Phylogeny and Structure of Fatty Acid Photodecarboxylases and Glucose-Methanol-Choline Oxidoreductases
Abstract
1. Introduction
2. Results
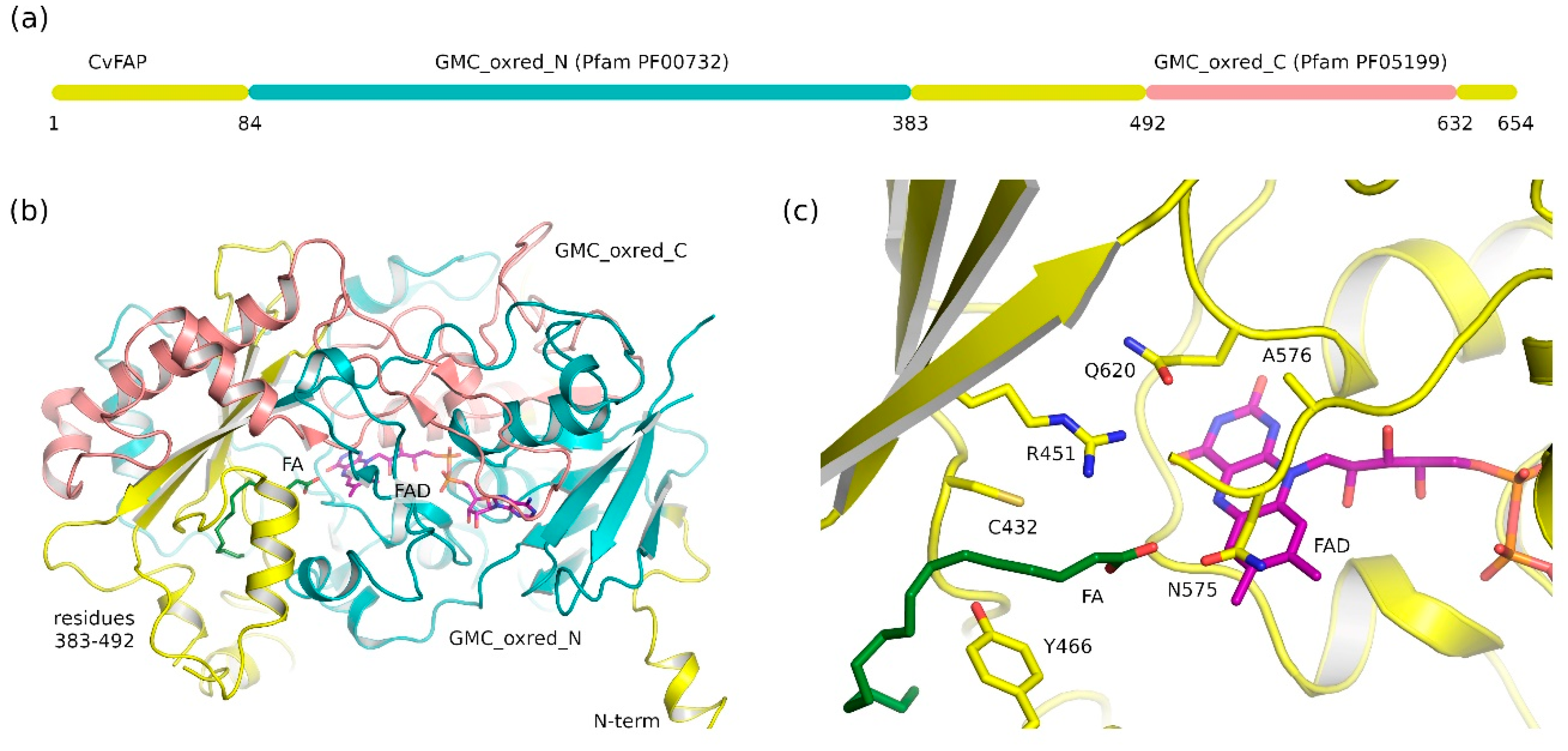
2.1. Fatty Acid Photodecarboxylases (FAP) Domain Annotation and Structure
2.2. Common Features of Known Glucose-Methanol-Choline (GMC) Proteins
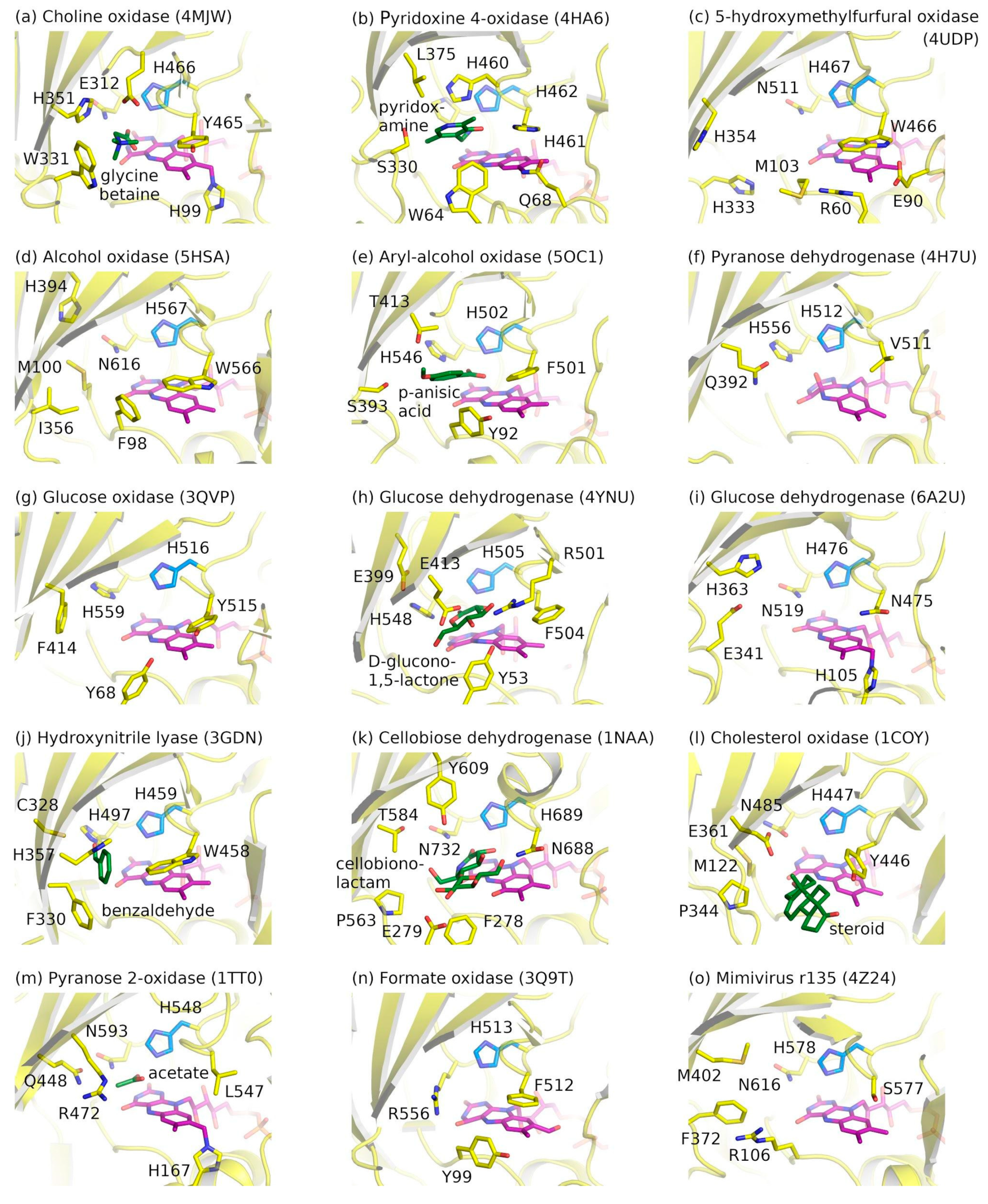
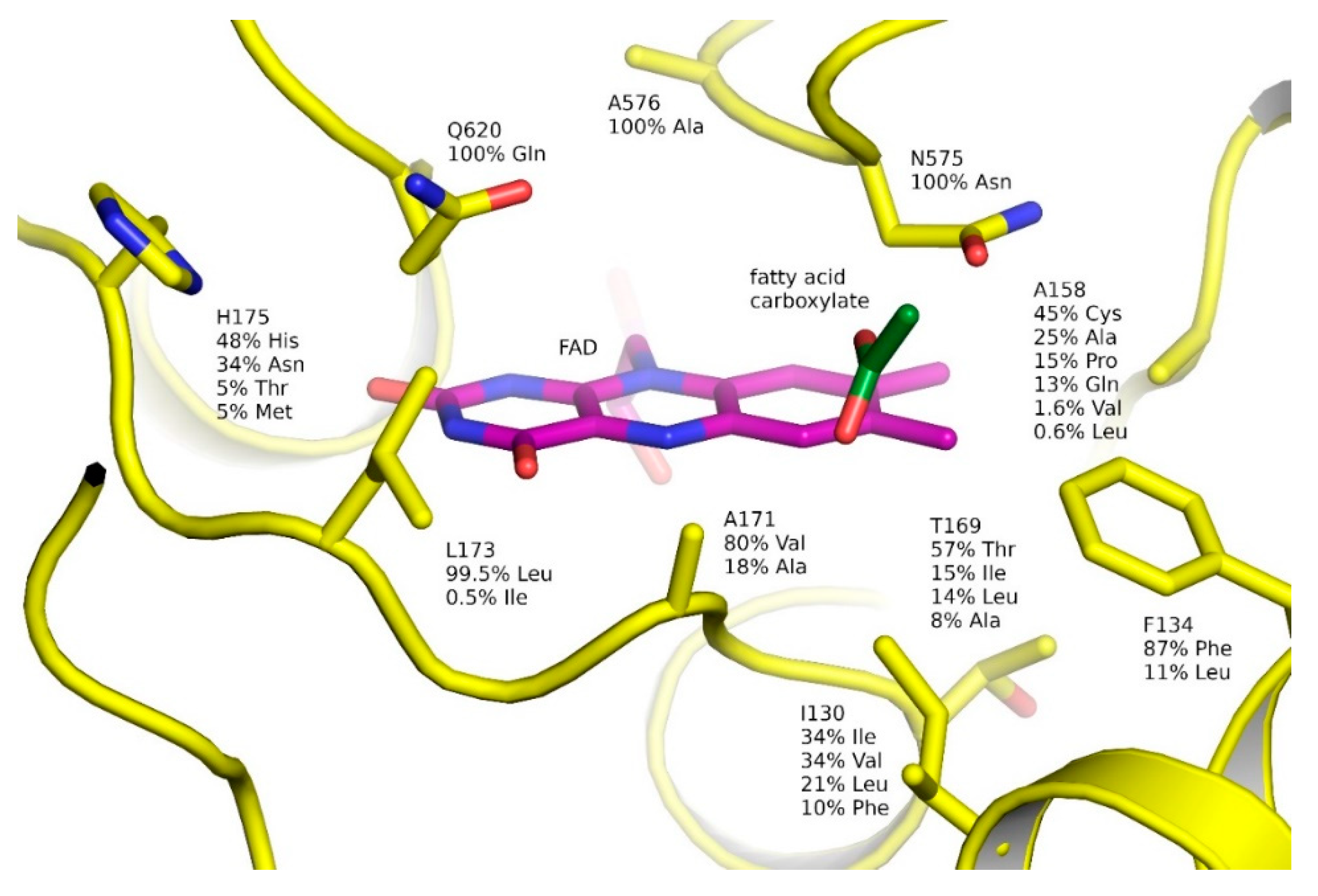
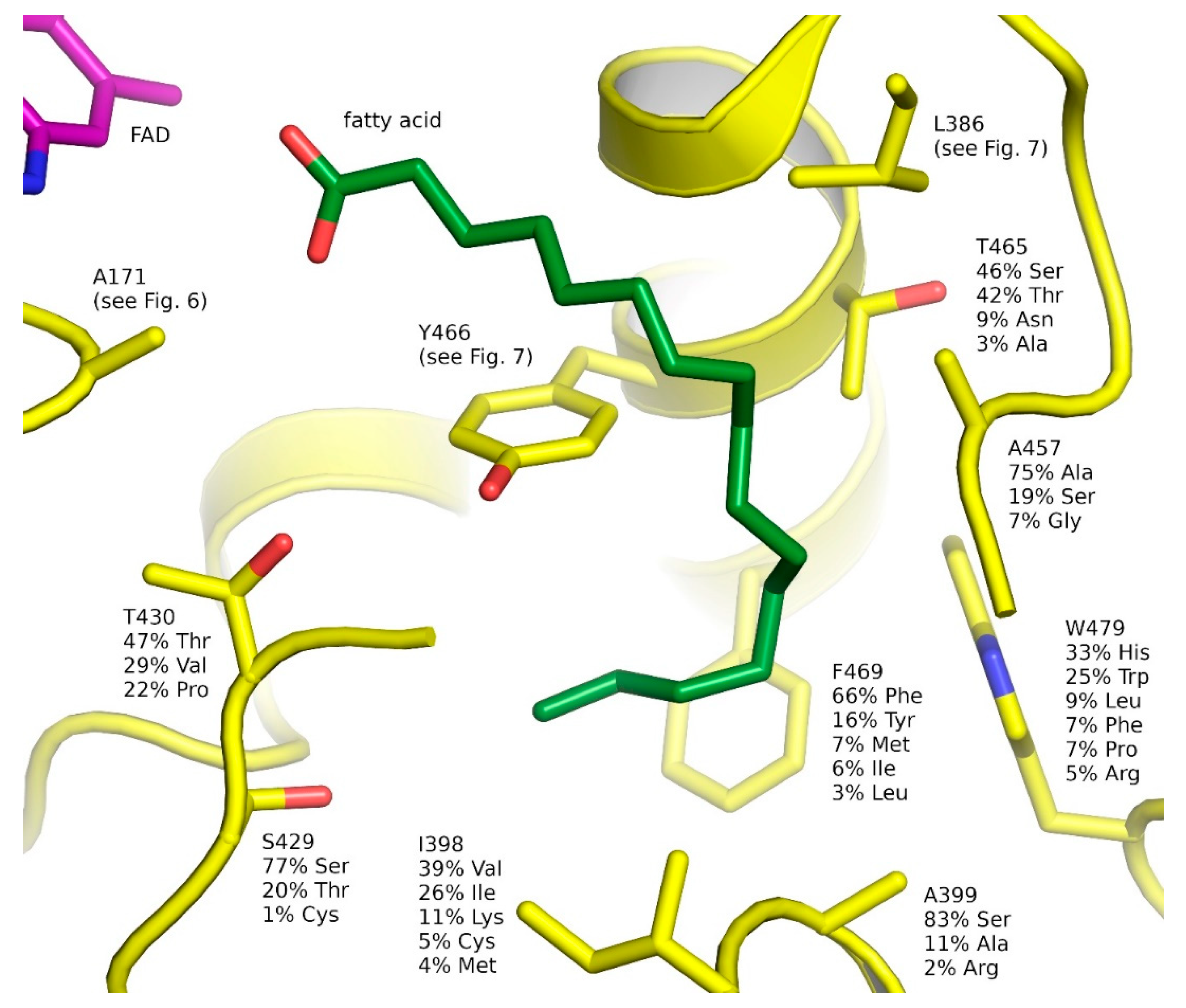
2.3. Ligand-Binding Pockets of GMC Family Proteins
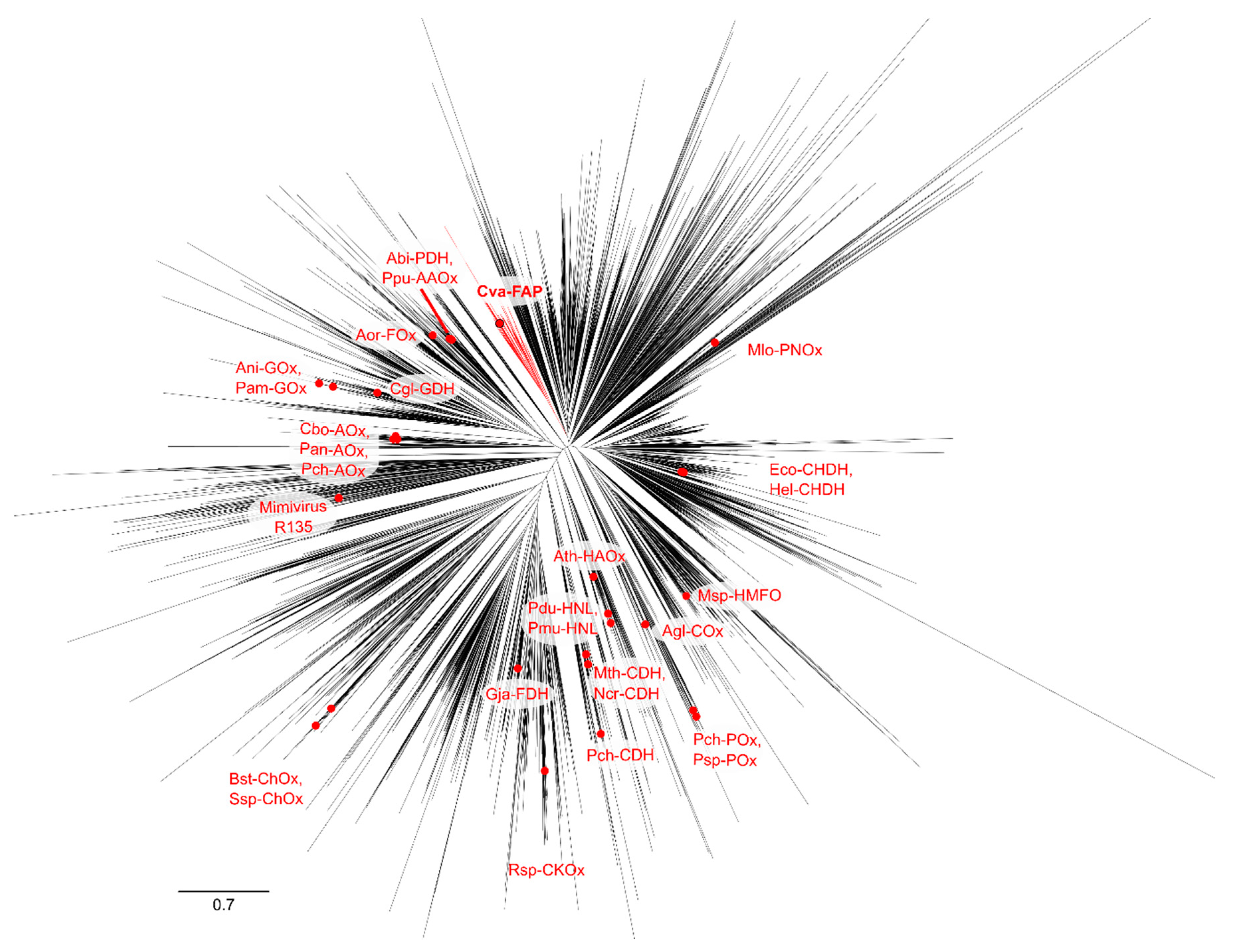
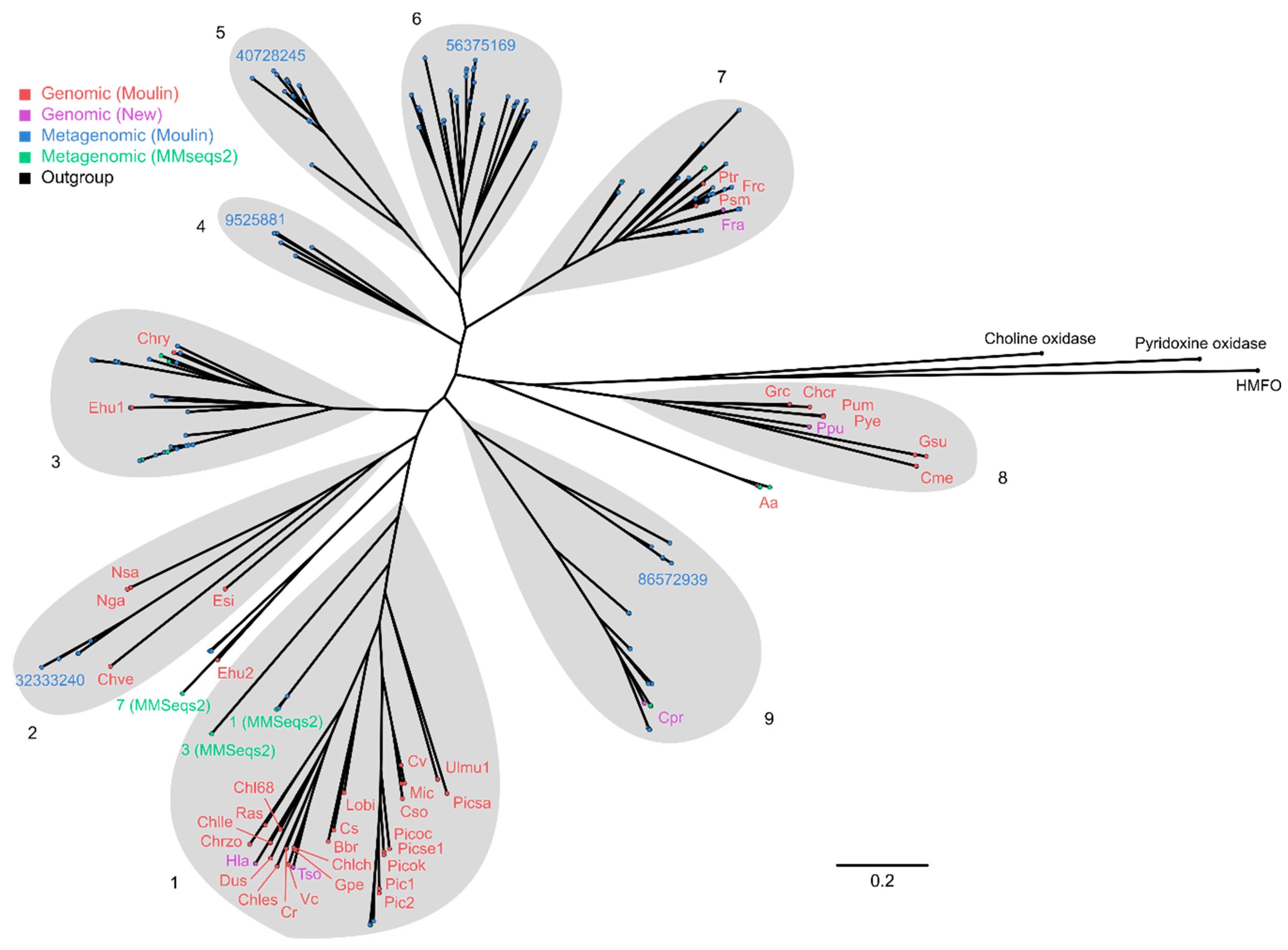
2.4. Phylogenetic Analysis of GMC Proteins
2.5. Phylogenetic Analysis of Putative FAP Proteins
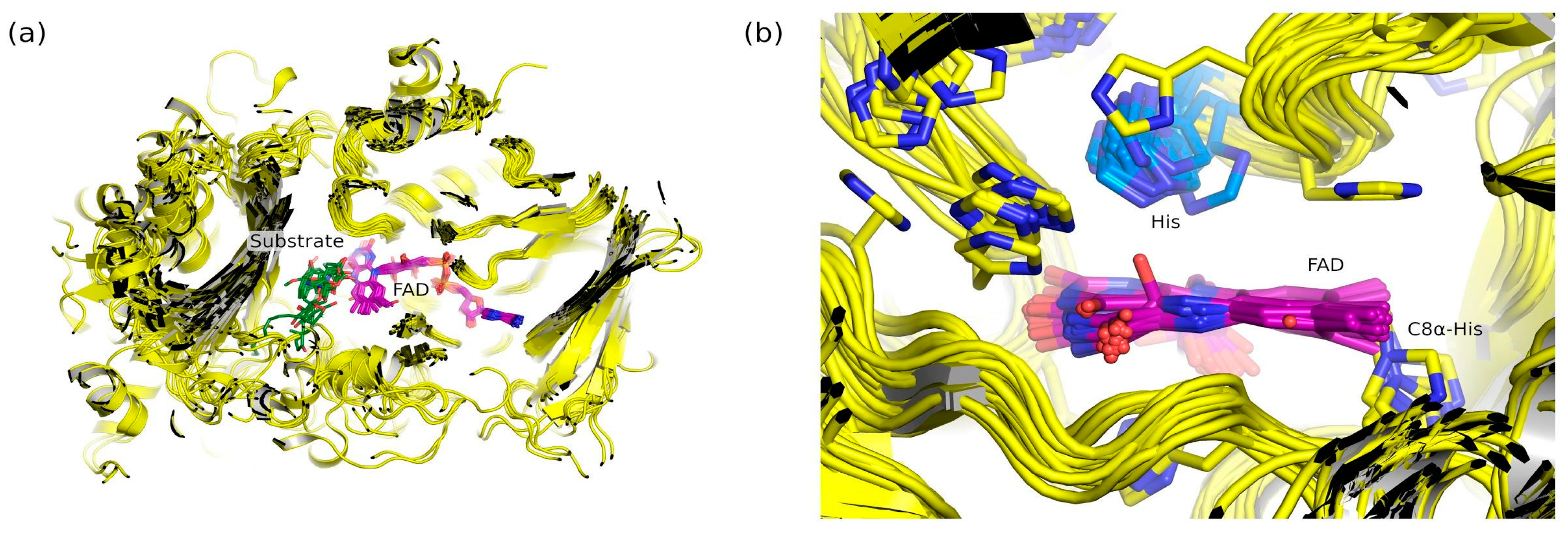
2.6. Natural Diversity of FAP Active Sites
3. Materials and Methods
3.1. Structural Analysis
3.2. Sequence Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rueda-Marquez, J.J.; Levchuk, I.; Fernández Ibañez, P.; Sillanpää, M. A critical review on application of photocatalysis for toxicity reduction of real wastewaters. J. Clean. Prod. 2020, 258, 120694. [Google Scholar] [CrossRef]
- Fujishima, A.; Honda, K. Electrochemical Photolysis of Water at a Semiconductor Electrode. Nature 1972, 238, 37–38. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jin, H.; Xu, P.; Zhu, C. When C–H bond functionalization meets visible-light photoredox catalysis. Tetrahedron Lett. 2014, 55, 36–48. [Google Scholar] [CrossRef]
- Ding, W.; Lu, L.-Q.; Liu, J.; Liu, D.; Song, H.-T.; Xiao, W.-J. Visible Light Photocatalytic Radical–Radical Cross-Coupling Reactions of Amines and Carbonyls: A Route to 1,2-Amino Alcohols. J. Org. Chem. 2016, 81, 7237–7243. [Google Scholar] [CrossRef]
- Primer, D.N.; Karakaya, I.; Tellis, J.C.; Molander, G.A. Single-Electron Transmetalation: An Enabling Technology for Secondary Alkylboron Cross-Coupling. J. Am. Chem. Soc. 2015, 137, 2195–2198. [Google Scholar] [CrossRef]
- Srivastava, V.P.; Yadav, L.D.S. Visible-Light-Triggered Oxidative C–H Aryloxylation of Phenolic Amidines; Photocatalytic Preparation of 2-Aminobenzoxazoles. Synlett 2013, 24, 2758–2762. [Google Scholar] [CrossRef]
- Keshari, T.; Srivastava, V.P.; Yadav, L.D.S. Visible-light-initiated photo-oxidative cyclization of phenolic amidines using CBr4—A metal free approach to 2-aminobenzoxazoles. RSC Adv. 2014, 4, 5815–5818. [Google Scholar] [CrossRef]
- Ischay, M.A.; Anzovino, M.E.; Du, J.; Yoon, T.P. Efficient Visible Light Photocatalysis of [2 + 2] Enone Cycloadditions. J. Am. Chem. Soc. 2008, 130, 12886–12887. [Google Scholar] [CrossRef]
- Lin, S.; Ischay, M.A.; Fry, C.G.; Yoon, T.P. Radical Cation Diels–Alder Cycloadditions by Visible Light Photocatalysis. J. Am. Chem. Soc. 2011, 133, 19350–19353. [Google Scholar] [CrossRef]
- Du, J.; Skubi, K.L.; Schultz, D.M.; Yoon, T.P. A Dual-Catalysis Approach to Enantioselective [2 + 2] Photocycloadditions Using Visible Light. Science 2014, 344, 392–396. [Google Scholar] [CrossRef]
- Tu, H.; Zhu, S.; Qing, F.-L.; Chu, L. Visible-light-induced halogenation of aliphatic CH bonds. Tetrahedron Lett. 2018, 59, 173–179. [Google Scholar] [CrossRef]
- Marzo, L.; Pagire, S.K.; Reiser, O.; König, B. Visible-Light Photocatalysis: Does It Make a Difference in Organic Synthesis? Angew. Chem. Int. Ed. 2018, 57, 10034–10072. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.; Lewis, J. Introduction: Biocatalysis in Industry. Chem. Rev. 2018, 118, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Truppo, M.D. Biocatalysis in the Pharmaceutical Industry: The Need for Speed. ACS Med. Chem. Lett. 2017, 8, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef]
- Bloom, J.D.; Arnold, F.H. In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 9995–10000. [Google Scholar] [CrossRef]
- Hellinga, H.W. Rational protein design: Combining theory and experiment. Proc. Natl. Acad. Sci. USA 1997, 94, 10015–10017. [Google Scholar] [CrossRef]
- Huang, P.-S.; Boyken, S.E.; Baker, D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef]
- Korendovych, I.V.; DeGrado, W.F. De novo protein design, a retrospective. Q. Rev. Biophys. 2020, 53. [Google Scholar] [CrossRef]
