Abstract
Aminotransferases are employed as industrial biocatalysts to produce chiral amines with high enantioselectivity and yield. BpTA-1 and BpTA-2 are the only two pyridoxal 5′-phosphate-dependent fold type IV transaminase enzymes in Bacillus altitudinis W3. Herein, we compared the structures and biochemical characteristics of BpTA-1 and BpTA-2 using bioinformatic analysis, circular dichroism spectroscopy, atomic force microscopy and other approaches. BpTA-1 and BpTA-2 are similar overall; both form homodimers and utilize a catalytic lysine. However, there are distinct differences in the substrate cofactor-binding pocket, molecular weight and the proportion of the secondary structure. Both enzymes have the same stereoselectivity but different enzymatic properties. BpTA-2 is more active under partial alkaline and ambient temperature conditions and BpTA-1 is more sensitive to pH and temperature. BpTA-2 as novel enzyme not only fills the building blocks of transaminase but also has broader industrial application potential for (R)-α-phenethylamines than BpTA-1. Structure-function relationships were explored to assess similarities and differences. The findings lay the foundation for modifying these enzymes via protein engineering to enhance their industrial application potential.
1. Introduction
Aminotransferases (ATAs), also known as transaminases (TAms), are a class of biological enzymes that can transfer amino groups from amino donors to amino acceptors in the presence of the coenzyme pyridoxal 5′-phosphate (PLP, a derivative of vitamin B6) to produce chiral amines with high enantioselectivity and yield [1,2,3]. Broad substrate specificity and high activity under mild, environmentally friendly conditions make these enzymes attractive as industrial biocatalysts [4,5,6,7,8,9]. TAms have been employed for the full-scale production of chiral amines represented by phenylpropylamines, methylbenzylamines, aminotetralins and various polyfunctional and aromatic amines [10,11,12,13]. Savile and his colleagues reported the successful synthesis of Sitagliptin with a large substrate precursor using ATA-117, an (R)-selective variant of the ω-TAm from Arthrobacter sp. KNK 168 [14]. Hence, TAms are one of the most valuable enzymes in the field of the asymmetric synthesis of the chiral pharmaceutical intermediates. Obviously, the discovery and research of TAms [15,16,17,18], not only fill the amine transaminase building blocks but also have important implications for the industrial production of chiral amines.
TAms belong to PLP-dependent enzymes. According to the different structure, ATAs can be divided into two categories, namely PLP-dependent enzymes of fold type Ⅰ and Ⅳ [19,20]. Although fold type Ⅰ ATAs (aspartate ATA superfamily) can vary significantly in terms of substrate and reaction specificities, they are all (S)-selective enzymes [21,22]. Fold type IV ATAs (D-alanine ATA superfamily) are represented by three different subfamilies; (S)-selective branched-chain TAms (BCATs), (R)-selective D-amino acid ATAs (DAATs) and R-TAms [23,24,25,26]. Thus, fold type IV ATAs include both (S)-selective enzymes [27] and (R)-selective enzymes [28]. Fold type IV ATAs are more advantageous in terms of substrate specificity and stereoselectivity and have greater industrial application potential than fold type I ATAs. Furthermore, compared to (S)-selective TAms [29,30], only a few (R)-selective counterparts have been studied [31,32,33]. Therefore, the mining and research of new (R)-selective TAms still have important significance.
At present, various studies have been conducted on fold type IV ATAs but direct comparisons of related enzymes from the same organisms remain scarce. In the present work, we compared Bacillus altitudinis W3 (originally known as Bacillus pumilius W3) fold type IV ATAs BpTA-1 and BpTA-2. Amino acid sequence alignment and domain analysis were performed, genes were overexpressed and enzymes were purified to homogeneity. Structural and functional features of the enzymes were also explored.
