROL and CRL1, respectively. When the self-immobilized lipases were used as biocatalysts to produce biodiesel from tallow seed oil, the rate of methyl ester production from the codisplayed recombinant strains GS115/pRCS was 1.4-fold higher than that from single-displayed ROL. Our preliminary results indicated that the codisplayed ROL and CRL1 lipases act synergistically in biodiesel production and overcame the limitations of single-enzyme system catalysis. Triglyceride transesterification with short-chain alcohols and codisplayed synergetic enzymes is an alternative strategy for sustainable biodiesel production. The present study demonstrated a convenient method for preparing stable whole-cell catalysts and a viable approach for self-immobilizing synergetic enzymes on a single cell. The codisplayed synergetic enzymes on P. pastoris are promising as whole-cell catalysts for producing biodiesel from vegetable and waste oils.
Author Contributions
Conceptualization, L.X. and Y.Y.; methodology, Y.Y.; supervision, L.X. and Y.Y.; investigation, Y.Z. and L.X.; formal analysis and data curation, Y.Z., H.P., S.Y., Z.H., Q.Z., Y.H., and L.X.; writing—original draft preparation, L.X. and X.Y.; writing—review and editing, L.X., X.W., and H.P.; funding acquisition, L.X. All authors read and agreed to the final draft of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Nos. 31971206 and 31170078).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
The authors thank the Analytical and Testing Center of Huazhong University of Science and Technology for their assistance with the flow cytometric analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CRL1 | Candida rugosa lipase 1 |
| FITC | Fluorescein Isothiocyanate-Conjugated |
| KpRS | pPIC9K-ROL-Sed1 |
| pNp | p-Nitrophenol Ester |
| ROL | Rhizopus oryzae Lipase |
| ZCS | pPICZα-CRL1-Sed1 |
References
- Reetz, M.T. Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 2002, 6, 145–150. [Google Scholar] [CrossRef]
- Ribeiro, B.D.; de Castro, A.M.; Coelho, M.A.Z.; Freire, D.M.G. Production and use of lipases in bioenergy: A review from the feedstocks to biodiesel production. Enzym. Res. 2011, 2011, 1–16. [Google Scholar] [CrossRef]
- Lee, N.H.; Kim, J.M.; Shin, H.Y.; Kang, S.W.; Kim, S.W. Biodiesel production using a mixture of immobilized Rhizopus oryzae and Candida rugosa lipases. Biotechnol. Bioprocess Eng. 2006, 11, 522–525. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Park, C.; Tae, B.; Han, S.O.; Kim, S.W. Development of batch and continuous processes on biodiesel production in a packed-bed reactor by a mixture of immobilized Candida rugosa and Rhizopus oryzae lipases. Appl. Biochem. Biotechnol. 2009, 161, 365–371. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Kang, S.W.; Song, Y.S.; Park, C.; Han, S.O.; Kim, S.W. Biodiesel production by a mixture of Candida rugosa and Rhizopus oryzae lipases using a supercritical carbon dioxide process. Bioresour. Technol. 2011, 102, 2105–2108. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Han, S.O.; Park, C.; Kim, S.W. Co-immobilization of Candida rugosa and Rhyzopus oryzae lipases and biodiesel production. Korean J. Chem. Eng. 2013, 30, 1335–1338. [Google Scholar] [CrossRef]
- Sirisha, V.; Jain, A.; Jain, A. Enzyme immobilization. Adv. Food Nutr. Res. 2016, 79, 179–211. [Google Scholar] [CrossRef] [PubMed]
- Hanes, J.; Jermutus, L.; Weber-Bornhauser, S.; Bosshard, H.R.; Plückthun, A. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc. Natl. Acad. Sci. USA 1998, 95, 14130–14135. [Google Scholar] [CrossRef]
- Grabherr, R.; Ernst, W.; Oker-Blom, C.; Jones, I. Developments in the use of baculoviruses for the surface display of complex eukaryotic proteins. Trends Biotechnol. 2001, 19, 231–236. [Google Scholar] [CrossRef]
- Shibasaki, S.; Ueda, M.; Ye, K.; Shimizu, K.; Kamasawa, N.; Osumi, M.; Tanaka, A. Creation of cell surface-engineered yeast that display different fluorescent proteins in response to the glucose concentration. Appl. Microbiol. Biotechnol. 2001, 57, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, P.; Gunneriusson, E.; Nygren, P.-Å.; Ståhl, S. Display of proteins on bacteria. J. Biotechnol. 2002, 96, 129–154. [Google Scholar] [CrossRef]
- Pan, X.X.; Xu, L.; Zhang, Y.; Xiao, X.; Wang, X.F.; Liu, Y.; Zhang, H.J.; Yan, Y.-J. Efficient display of active Geotrichum sp. lipase on Pichia pastoris cell wall and its application as a whole-cell biocatalyst to enrich EPA and DHA in fish oil. J. Agric. Food Chem. 2012, 60, 9673–9679. [Google Scholar] [CrossRef]