- Bloom, J.D.; Meyer, M.M.; Meinhold, P.; Otey, C.R.; MacMillan, D.; Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005, 15, 447–452. [Google Scholar] [CrossRef]
- Steiner, K.; Schwab, H. Recent advances in rational approaches for enzyme engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209010. [Google Scholar] [CrossRef] [PubMed]
- Otte, K.B.; Hauer, B. Enzyme engineering in the context of novel pathways and products. Curr. Opin. Struct. Biotechnol. 2015, 35, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.; Tawfik, D.S. Enzyme engineering: Reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 2017, 47, 140–150. [Google Scholar] [CrossRef]
- Losi, A.; Gärtner, W. Old chromophores, new photoactivation paradigms, trendy applications: Flavins in blue light-sensing photoreceptors. Photochem. Photobiol. 2011, 87, 491–510. [Google Scholar] [CrossRef]
- Conrad, K.S.; Manahan, C.C.; Crane, B.R. Photochemistry of flavoprotein light sensors. Nat. Chem. Biol. 2014, 10, 801–809. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Shemetov, A.A.; Kaberniuk, A.A.; Verkhusha, V.V. Natural Photoreceptors as a Source of Fluorescent Proteins, Biosensors, and Optogenetic Tools. Annu. Rev. Biochem. 2015, 84, 519–550. [Google Scholar] [CrossRef]
- Björn, L.O. Photoenzymes and Related Topics: An Update. Photochem. Photobiol. 2018, 94, 459–465. [Google Scholar] [CrossRef]
- Kaschner, M.; Loeschcke, A.; Krause, J.; Minh, B.Q.; Heck, A.; Endres, S.; Svensson, V.; Wirtz, A.; von Haeseler, A.; Jaeger, K.-E.; et al. Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria. Mol. Microbiol. 2014, 93, 1066–1078. [Google Scholar] [CrossRef]
- Shen, G.; Canniffe, D.P.; Ho, M.-Y.; Kurashov, V.; van der Est, A.; Golbeck, J.H.; Bryant, D.A. Characterization of chlorophyll f synthase heterologously produced in Synechococcus sp. PCC 7002. Photosynth. Res. 2019, 140, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Massey, V.; Hemmerich, P. Active-site probes of flavoproteins. Biochem. Soc. Trans. 1980, 8, 246–257. [Google Scholar] [CrossRef]
- Heuts, D.P.H.M.; Scrutton, N.S.; McIntire, W.S.; Fraaije, M.W. What’s in a covalent bond? FEBS J. 2009, 276, 3405–3427. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; Mattevi, A. Flavoenzymes: Diverse catalysts with recurrent features. Trends Biochem. Sci. 2000, 25, 126–132. [Google Scholar] [CrossRef]
- Mansoorabadi, S.O.; Thibodeaux, C.J.; Liu, H. The Diverse Roles of Flavin Coenzymes Nature’s Most Versatile Thespians. J. Org. Chem. 2007, 72, 6329–6342. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Gómez Castellanos, J.R.; Gadda, G.; Fraaije, M.W.; Mattevi, A. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. Chem. Rev. 2018, 118, 1742–1769. [Google Scholar] [CrossRef] [PubMed]
- Hall, M. Flavoenzymes for biocatalysis. In The Enzymes; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Martin, C.; Binda, C.; Fraaije, M.; Mattevi, A. The multipurpose family of flavoprotein oxidases. In The Enzymes; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Macheroux, P.; Kappes, B.; Ealick, S.E. Flavogenomics—A genomic and structural view of flavin-dependent proteins. FEBS J. 2011, 278, 2625–2634. [Google Scholar] [CrossRef]
- Sützl, L.; Foley, G.; Gillam, E.M.J.; Bodén, M.; Haltrich, D. The GMC superfamily of oxidoreductases revisited: Analysis and evolution of fungal GMC oxidoreductases. Biotechnol. Biofuels 2019, 12, 118. [Google Scholar] [CrossRef]
- Dijkman, W.P.; de Gonzalo, G.; Mattevi, A.; Fraaije, M.W. Flavoprotein oxidases: Classification and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5177–5188. [Google Scholar] [CrossRef]
- Sampson, N.S.; Vrielink, A. Cholesterol Oxidases: A Study of Nature’s Approach to Protein Design. Acc. Chem. Res. 2003, 36, 713–722. [Google Scholar] [CrossRef]