2. Results and Discussion
2.1. Bioinformatic Analysis
B. altitudinis W3 aminotransferases BpTA-2 and BpTA-1 are the only two PLP-dependent fold type Ⅳ TAms in this strain [34]. And ATA-117 is an (R)-selective TAm mutant from Arthrobacter sp. KNK 168. Phylogenetic tree analysis [35] and alignment of the amino acid sequences of aminotransferases ATA-117, BpTA-2 and BpTA-1 were performed and the results revealed high homology but low similarity (Figure 1). The sequence of BpTA-2 shares 18.4% sequence identity with that of BpTA-1. Moreover, the sequence identity between the sequences BpTA-2 and ATA-117 is even lower, only 14.6%. Although the amino acid sequence of BpTA-2 is longer than that of BpTA-1, the length of their conserved domains is similar; BpTA-2 spans residues 57–344 and BpTA-1 spans residues 6–292. As typical PLP-dependent fold type IV TAms, both BpTA-2 and BpTA-1 utilize a catalytic lysine. Homology modelling of the above two enzymes showed that both form homodimers (Figure 2), with active sites shielded from solvent by lid residues in the liganded form. However, they also have differences. The substrate cofactor-binding pocket of BpTA-2 is composed of 10 amino acid residues (Y66, F71, R95, V151, K197, Y202, E233, I260, T261 and T303), while that of BpTA-1 is composed of 9 amino acid residues [35]. Compared with the amino acid residues that constitute the substrate cofactor-binding pocket in BpTA-1, BpTA-2 has an additional amino acid residue V151 in addition to the different positions of the residues. Therefore, this residue V151 may be the key reason for the difference in enzyme activity between BpTA-2 and BpTA-1.
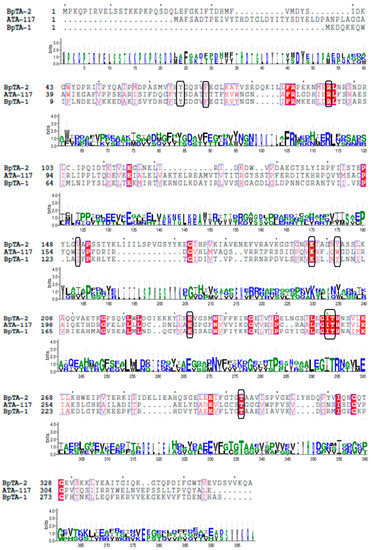
Figure 1.
Amino acid sequence alignment of the transaminase BpTA-2, BpTA-1 and ATA-117. The amino acid residues in the black box are the active site residues of these three enzymes.
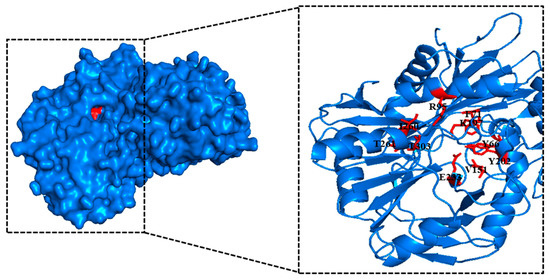
Figure 2.
Homology modelling structure of BpAT-2 showing conserved domains. The 10 residues in red (Y66, F71, R95, V151, K197, Y202, E233, I260, T261 and T303) constitute the catalytic site and the cofactor-binding pocket of BpAT-2.
2.2. SDS-PAGE and Gel Filtration Chromatography Analyses
BpTA-1 or BpTA-2, encoded by ota3 and ota8, were mainly expressed in soluble form and the recombinant proteins were both subsequently purified by immobilized metal affinity chromatography. The crude and pure enzymes are shown in Figure 3, respectively. Under the same experimental conditions, BpTA-1 was expressed at higher levels than BpTA-2. The molecular masses of BpTA-1 and BpTA-2 were ~33.4 kDa and ~40.0 kDa based on sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), in accordance with the theoretical values and both yielded a single band following purification. To further confirm the molecular weight of BpTA-1 and BpTA-2, pure enzymes were loaded onto a size-exclusion chromatography column (Figure 4). Based on protein standards and the associated standard curve, the molecular weights of BpTA-1 and BpTA-2 were determined to be 65.84 kDa and 79.98 kDa, respectively. The results of SDS-PAGE and gel filtration chromatography indicated that BpTA-1 and BpTA-2 generally exist in the form of dimers. This result is consistent with the above bioinformatics analysis; BpTA-1 and BpTA-2 are typical PLP-dependent fold type IV TAms that are active in dimeric form.
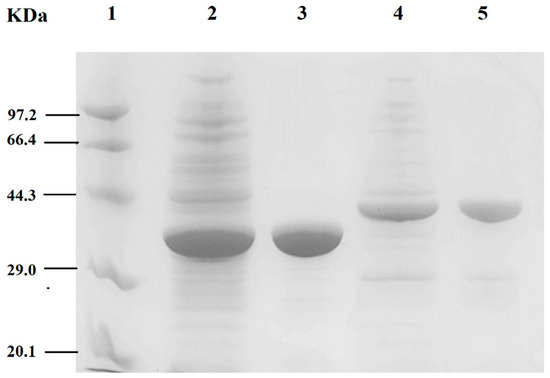
Figure 3.