- Tabañag, I.D.F.; Chu, I.-M.; Wei, Y.-H.; Tsai, S.-L. The Role of yeast-surface-display techniques in creating biocatalysts for consolidated bioprocessing. Catalysts 2018, 8, 94. [Google Scholar] [CrossRef]
- Moura, M.V.H.; Da Silva, G.P.; Machado, A.C.D.O.; Torres, F.A.G.; Freire, D.M.G.; Almeida, R.V. Displaying lipase B from Candida antarctica in Pichia pastoris using the yeast surface display approach: Prospection of a new anchor and characterization of the whole cell biocatalyst. PLoS ONE 2015, 10, e0141454. [Google Scholar] [CrossRef] [PubMed]
- Rockberg, J.; Löfblom, J.; Hjelm, B.; Uhlén, M.; Ståhl, S. Epitope mapping of antibodies using bacterial surface display. Nat. Chem. Biol. 2008, 5, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Cherf, G.M.; Cochran, J.R. Applications of yeast surface display for protein engineering. Methods Mol. Biol. 2015, 1319, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Andreu, C.; del Olmo, M.L. Yeast arming systems: Pros and cons of different protein anchors and other elements required for display. Appl. Microbiol. Biotechnol. 2018, 102, 2543–2561. [Google Scholar] [CrossRef]
- Linciano, S.; Pluda, S.; Bacchin, A.; Angelini, A. Molecular evolution of peptides by yeast surface display technology. MedChemComm 2019, 10, 1569–1580. [Google Scholar] [CrossRef]
- Xu, L.; Xiao, X.; Wang, F.; He, Y.; Yang, X.; Hu, J.; Feng, Z.; Yan, Y. Surface-displayed thermostable Candida rugosa Lipase 1 for docosahexaenoic acid enrichment. Appl. Biochem. Biotechnol. 2019, 190, 218–231. [Google Scholar] [CrossRef]
- Rizos, K.; Lattemann, C.T.; Bumann, D.; Meyer, T.F.; Aebischer, T. Autodisplay. Infect. Immun. 2003, 71, 6320–6328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vandormael, P.; Verschueren, P.; de Winter, L.; Somers, V. cDNA phage display for the discovery of theranostic autoantibodies in rheumatoid arthritis. Immunol. Res. 2017, 65, 307–325. [Google Scholar] [CrossRef]
- Park, T.J.; Zheng, S.; Kang, Y.J.; Lee, S.Y. Development of a whole-cell biosensor by cell surface display of a gold-binding polypeptide on the gold surface. FEMS Microbiol. Lett. 2009, 293, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ho, S.-H.; Hasunuma, T.; Chang, J.-S.; Ren, N.-Q.; Kondo, A. Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresour. Technol. 2016, 215, 324–333. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, R.; Lian, Z.; Wang, S.; Wright, A.T. Yeast cell surface display for lipase whole cell catalyst and its applications. J. Mol. Catal. B Enzym. 2014, 106, 17–25. [Google Scholar] [CrossRef]
- Hama, S.; Tamalampudi, S.; Fukumizu, T.; Miura, K.; Yamaji, H.; Kondo, A.; Fukuda, H. Lipase localization in Rhizopus oryzae cells immobilized within biomass support particles for use as whole-cell biocatalysts in biodiesel-fuel production. J. Biosci. Bioeng. 2006, 101, 328–333. [Google Scholar] [CrossRef]
- Nogueira, L.A. Does biodiesel make sense? Energy 2011, 36, 3659–3666. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, L.; Dai, M. A synergetic whole-cell biocatalyst for biodiesel production. RSC Adv. 2012, 2, 6170–6173. [Google Scholar] [CrossRef]
- Minning, S.; Schmidt-Dannert, C.; Schmid, R.D. Functional expression of Rhizopus oryzae lipase in Pichia pastoris: High-level production and some properties. J. Biotechnol. 1998, 66, 147–156. [Google Scholar] [CrossRef]
- Domínguez de María, P.; Sánchez-Montero, J.M.; Sinisterra, J.V.; Alcántara, A.R. Understanding Candida rugosa lipases: An overview. Biotechnol. Adv. 2006, 24, 180–196. [Google Scholar] [CrossRef]
- Gardossi, L.; Poulsen, P.B.; Ballesteros, A.; Hult, K.; Švedas, V.K.; Vasić-Rački, Đ.; Carrea, G.; Magnusson, A.; Schmid, A.; Wohlgemuth, R.; et al. Guidelines for reporting of biocatalytic reactions. Trends Biotechnol. 2010, 28, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Shimizu, S. Purification and characterization of the lipase from Pseudomonas fluorescens HU380. J. Biosci. Bioeng. 2003, 96, 219–226. [Google Scholar] [CrossRef]
- Bin Ibrahim, N.A.; Guo, Z.; Xu, X. Enzymatic interesterification of palm stearin and coconut oil by a dual lipase system. J. Am. Oil Chem. Soc. 2007, 85, 37–45. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, H.; Yan, Y. Optimization of lipase-catalyzed transesterification of lard for biodiesel production using response surface methodology. Appl. Biochem. Biotechnol. 2008, 160, 504–515. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).