- Cavener, D.R. GMC oxidoreductases: A newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 1992, 223, 811–814. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Ferri, S.; Kojima, K.; Sode, K. Review of glucose oxidases and glucose dehydrogenases: A bird’s eye view of glucose sensing enzymes. J. Diabetes Sci. Technol. 2011, 5, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Mano, N. Engineering glucose oxidase for bioelectrochemical applications. Bioelectrochemistry 2019, 128, 218–240. [Google Scholar] [CrossRef] [PubMed]
- Okuda-Shimazaki, J.; Yoshida, H.; Sode, K. FAD dependent glucose dehydrogenases—Discovery and engineering of representative glucose sensing enzymes. Bioelectrochemistry 2020, 132, 107414. [Google Scholar] [CrossRef]
- Ozimek, P.; Veenhuis, M.; van der Klei, I.J. Alcohol oxidase: A complex peroxisomal, oligomeric flavoprotein. FEMS Yeast Res. 2005, 5, 975–983. [Google Scholar] [CrossRef]
- Pickl, M.; Fuchs, M.; Glueck, S.M.; Faber, K. The substrate tolerance of alcohol oxidases. Appl. Microbiol. Biotechnol. 2015, 99, 6617–6642. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Gadda, G. Alcohol oxidation by flavoenzymes. Biomol. Concepts 2014, 5, 299–318. [Google Scholar] [CrossRef]
- Serrano, A.; Carro, J.; Martínez, A.T. Reaction mechanisms and applications of aryl-alcohol oxidase. In The Enzymes; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Gadda, G. Choline oxidases. In The Enzymes; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Ikuta, S.; Imamura, S.; Misaki, H.; Horiuti, Y. Purification and characterization of choline oxidase from Arthrobacter globiformis. J. Biochem. 1977, 82, 1741–1749. [Google Scholar] [CrossRef]
- Tani, Y.; Mori, N.; Ogata, K.; Yamada, H. Production and Purification of Choline Oxidase from Cylindrocarpon didymum M-1. Agric. Biol. Chem. 1979, 43, 815–820. [Google Scholar] [CrossRef]
- Salvi, F.; Gadda, G. Human choline dehydrogenase: Medical promises and biochemical challenges. Arch. Biochem. Biophys. 2013, 537, 243–252. [Google Scholar] [CrossRef]
- Nishimura, I.; Okada, K.; Koyama, Y. Cloning and expression of pyranose oxidase cDNA from Coriolus versicolor in Escherichia coli. J. Biotechnol. 1996, 52, 11–20. [Google Scholar] [CrossRef]
- Mendes, S.; Banha, C.; Madeira, J.; Santos, D.; Miranda, V.; Manzanera, M.; Ventura, M.R.; van Berkel, W.J.H.; Martins, L.O. Characterization of a bacterial pyranose 2-oxidase from Arthrobacter siccitolerans. J. Mol. Catal. B Enzym. 2016, 133, S34–S43. [Google Scholar] [CrossRef]
- Herzog, P.L.; Sützl, L.; Eisenhut, B.; Maresch, D.; Haltrich, D.; Obinger, C.; Peterbauer, C.K. Versatile Oxidase and Dehydrogenase Activities of Bacterial Pyranose 2-Oxidase Facilitate Redox Cycling with Manganese Peroxidase In Vitro. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Peterbauer, C.K. Pyranose dehydrogenases: Rare enzymes for electrochemistry and biocatalysis. Bioelectrochemistry 2020, 132, 107399. [Google Scholar] [CrossRef]
- Willot, S.J.-P.; Hoang, M.D.; Paul, C.E.; Alcalde, M.; Arends, I.W.C.E.; Bommarius, A.S.; Bommarius, B.; Hollmann, F. FOx News: Towards Methanol-driven Biocatalytic Oxyfunctionalisation Reactions. ChemCatChem 2020, 12, 2713–2716. [Google Scholar] [CrossRef]
- Kondo, T.; Morikawa, Y.; Hayashi, N.; Kitamoto, N. Purification and characterization of formate oxidase from a formaldehyde-resistant fungus. FEMS Microbiol. Lett. 2002, 214, 137–142. [Google Scholar] [CrossRef]
- Uchida, H.; Hojyo, M.; Fujii, Y.; Maeda, Y.; Kajimura, R.; Yamanaka, H.; Sakurai, A.; Sakakibara, M.; Aisaka, K. Purification, characterization, and potential applications of formate oxidase from Debaryomyces vanrijiae MH201. Appl. Microbiol. Biotechnol. 2007, 74, 805–812. [Google Scholar] [CrossRef]
- Maeda, Y.; Doubayashi, D.; Oki, M.; Nose, H.; Sakurai, A.; Isa, K.; Fujii, Y.; Uchida, H. Expression in Escherichia coli of an unnamed protein gene from Aspergillus oryzae RIB40 and cofactor analyses of the gene product as formate oxidase. Biosci. Biotechnol. Biochem. 2009, 73, 2645–2649. [Google Scholar] [CrossRef]
- Bollella, P.; Gorton, L.; Antiochia, R. Direct Electron Transfer of Dehydrogenases for Development of 3rd Generation Biosensors and Enzymatic Fuel Cells. Sensors 2018, 18, 1319. [Google Scholar] [CrossRef]