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of BpTA-1 and BpTA-2. Lane 1. Protein marker (14.3–97.2 kDa); Lane 2. Crude fractions of BpTA-1; Lane 3. Purification fractions of BpTA-1; Lane 4. Crude fractions of BpTA-2; Lane 5. Purification fractions of BpTA-2.
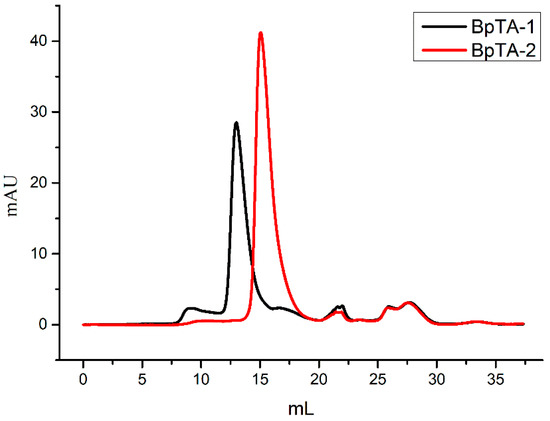
Figure 4.
The size-exclusion chromatography elution curves of BpTA-1 and BpTA-2.
2.3. Secondary Structural Analysis
Secondary structure analysis of BpTA-1 and BpTA-2 was performed using circular dichroism (CD) spectroscopy and the results are shown in Figure 5. The CD spectrum has one positive peak near 198 nm and one negative trough near 218 nm, which are typical characteristics of α-helical and β-sheet structures, respectively. Based on experiments and theoretical calculations, BpTA-1 is composed of ~20% α-helix, 47% β-sheet and 33% random coil, whereas BpTA-2 is composed of ~30.5% α-helix, 14% β-sheet and 55.5% random coil. Although the two enzymes are similar overall, their secondary structural elements differ somewhat under the same experimental conditions. Compared with BpTA-1, BpTA-2 has a higher proportion of α-helical and random coil content and a smaller proportion of β-sheet content. From the perspective of secondary structure, α-helix structure shows the orderliness of protein, while its structure, such as β-sheet, β-turn and random coil, reflects the looseness of protein. Therefore, the higher α-helical content of BpTA-2 indicates that it has lower sensitivity to the surrounding environment than BpTA-1.
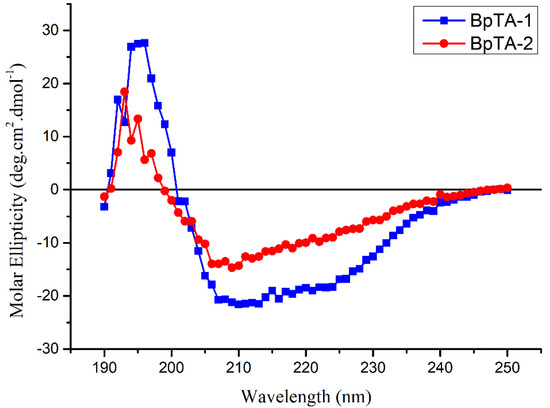
Figure 5.
Ultraviolet circular dichroism (UV-CD) spectrum of purified BpTA-1 and BpTA-2 at room temperature and pH 7.0.
2.4. Atomic Force Microscopy Imaging
Atomic force microscopy (AFM) can yield high-resolution images of enzyme molecules under physiological conditions that can reveal details of the morphology, particle diameter and height distribution. Figure 6 shows the 2D and 3D AFM imaging results for BpTA-1 and BpTA-2 at different dilutions. At a 20-fold dilution, BpTA-1 (Figure 6A) and BpTA-2 (Figure 6B) displayed aggregation. Although both enzyme conglomerates are spherical, BpTA-1 is flatter than that of BpTA-2. At a 200-fold dilution, BpTA-1 conglomerates (Figure 6C) and BpTA-2 conglomerates (Figure 6D) were dispersed. BpTA-1 conglomerates were observed as discrete particles with an average diameter of 125.3 nm and an average height of 5.45 nm. Similarly, BpTA-2 conglomerates formed discrete particles with an average diameter of 176.6 nm and an average height of 7.92 nm. Subsequently, even though the enzyme solution was further diluted, the average size of the protein aggregates did not change. The above results indicate that BpTA-2 and BpTA-1 may exist in nature as aggregations and perform the related transamination reactions in the same form.
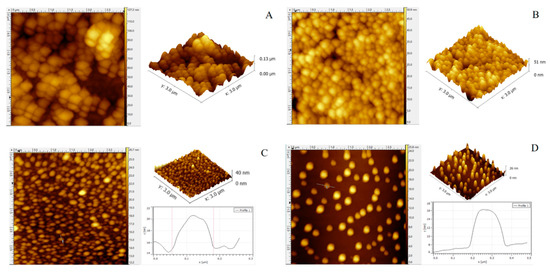
Figure 6.