- Adachi, T.; Kaida, Y.; Kitazumi, Y.; Shirai, O.; Kano, K. Bioelectrocatalytic performance of d-fructose dehydrogenase. Bioelectrochemistry 2019, 129, 1–9. [Google Scholar] [CrossRef]
- Ramkissoon, K.R.; Miller, J.K.; Ojha, S.; Watson, D.S.; Bomar, M.G.; Galande, A.K.; Shearer, A.G. Rapid Identification of Sequences for Orphan Enzymes to Power Accurate Protein Annotation. PLoS ONE 2013, 8, e84508. [Google Scholar] [CrossRef]
- Sundaram, T.K.; Snell, E.E. The bacterial oxidation of vitamin B6. V. The enzymatic formation of pyridoxal and isopyridoxal from pyridoxine. J. Biol. Chem. 1969, 244, 2577–2584. [Google Scholar] [PubMed]
- Kaneda, Y.; Ohnishi, K.; Yagi, T. Purification, Molecular Cloning, and Characterization of Pyridoxine 4-Oxidase from Microbacterium luteolum. Biosci. Biotechnol. Biochem. 2002, 66, 1022–1031. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, B.; Yoshikane, Y.; Yokochi, N.; Ohnishi, K.; Yagi, T. The nitrogen-fixing symbiotic bacterium Mesorhizobium loti has and expresses the gene encoding pyridoxine 4-oxidase involved in the degradation of vitamin B6. FEMS Microbiol. Lett. 2006, 234, 225–230. [Google Scholar] [CrossRef]
- Devi, S.; Kanwar, S.S. Cholesterol Oxidase: Source, Properties and Applications. Insights Enzym. Res. 2017, 1. [Google Scholar] [CrossRef]
- Csarman, F.; Wohlschlager, L.; Ludwig, R. Cellobiose dehydrogenase. In The Enzymes; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Bao, W.J.; Usha, S.N.; Renganathan, V. Purification and Characterization of Cellobiose Dehydrogenase, a Novel Extracellular Hemoflavoenzyme from the White-Rot Fungus Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1993, 300, 705–713. [Google Scholar] [CrossRef]
- Zamocky, M.; Ludwig, R.; Peterbauer, C.; Hallberg, B.M.; Divne, C.; Nicholls, P.; Haltrich, D. Cellobiose dehydrogenase—A flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr. Protein Pept. Sci. 2006, 7, 255–280. [Google Scholar] [CrossRef]
- Hickel, A.; Hasslacher, M.; Griengl, H. Hydroxynitrile lyases: Functions and properties. Physiol. Plant. 1996, 98, 891–898. [Google Scholar] [CrossRef]
- Dadashipour, M.; Asano, Y. Hydroxynitrile Lyases: Insights into Biochemistry, Discovery, and Engineering. ACS Catal. 2011, 1, 1121–1149. [Google Scholar] [CrossRef]
- Dijkman, W.P.; Fraaije, M.W. Discovery and characterization of a 5-hydroxymethylfurfural oxidase from Methylovorus sp. strain MP688. Appl. Environ. Microbiol. 2014, 80, 1082–1090. [Google Scholar] [CrossRef]
- Kim, E.-M.; Kim, J.; Seo, J.-H.; Park, J.-S.; Kim, D.-H.; Kim, B.-G. Identification and Characterization of the Rhizobium sp. Strain GIN611 Glycoside Oxidoreductase Resulting in the Deglycosylation of Ginsenosides. Appl. Environ. Microbiol. 2012, 78, 242–249. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Faust, A.; Trenkamp, S.; Bär, S.; Franke, R.; Efremova, N.; Tietjen, K.; Schreiber, L.; Saedler, H.; Yephremov, A. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 2006, 224, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Sorigué, D.; Légeret, B.; Cuiné, S.; Blangy, S.; Moulin, S.; Billon, E.; Richaud, P.; Brugière, S.; Couté, Y.; Nurizzo, D.; et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science 2017, 357, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Moulin, S.; Beyly, A.; Blangy, S.; Légeret, B.; Floriani, M.; Burlacot, A.; Sorigué, D.; Li-Beisson, Y.; Peltier, G.; Beisson, F. Fatty acid photodecarboxylase is an ancient photoenzyme responsible for hydrocarbon formation in the thylakoid membranes of algae. bioRxiv 2020. [Google Scholar] [CrossRef]
- Heyes, D.J.; Lakavath, B.; Hardman, S.J.O.; Sakuma, M.; Hedison, T.M.; Scrutton, N.S. Photochemical Mechanism of Light-Driven Fatty Acid Photodecarboxylase. ACS Catal. 2020, 10, 6691–6696. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, F.M.; Wadforth, C. Purification and some properties of alcohol oxidase from alkane-grown Candida tropicalis. Biochem. J. 1992, 282, 325–331. [Google Scholar] [CrossRef][Green Version]