2D/3D Atomic force microscopy (AFM) images of BpTA-1 and BpTA-2. (A) Enzyme BpTA-1 diluted 20-fold; (B) Enzyme BpTA-2 diluted 20-fold; (C) Enzyme BpTA-1 diluted 200-fold and the size of its single particles; (D) Enzyme BpTA-2 diluted 200-fold and the size of its single particles.
2.5. Enzyme Activity Analysis
BpTA-2 displayed high activity with the substrate (R)-α-phenethylamine but activity with (S)-α-phenethylamine was negligible, suggesting that BpTA-2 has (R)-selectivity and can accept specific chiral amines (Table 1). As the only two PLP-dependent fold type Ⅳ TAms in B. altitudinis W3, the stereoselectivity of BpTA-2 was the same as that of BpTA-1. However, the activity of BpTA-2 with (R)-α-phenethylamine was higher than that of BpTA-1 [35], which may be due to differences in their structures. Thus, in terms of industrial applications using (R)-α-phenethylamine, BpTA-2 is more valuable than BpTA-1.

Table 1.
BpTA-2 and other TAms specificity with (R)-α-phenethylamine or (S)-α-phenethylamine as an amino donor.
Also, as a newly discovered wild TAm, the (R)-selective TAm from Fusarium oxysporum [36] has higher activity on (R)-α-phenethylamine than BpTA-1 but lower than BpTA-2. The enzyme activities of the TAms from Thermobaculum terrenum and Haliangium ochraceum to (R)-α-phenethylamine are 0.15 and 0.10 U/mg [37], respectively, which are lower than purified BpTA-2 and BpTA-1. Furthermore, compared with the purified BpTA-2 activity on (R)-α-phenethylamine, the transaminase HEWT from Halomonas elongate [38], Chromobacterium violaceum ω-TAm [39] and Vibrio fluvialis ω-TAm [40] as the (S)-selective TAm, have weaker enzyme activity on the substrate (S)-α-phenethylamine. The above results show that BpTA-2 has higher activity on (R)-α-phenethylamine and the industrial production potential of this substrate.
2.6. Effects of pH and Temperature on Enzyme Activity
The effect of pH on the activity of BpTA-2 was investigated with (R)-α-phenethylamine and sodium pyruvate as amino donors and acceptors, respectively, at 35 °C and pH ranging from 5.0 to 13.0 (Figure 7A). The results showed that BpTA-2 was maximally active at pH 9.0. In the range of pH 5.0 to 9.0, enzyme activity increased with increasing pH value. In the range of pH 9.0 to 13.0, enzyme activity decreased with increasing pH value. At pH values below 5.0 or above 13.0, BpTA-2 almost lost its activity.
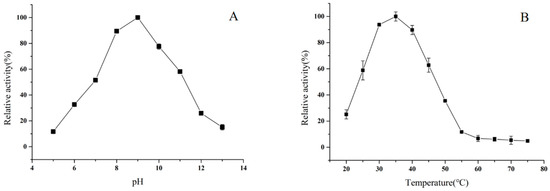
Figure 7.
Effects of pH (A) and temperature (B) on BpTA-2 activity. Maximum activity in A (3.53 ± 0.11 U/mg) and B (3.50 ± 0.15 U/mg) was set as 100% and relative activity was calculated by comparison with the maximal activity.
The effect of temperature on the activity of BpTA-2 was investigated at pH 7.0 across a temperature range from 20 °C to 75 °C (Figure 7B). The results revealed that the optimal temperature for BpTA-2 was 35 °C. Enzyme activity was increased with increasing temperature between 20 °C and 35 °C. When the temperature was above 35 °C, enzyme activity was decreased with the increase of temperature. Until the temperature was higher than 55 °C, enzyme activity was inactivated due to the high temperature. However, at temperatures below 20 °C, the enzyme remained weakly active, about 20% of the highest activity. To sum up, BpTA-2 exhibited the maximum activity under slightly alkaline and ambient temperature conditions, indicating that this condition would promote the enzymatic reaction.
By contrast, BpTA-1 was more active under neutral and high temperature conditions, which exhibits the best catalytic efficiency at pH 7.0 and 45 °C [35]. Both pH and temperature had a more pronounced effect on BpTA-1 than on BpTA-2. BpTA-2 retained approximately 90% and 80% activity at pH 8.0 and 10.0, respectively and 90% activity at 30 °C and 40 °C. By comparison, BpTA-1 retained only about 30% activity at pH 6.0 and 9.0 and activity was <30% at 40 °C and 50 °C. This demonstrates that BpTA-1 is more sensitive to pH and temperature than BpTA-2, so that the catalytic reactions involved in BpTA-1 have higher requirements on the surrounding environment. Moreover, BpTA-1 and BpTA-2 are the only two PLP-dependent fold type Ⅳ TAms in B. altitudinis W3 and their optimal reaction conditions are so complementary, which may be important for the survival and proliferation of this strain.
2.7. Effects of pH and Temperature on Enzyme Stability
BpTA-2 was incubated in 50 mM phosphate buffer (pH 7.0) for 30 min at temperatures from 10 °C to 70 °C and its residual enzyme activity was measured (Figure 8A). Exposed to the temperature below 20 °C, BpTA-2 has always maintained the best catalytic activity. Nevertheless, at the temperature above 20 °C, the residual activity of the enzyme is decreased sharply as temperature increases until it is completely lost at 60 °C.
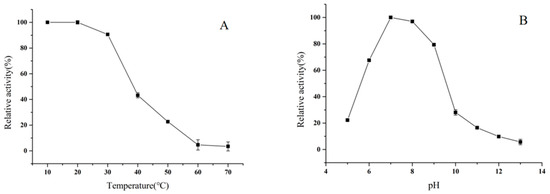
Figure 8.
Effects of temperature (A) and pH (B) on BpTA-2 stability. Maximum activity in A (3.62 ± 0.19 U/mg) and B (3.30 ± 0.17 U/mg) was set as 100% and relative activity was calculated by comparison with the maximal activity.
Similarly, the effect of pH on BpTA-2 stability was executed (Figure 8B). BpTA-2 was stored in the buffer solutions with pH ranging from 5.0 to 13.0 at 4 °C for 30 min and residual enzyme activity was measured. Between pH 5.0 and 7.0, enzyme activity increased rapidly with increasing pH value. At the pH above 7.0, the residual activity of the enzyme is decreased continuously as pH value increases until it is completely lost at pH 13.0. Between pH 8.0 and 10.0, the enzyme activity decreased sharply.
The above results indicate that both temperature and pH affected the stability of BpTA-2 and low temperature and neutral pH were more conducive for preserving enzyme activity. Moreover, both pH and temperature also affected the stability of BpTA-1 [35]. Although the residual enzyme activity is different, the variation trend of BpTA-1 is similar to BpTA-2 under different variables.
3. Materials and Methods
3.1. Bioinformatic Analysis of the ota8 Gene and the BpTA-2 Protein
The ota8 gene (GeneBank No: MN057780) was identified in the genome of Bacillus altitudinis W3 (GeneBank No: CP011150.1), which was isolated by our laboratory [41] and deposited in the China Center for Type Culture Collection (CCTCC No. M2015018). The deduced amino acid sequence of BpTA-2 (encoded by ota8) was submitted to the Swiss-Model online server (http://swissmodel.expasy.org/interactive) for homology modelling. Among the TAms with a known 3D structure, an enzyme in Mycobacterium smegmatis (PDB ID: 3JZ6) shares 48.57% sequence identity with BpAT-2. The PDB file for this enzyme was obtained from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) and assessed using a PDB Viewer, a powerful protein 3D-structure-viewing software. Meanwhile, the NCBI conserved domain database (CDD; http://www.ncbi.nlm.nih.gov/cdd/), sequence analysis software DNAMAN 5.0 and online software Weblogo3 (http://weblogo.threeplusone.com/) were employed to analyze the conserved domain sequence of BpTA-2 [42]. The amino acid sequence of BpTA-2 was compared with the sequences of aminotransferase ATA-117 (GeneBank No: AB638718.1) from Arthrobacter sp. KNK 168 and aminotransferase BpTA-1 (encoded by ota3; GeneBank No: MH196528) from B. altitudinis W3.
3.2. Materials
A MiniBEST Bacterial Genomic DNA Extraction Kit Version 3.0, PrimeSTAR HS DNA polymerase, 5 × PrimeSTAR buffer (Mg2+ plus), protein markers and other molecular experimental products were purchased from TaKaRa (Otsu, Japan). PCR primers production and gene sequencing were performed by Hongxun (Suzhou, China). The pCold II vector was purchased from TaKaRa (Dalian, China). Ampicillin sodium salt was purchased from Molekula Ltd. (Gillingham, UK). All analytical grade chemical reagents, unless stated otherwise, were purchased from Sigma-Aldrich (Shanghai, China).