- Robbins, J.M.; Geng, J.; Barry, B.A.; Gadda, G.; Bommarius, A.S. Photoirradiation Generates an Ultrastable 8-Formyl FAD Semiquinone Radical with Unusual Properties in Formate Oxidase. Biochemistry 2018, 57, 5818–5826. [Google Scholar] [CrossRef]
- Su, D.; Smitherman, C.; Gadda, G. A Metastable Photoinduced Protein–Flavin Adduct in Choline Oxidase, an Enzyme Not Involved in Light-Dependent Processes. J. Phys. Chem. B 2020, 124, 3936–3943. [Google Scholar] [CrossRef]
- Huijbers, M.M.E.; Zhang, W.; Tonin, F.; Hollmann, F. Light-Driven Enzymatic Decarboxylation of Fatty Acids. Angew. Chem. Int. Ed. Engl. 2018, 57, 13648–13651. [Google Scholar] [CrossRef]
- Xu, J.; Hu, Y.; Fan, J.; Arkin, M.; Li, D.; Peng, Y.; Xu, W.; Lin, X.; Wu, Q. Light-Driven Kinetic Resolution of α-Functionalized Carboxylic Acids Enabled by an Engineered Fatty Acid Photodecarboxylase. Angew. Chem. 2019, 131, 8562–8566. [Google Scholar] [CrossRef]
- Zhang, W.; Ma, M.; Huijbers, M.M.E.; Filonenko, G.A.; Pidko, E.A.; van Schie, M.; de Boer, S.; Burek, B.O.; Bloh, J.Z.; van Berkel, W.J.H.; et al. Hydrocarbon Synthesis via Photoenzymatic Decarboxylation of Carboxylic Acids. J. Am. Chem. Soc. 2019, 141, 3116–3120. [Google Scholar] [CrossRef]
- Cha, H.-J.; Hwang, S.-Y.; Lee, D.-S.; Kumar, A.R.; Kwon, Y.-U.; Voß, M.; Schuiten, E.; Bornscheuer, U.T.; Hollmann, F.; Oh, D.-K.; et al. Whole-Cell Photoenzymatic Cascades to Synthesize Long-Chain Aliphatic Amines and Esters from Renewable Fatty Acids. Angew. Chem. 2020, 132, 7090–7094. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Janin, J.; Chothia, C. Domains in proteins: Definitions, location, and structural principles. In Methods in Enzymology; Diffraction Methods for Biological Macromolecules Part B; Academic Press: Cambridge, MA, USA, 1985; Volume 115, pp. 420–430. [Google Scholar]
- Moore, A.D.; Björklund, Å.K.; Ekman, D.; Bornberg-Bauer, E.; Elofsson, A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008, 33, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Bateman, A. The evolution of protein domain families. Biochem. Soc. Trans. 2009, 37, 751–755. [Google Scholar] [CrossRef]
- Salvi, F.; Wang, Y.-F.; Weber, I.T.; Gadda, G. Structure of choline oxidase in complex with the reaction product glycine betaine. Acta Cryst. D 2014, 70, 405–413. [Google Scholar] [CrossRef]
- Mugo, A.N.; Kobayashi, J.; Yamasaki, T.; Mikami, B.; Ohnishi, K.; Yoshikane, Y.; Yagi, T. Crystal structure of pyridoxine 4-oxidase from Mesorhizobium loti. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2013, 1834, 953–963. [Google Scholar] [CrossRef]
- Dijkman, W.P.; Binda, C.; Fraaije, M.W.; Mattevi, A. Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase. ACS Catal. 2015, 5, 1833–1839. [Google Scholar] [CrossRef]
- Koch, C.; Neumann, P.; Valerius, O.; Feussner, I.; Ficner, R. Crystal Structure of Alcohol Oxidase from Pichia pastoris. PLoS ONE 2016, 11, e0149846. [Google Scholar] [CrossRef]
- Carro, J.; Martínez-Júlvez, M.; Medina, M.; Martínez, A.T.; Ferreira, P. Protein dynamics promote hydride tunnelling in substrate oxidation by aryl-alcohol oxidase. Phys. Chem. Chem. Phys. 2017, 19, 28666–28675. [Google Scholar] [CrossRef]
- Tan, T.C.; Spadiut, O.; Wongnate, T.; Sucharitakul, J.; Krondorfer, I.; Sygmund, C.; Haltrich, D.; Chaiyen, P.; Peterbauer, C.K.; Divne, C. The 1.6 Å Crystal Structure of Pyranose Dehydrogenase from Agaricus meleagris Rationalizes Substrate Specificity and Reveals a Flavin Intermediate. PLoS ONE 2013, 8, e53567. [Google Scholar] [CrossRef]
- Kommoju, P.-R.; Chen, Z.; Bruckner, R.C.; Mathews, F.S.; Jorns, M.S. Probing Oxygen Activation Sites in Two Flavoprotein Oxidases Using Chloride as an Oxygen Surrogate. Biochemistry 2011, 50, 5521–5534. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Sakai, G.; Mori, K.; Kojima, K.; Kamitori, S.; Sode, K. Structural analysis of fungus-derived FAD glucose dehydrogenase. Sci. Rep. 2015, 5, 13498. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Kojima, K.; Shiota, M.; Yoshimatsu, K.; Yamazaki, T.; Ferri, S.; Tsugawa, W.; Kamitori, S.; Sode, K. X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein. Acta Cryst. D 2019, 75, 841–851. [Google Scholar] [CrossRef]