3.3. Gene Expression and Enzyme Purification
The ota8 gene was obtained by polymerase chain reaction (PCR) amplification from B. altitudinis W3 genomic DNA using PrimeSTAR HS DNA polymerase and oligonucleotide primers FWD-ota8 (5′-GCCG CTCGAGATGCCAAAACAACCCATCCGCGTTG-3′) and REW-ota8 (5′-GCCG TGCAGTTTATGCCTGCTTGACGACCGAATCG-3′) incorporating XhoI and PstI restriction sites, respectively (underlined), according to the manufacturer’s instructions. The resulting recombinant plasmid (pCold II-ota8) was verified by sequencing, transformed into Escherichia coli BL21 (DE3) competent cells and transformants were culture in LB broth for protein expression [35]. The crude extract or lysate was obtained after induced and expressed.
Recombinant proteins were purified by chromatography using an Avant purifier (GE Healthcare, Little Chalfont, UK). The supernatant containing the target protein was passed through a 0.22_μm cellulose filter, loaded onto a 1 mL Ni-HisTrap TM column (GE Healthcare, Shanghai, China) and purified according to the manufacturer’s instructions. Enzyme-containing fractions were desalted using a HiTrap TM Desalting column (5 mL) and an ultra-filtration centrifuge tube. Purified enzymes were stored at 4 °C for short periods or at −20 °C for long periods. Both BpTA-2 and BpTA-1 were purified using these experimental procedures. Enzymatic activity was measured by high-performance liquid chromatography (HPLC) assay and crude and purified enzymes were analyzed by SDS-PAGE (see below).
3.4. SDS-PAGE and Gel Filtration Chromatography
The molecular mass of pure enzymes was determined by 12% SDS-PAGE [43] with staining by Coomassie Brilliant Blue R250 and broad range protein markers (14.3–97.2 kDa). The molecular mass of pure enzymes was further investigated by size-exclusion chromatography with a Superdex G-200 10/300GL column eluted using 50 mM phosphate buffer pH 7.0 [44]. Myoglobin (17 kDa), ovalbumin (44 kDa), bovine serum albumin (67 kDa) and γ-globulin (158 kDa) served as standards for calibrating molecular masses.
3.5. Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy measurements were performed on a Chirascan qCD from Applied Photophysics. Pure enzyme samples (0.11 mg/mL) were prepared in sodium phosphate buffer (2.5 mM, pH 7.0) and stored at 4 °C. A 1 mm path length cuvette was used for CD measurements. Each reported spectrum is the average of 10 measurements scanned between 190 and 250 nm. CD spectra of proteins was analyzed using CDtool software and the DichroWeb (http://dichroweb.cryst.bbk.ac.uk) online server [45,46,47].
3.6. Atomic Force Microscopy
Imaging experiments were carried out using a Nanoscope II atomic force microscopy equipped with a type-D head (Digital Instruments, Santa Barbara, CA, USA). Samples were spread on the mica surface and silicon nitride triangular cantilevers with wide legs (2 mm length; spring constant 0.12 N/m) were used for scanning the mica surfaces. Images were scanned from 80 to 14,000 nm at a 5.78 Hz scan speed and 0 V setpoint voltage and they included 400 sample points per line and 400 lines.
3.7. Enzyme Activity Assay
Measurement of enzyme activity was performed by incubating (R)-α-phenethylamine (20 mM) or (S)-α-phenethylamine (20 mM) with sodium pyruvate (20 mM) in purified (or crude) enzyme and coenzyme PLP at 40 °C and pH 7.0. The reaction was terminated after 15 min by adding an appropriate amount of ethyl acetate, centrifuged (12,000× g, 1min) and the organic phase was analyzed by HPLC using an Agilent C18 column (250 × 4.6 mm, Agilent, Santa Clara, CA, USA) at a UV absorbance of 254 nm with a mobile phase of acetonitrile/ultrapure water (50/50, v/v) and a flow rate of 0.6 mL/min. Enzyme activity was estimated based on the acetophenone yield and all the experiments were conducted with three replicates.
3.8. Effects of pH and Temperature on Enzyme Activity and Stability
The reaction between (R)-α-phenethylamine and sodium pyruvate was designedly carried out at temperatures from 20 to 75 °C at pH 9.0 aimed to determine the optimal temperature for pure enzymes. At the optimum temperatures, the specific buffers with pH values from 5.0 to 13.0 were prepared to determine the optimal pH of purified enzymes, which were respectively and accurately controlled with different pH values using sodium phosphate buffer (pH 5.0 to 8.0), glycine-NaOH buffer (pH 9.0 to 11.0) and KCL-NaOH buffer (pH 12.0 to 13.0). Aiming to evaluate the thermostability and pH stability of BpTA-2, the residual enzyme activities were measured after incubating the enzyme for 30 min with the optimal reaction conditions by separate single controlling the temperatures (varying from 10 to 70 °C) or pH values (varying from 5.0 to 13.0). Relative activities (%) were calculated using the maximal activity as 100%. All the experiments were conducted with three replicates.