- Dreveny, I.; Andryushkova, A.S.; Glieder, A.; Gruber, K.; Kratky, C. Substrate Binding in the FAD-Dependent Hydroxynitrile Lyase from Almond Provides Insight into the Mechanism of Cyanohydrin Formation and Explains the Absence of Dehydrogenation Activity. Biochemistry 2009, 48, 3370–3377. [Google Scholar] [CrossRef]
- Hallberg, B.M.; Henriksson, G.; Pettersson, G.; Vasella, A.; Divne, C. Mechanism of the Reductive Half-reaction in Cellobiose Dehydrogenase. J. Biol. Chem. 2003, 278, 7160–7166. [Google Scholar] [CrossRef]
- Li, J.; Vrielink, A.; Brick, P.; Blow, D.M. Crystal structure of cholesterol oxidase complexed with a steroid substrate: Implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry 1993, 32, 11507–11515. [Google Scholar] [CrossRef]
- Martin Hallberg, B.; Leitner, C.; Haltrich, D.; Divne, C. Crystal Structure of the 270 kDa Homotetrameric Lignin-degrading Enzyme Pyranose 2-Oxidase. J. Mol. Biol. 2004, 341, 781–796. [Google Scholar] [CrossRef]
- Doubayashi, D.; Ootake, T.; Maeda, Y.; Oki, M.; Tokunaga, Y.; Sakurai, A.; Nagaosa, Y.; Mikami, B.; Uchida, H. Formate oxidase, an enzyme of the glucose-methanol-choline oxidoreductase family, has a His-Arg pair and 8-formyl-FAD at the catalytic site. Biosci. Biotechnol. Biochem. 2011, 75, 1662–1667. [Google Scholar] [CrossRef]
- Klose, T.; Herbst, D.A.; Zhu, H.; Max, J.P.; Kenttämaa, H.I.; Rossmann, M.G. A Mimivirus Enzyme that Participates in Viral Entry. Structure 2015, 23, 1058–1065. [Google Scholar] [CrossRef]
- Wongnate, T.; Chaiyen, P. The substrate oxidation mechanism of pyranose 2-oxidase and other related enzymes in the glucose–methanol–choline superfamily. FEBS J. 2013, 280, 3009–3027. [Google Scholar] [CrossRef]
- Robbins, J.M.; Souffrant, M.G.; Hamelberg, D.; Gadda, G.; Bommarius, A.S. Enzyme-Mediated Conversion of Flavin Adenine Dinucleotide (FAD) to 8-Formyl FAD in Formate Oxidase Results in a Modified Cofactor with Enhanced Catalytic Properties. Biochemistry 2017, 56, 3800–3807. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.-Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017—Beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Carradec, Q.; Pelletier, E.; Da Silva, C.; Alberti, A.; Seeleuthner, Y.; Blanc-Mathieu, R.; Lima-Mendez, G.; Rocha, F.; Tirichine, L.; Labadie, K.; et al. A global ocean atlas of eukaryotic genes. Nat. Commun. 2018, 9, 373. [Google Scholar] [CrossRef]
- Steinegger, M.; Mirdita, M.; Söding, J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 2019, 16, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Lemieux, C.; Turmel, M.; Otis, C.; Pombert, J.-F. A streamlined and predominantly diploid genome in the tiny marine green alga Chloropicon primus. Nat. Commun. 2019, 10, 4061. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Rambaut, A. FigTree v1.4.4; The Institute of Evolutionary Biology: Edinburgh, UK, 2018. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]

| Short Name, EC, CAZy | Name | Catalyzed Reaction | Hosts |
|---|---|---|---|
| GOx 1.1.3.4 AA3_2 | Glucose oxidases | Oxidation of β-d-glucose at the C1 hydroxyl group utilizing oxygen as electron acceptor with the concomitant production of d-glucono-delta-lactone and hydrogen peroxide [43]. GOx are highly specific for β-d-glucose as a substrate, although some of the species can also oxidize other sugars, such as d-galactose, d-mannose or d-xylose [43,44]. | Mainly found in fungi, e.g., Aspergillus niger and Penicillium species, but also found in insects, algae and fruits [44]. |
| GDH 1.1.5.9 AA3_2 | FAD-dependent glucose dehydrogenases | Transformation of glucose at the first hydroxyl group into glucono-1,5-lactone and does not utilize oxygen as the electron acceptor. | Found in Gram-negative bacteria, fungi, and in some insects [43,45]. |
| AOx 1.1.3.13 AA3_3 | Alcohol oxidases(also known as methanol oxidases) | Oxidation of methanol as well as other short aliphatic alcohols with two to four carbon atoms [46,47,48] to the corresponding carbonyl compounds accompanied by a release of hydrogen peroxide. | Mainly found in yeasts and filamentous fungi [48]. |