4. Conclusions
Aminotransferases BpTA-2 and BpTA-1 are the only two PLP-dependent fold type Ⅳ TAms in B. altitudinis W3. Through bioinformatics analysis, SDS-PAGE and gel filtration chromatography analyses, both enzymes are typical PLP-dependent fold type Ⅳ TAms that form homodimers and utilize catalytic lysine. The catalytic characteristics of enzymes BpTA-2 and BpTA-1 were compared and they shared the same stereoselectivity but differed in terms of enzymatic properties. Combined with the comparison and analysis of the two enzyme substrate cofactor-binding pockets, we found that this residue V151 may be the key reason for the difference in enzyme activity between BpTA-2 and BpTA-1. Compared to BpTA-1, BpTA-2 was more active under slightly alkaline and ambient temperature conditions. And BpTA-1 was more sensitive to pH and temperature than BpTA-2, so that the catalytic reactions involved have higher requirements on the surrounding environment. Meanwhile, the results of CD spectroscopy measurements indicate that BpTA-2 has lower sensitivity to the surrounding environment than BpTA-1 due to its higher α-helical content. BpTA-2 as novel enzyme not only fills the building blocks of transaminase but also has more industrial application potential with (R)-α-phenethylamine than BpTA-1. The overall features of the two enzymes were characterized by AFM. And the 2D and 3D AFM imaging results indicate that BpTA-2 and BpTA-1 may exist in nature as aggregations and perform the related transamination reactions in the same form. The above results reveal that structural features and catalytic characteristics can differ between similar transaminases and differences can be investigated by probing structure-activity relationships. The findings will be of irreplaceable significance for future research on transaminases, especially protein engineering and industrial applications.
Author Contributions
Conceptualization, L.Z. and X.L.; methodology, L.Z.; software, L.Z. and Z.X.; validation, Z.X., Q.T. and Z.G.; formal analysis, X.L.; investigation, L.Z.; resources, Y.C.; data curation, Z.X.; writing—original draft preparation, L.Z.; writing—review and editing, L.Z.; visualization, L.Z.; supervision, X.L.; project administration, X.L.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Collaborative Innovation Involving Production, Teaching & Research Funds of Jiangsu Province (BY2014023-28) and the Agricultural Support Project, Wuxi Science & Technology Development (CLE01N1310).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schmidt, N.G.; Simon, R.C.; Kroutil, W. Biocatalytic asymmetric synthesis of optically pure aromatic propargylic amines employing ω-transaminases. Adv. Synth. Catal. 2015, 357, 1815–1821. [Google Scholar] [CrossRef]
- Svedendahl, M.; Branneby, C.; Lindberg, L.; Berglund, P. Reversed enantiopreference of an ω-transaminase by a single-point mutation. ChemCatChem 2010, 2, 976–980. [Google Scholar] [CrossRef]
- Wu, X.; Fei, M.; Chen, Y.; Wang, Z.; Chen, Y. Enzymatic synthesis of L-norephedrine by coupling recombinant pyruvate decarboxylase and ω-transaminase. Appl. Microbiol. Biotechnol. 2014, 98, 7399–7408. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Berglund, P. Transaminase biocatalysis: Optimization and application. Green Chem. 2017, 19, 333–360. [Google Scholar] [CrossRef]
- Ghislieri, D.; Turner, N.J. Biocatalytic approaches to the synthesis of enantiomerically pure chiral amines. Top. Catal. 2014, 57, 284–300. [Google Scholar] [CrossRef]
- Kohls, H.; Steffen-Munsberg, F.; Höhne, M. Recent achievements in developing the biocatalytic toolbox for chiral amine synthesis. Curr. Opin. Chem. Biol. 2014, 19, 180–192. [Google Scholar] [CrossRef]
- Fuchs, M.; Farnberger, J.E.; Kroutil, W. The industrial age of biocatalytic transamination. Eur. J. Org. Chem. 2015, 32, 6965–6982. [Google Scholar] [CrossRef]