| AAOx 1.1.3.7 AA3_2 | Aryl-alcohol oxidases | Oxidation of a plethora of aromatic, and some aliphatic, polyunsaturated alcohols bearing conjugated primary hydroxyl groups [49] accompanied by the formation of hydrogen peroxide at the expense of dioxygen [48]. | Commonly found in fungi such as Pleurotus eryngii [49]. |
| COx 1.1.3.17 | Choline oxidases | Four-electron oxidation of choline to glycine betaine (N,N,N-trimethylglycine; betaine) via betaine aldehyde as intermediate [50]. | Identified in Gram-negative bacterium Arthrobacter globiformis [51] and in the fungus Cylindrocarpon didymum [52], among others. |
| CHDH 1.1.99.1 | Choline dehydrogenases | Formation of betaine aldehyde from choline. | Found in humans as well as in other animals, bacteria and fungi [53]. |
| POx 1.1.3.10 AA3_4 | Pyranose oxidases | C-2 oxidation of common monosaccharides including d-glucose, d-galactose, and d-xylose to the corresponding 2-keto sugars. The preferred substrate of pyranose oxidases is D-glucose which is converted to 2-keto-d-glucose [54]. | Typically found in lignin-degrading white rot fungi as well as in actinobacteria, proteobacteria and bacilli [55,56]. |
| PDH 1.1.99.29 AA3_2 | Pyranose dehydrogenases | Monooxidations at C1, C2, C3 or dioxidations at C2, 3 or C3, 4, depending on the pyranose sugar form (mono-/di-/oligo-saccharide or glycoside) and the enzyme source [57]. | The spread appears to be limited to a narrow group of fungi (Agaricaceae) [57]. |
| FOx 1.2.3.1 | Formate oxidases | Oxidation of formate to carbon dioxide and utilization of oxygen as an electron acceptor. They may also exhibit a low methanol oxidase activity [58]. | Identified in formaldehyde-resistant fungi as Aspergillus nomius IRI013 [59], Debaryomyces vanrijiae MH201 [60] and Aspergillus oryzae RIB40 [61]. |
| FDH 1.1.99.11 | Fructose dehydrogenases | Oxidation of d-fructose to produce 5-dehydro-d-fructose; their physiological electron acceptors are ubiquinones [62,63]. | Commonly present in acetic acid bacteria, such as Gluconobacter species [63]. Computational methods allowed identification of 160 different FDH genes [64]. |
| PNOx 1.1.3.12 | Pyridoxine 4-oxidases | Oxidation of pyridoxine by oxygen or other hydrogen acceptors to form pyridoxal and hydrogen peroxide or reduced forms of the acceptors, respectively. | Identified in bacteria Pseudomonas sp. MA-1 [65], Microbacterium luteolum [66], and Mesorhizobium loti [67]. |
| CHOx 1.1.3.6 | Cholesterol oxidases | Oxidation of cholesterol (5-cholesten-3β-ol) to 4-cholesten-3-one with the reduction of molecular oxygen to hydrogen peroxide. | GMC family CHOxs are found mostly in actinomycetes such as Streptomyces sp., Brevibcterium, Rhodococcus sp., as well as in bacteria Arthrobacter, Nocardia and Mycobacterium sp. [40,68]. |
| CDH 1.1.99.18 AA3_1 | Cellobiose dehydrogenases | Transformation of cellobiose into cellobiono-1,5-lactone [69]; oxygen serves as a poor electron acceptor in comparison with other acceptors such as cytochrome c, dichlorophenolindophenol, Mn3+ and benzoquinones [70]. | Found in numerous wood-degrading fungi, both in basidiomycetes and ascomycetes [69,71]. |
| HNL 4.1.2.10 | FAD-dependent hydroxynitrile lyases | Reversible cleavage of cyanohydrins such as (R)-mandelonitrile into the corresponding aldehyde or ketone and hydrogen cyanide. | Mainly found in plants [72,73]. |
| HMFO 1.1.3.47 | 5-(Hydroxymethyl)furfural oxidase | Oxidation of many aldehydes, primary alcohols, and thiols, in particular, oxidation of 5-hydroxymethylfurfural to 2,5-furandicarboxylic acid [74]. | Discovered in Methylovorus sp. strain MP688 [74]. |
| CKOx | Compound K oxidase | Oxidation of the ginsenoside compound K, which leads to its spontaneous deglycosylation, as well as oxidation of other ginsenoside compounds, such as Rb1, Rb2, Rb3, Rc, F2, CK, Rh2, Re, F1, and the isoflavone daidzin, at lower rates [75]. | Identified in α-proteobacterium Rhizobium sp. GIN611 [75]. |
| HAOx | Hydroxy fatty acid oxidase | Oxidation of long-chain ω-hydroxy fatty acids to ω-oxo fatty acids was ascribed to ACE/HTH [76], a 594 amino acid-long GMC family protein not related to other HAOxs. | Arabidopsis thaliana [76]. |
| № | PDB ID, Chain | UniProt ID | Protein Name | Organism | Reference |
|---|---|---|---|---|---|
| 1 | 5NCC, A | A0A248QE08 | Fatty acid photodecarboxylase | Chlorella variabilis | [77] |
| 2 | 4MJW, A | Q7X2H8 | Choline oxidase | Arthrobacter globiformis | [91] |
| 3 | 4HA6, A | Q5NT46 | Pyridoxine 4-oxidase | Rhizobium loti | [92] |
| 4 | 4UDP, B | E4QP00 | 5-(hydroxymethyl) furfural oxidase | Methylovorus sp. (strain MP688) | [93] |
| 5 | 5HSA, A | F2QY27 | Alcohol oxidase | Pichia pastoris | [94] |
| 6 | 5OC1, A | O94219 | Aryl-alcohol oxidase | Pleurotus eryngii | [95] |
| 7 | 4H7U, A | Q3L245 | Pyranose dehydrogenase | Agaricus meleagris | [96] |
| 8 | 3QVP, A | P13006 | Glucose oxidase | Aspergillus niger | [97] |
| 9 | 4YNU, A | B8MX95 | Glucose dehydrogenase | Aspergillus flavus | [98] |
| 10 | 6A2U, B | Q8GQE7 | Glucose dehydrogenase | Burkholderia cepacia | [99] |
| 11 | 3GDN, B | Q945K2 | Hydroxynitrile lyase | Prunus dulcis | [100] |
| 12 | 1NAA, B | Q01738 | Cellobiose dehydrogenase | Phanerochaete chrysosporium | [101] |
| 13 | 1COY, A | P22637 | Cholesterol oxidase | Brevibacterium sterolicum | [102] |
| 14 | 1TT0, A | Q7ZA32 | Pyranose 2-oxidase | Trametes multicolor | [103] |
| 15 | 3Q9T, A | Q2UD26 | Formate oxidase | Aspergillus oryzae | [104] |
| 16 | 4Z24, A | Q5UPL2 | Putative GMC-type oxidoreductase R135 | Acanthamoeba polyphaga mimivirus | [105] |
| Dataset ID | Dataset Contents | Number of Sequences |
|---|---|---|
| A1 | Pfam PF00732 GMC_oxred_N seed sequences | 20 |
| A2 | Pfam PF05199 GMC_oxred_C seed sequences | 76 |
| A3 | NCBI non-redundant sequences obtained using PSI-BLAST with GMC_oxred_N seed sequences | 147,949 |
| A4 | NCBI non-redundant sequences obtained using PSI-BLAST with GMC_oxred_C seed sequences | 150,593 |
| A5 | Sequences present in both A3 and A4 | 135,174 |
| A6 | Sequences present in A3 and/or in A4 | 163,368 |
| B1 | Centroid sequences from the A6 clusters (at 40% identity) | 5660 |
| B2 | All sequences from the A6 clusters that contain FAPs (centroids GBF88787.1, XP_005785285.1, OEU15591.1, QDZ18370.1, XP_005757666.1, XP_005537774.1, EWM27492.1) | 36 |
| B3 | 500 PSI-BLAST NCBI hits using B2 as a seed | 500 |
| B4 | Sequences from Sorigue et al., 2017 [77] | 50 |
| B5 | Sequences from Moulin et al., 2020 [78] | 381 |
| B6 | Putative FAPs from Tetrabaena socialis, Chloropicon primus, Porphyridium purpureum, Haematococcus lacustris, Fragilaria radians | 5 |
| B7 | Hits from Tara oceans obtained using MMseqs2 | 300 |
| B8 | Crystallized GMC proteins | 25 |
| B_unique | Unique sequences from B1-B8 | 6680 |
| B_FAP | Unique putative FAP sequences from B_unique 1 | 227 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleksenko, V.A.; Anand, D.; Remeeva, A.; Nazarenko, V.V.; Gordeliy, V.; Jaeger, K.-E.; Krauss, U.; Gushchin, I. Phylogeny and Structure of Fatty Acid Photodecarboxylases and Glucose-Methanol-Choline Oxidoreductases. Catalysts 2020, 10, 1072. https://doi.org/10.3390/catal10091072
Aleksenko VA, Anand D, Remeeva A, Nazarenko VV, Gordeliy V, Jaeger K-E, Krauss U, Gushchin I. Phylogeny and Structure of Fatty Acid Photodecarboxylases and Glucose-Methanol-Choline Oxidoreductases. Catalysts. 2020; 10(9):1072. https://doi.org/10.3390/catal10091072
Chicago/Turabian StyleAleksenko, Vladimir A., Deepak Anand, Alina Remeeva, Vera V. Nazarenko, Valentin Gordeliy, Karl-Erich Jaeger, Ulrich Krauss, and Ivan Gushchin. 2020. "Phylogeny and Structure of Fatty Acid Photodecarboxylases and Glucose-Methanol-Choline Oxidoreductases" Catalysts 10, no. 9: 1072. https://doi.org/10.3390/catal10091072
APA StyleAleksenko, V. A., Anand, D., Remeeva, A., Nazarenko, V. V., Gordeliy, V., Jaeger, K.-E., Krauss, U., & Gushchin, I. (2020). Phylogeny and Structure of Fatty Acid Photodecarboxylases and Glucose-Methanol-Choline Oxidoreductases. Catalysts, 10(9), 1072. https://doi.org/10.3390/catal10091072