- Simon, R.C.; Richter, N.; Busto, E.; Kroutil, W. Recent developments of cascade reactions involving ω-transaminases. ACS Catal. 2014, 4, 129–143. [Google Scholar] [CrossRef]
- Narancic, T.; Almahboub, S.A.; O’Connor, K.E. Unnatural amino acids: Production and biotechnological potential. World J. Microb. Biotechnol. 2019, 35, 67. [Google Scholar] [CrossRef]
- Martin, A.R.; DiSanto, R.; Plotnikov, I.; Kamat, S.; Shonnard, D.; Pannuri, S. Improved activity and thermostability of (S)-aminotransferase by error-prone polymerase chain reaction for the production of a chiral amine. Biochem. Eng. J. 2007, 37, 246–255. [Google Scholar] [CrossRef]
- Citoler, J.; Derrington, S.R.; Galman, J.L.; Bevinakatti, H.; Turner, N.J. A biocatalytic cascade for the conversion of fatty acids to fatty amines. Green Chem. 2019, 21, 4932–4935. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Stekhanova, T.N.; Popinako, A.V.; Rakitina, T.V.; Nikolaeva, A.Y.; Boyko, K.M.; Popov, V.O. Diaminopelargonic acid transaminase from Psychrobacter cryohalolentis is active towards (S)-(-)-1-phenylethylamine, aldehydes and α-diketones. Appl. Microbiol. Biotechnol. 2018, 102, 9621–9633. [Google Scholar] [CrossRef] [PubMed]
- Ferrandi, E.E.; Monti, D. Amine transaminases in chiral amines synthesis: Recent advances and challenges. World J. Microbiol. Biotechnol. 2018, 34, 13. [Google Scholar] [CrossRef] [PubMed]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Yang, S.; Lai, Y.; Meng, D.; Tian, Q.; Guan, Z.; Cai, Y.; Liao, X. Effect of residue substitution via site-directed mutagenesis on activity and steroselectivity of transaminase BpTA from Bacillus pumilus W3 for sitafloxacin hydrate intermediate. Int. J. Biol. Macromol. 2019, 137, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Markossian, K.A.; Golub, N.V.; Kleymenov, S.Y.; Muranov, K.O.; Sholukh, M.V.; Kurganov, B.I. Effect of α-crystallin on thermostability of mitochondrial aspartate aminotransferase. Int. J. Biol. Macromol. 2009, 44, 441–446. [Google Scholar] [CrossRef]
- Patil, M.D.; Grogan, G.; Bommarius, A.; Yun, H. Recent advances in ω-transaminase-mediated biocatalysis for the enantioselective synthesis of chiral amines. Catalysts 2018, 8, 254. [Google Scholar] [CrossRef]
- Molnár, Z.; Farkas, E.; Lakó, Á.; Erdélyi, B.; Kroutil, W.; Vértessy, B.G.; Paizs, C.; Poppe, L. Immobilized whole-cell transaminase biocatalysts for continuous-flow kinetic resolution of amines. Catalysts 2019, 9, 438. [Google Scholar] [CrossRef]
- Grishin, N.V.; Phillips, M.A.; Goldsmith, E.J. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 1995, 4, 1291–1304. [Google Scholar] [CrossRef]
- Mueser, T.C.; Drago, V.; Kovalevsky, A.; Dajnowicz, S. Pyridoxal 5′-phosphate dependent reactions: Analyzing the mechanism of aspartate aminotransferase. Method Enzymol. 2020, 634, 333–359. [Google Scholar] [CrossRef]
- Schneider, G.; Käck, H.; Lindqvist, Y. The manifold of vitamin B6 dependent enzymes. Structure 2000, 8, R1–R6. [Google Scholar] [CrossRef]
- Steffen-Munsberg, F.; Vickers, C.; Kohls, H.; Land, H.; Mallin, H.; Nobili, A.; Skalden, L.; Bergh, T.; Joosten, H.; Berglund, P.; et al. Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv. 2015, 33, 566–604. [Google Scholar] [CrossRef]
- Inoue, K.; Kuramitsu, S.; Aki, K.; Watanabe, Y.; Takagi, T.; Nishigai, M.; Ikai, A.; Kagamiyama, H. Branched-chain amino acid aminotransferase of Escherichia coli: Overproduction and properties1. J. Biochem. 1988, 104, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, K.; Masu, Y.; Asano, S.; Tanaka, H.; Soda, K. Thermostable D-amino acid aminotransferase from a thermophilic Bacillus species. J. Biol. Chem. 1989, 264, 2445–2449. [Google Scholar]
- Iwasaki, A.; Matsumoto, K.; Hasegawa, J.; Yasohara, Y. A novel transaminase, (R)-amine:pyruvate aminotransferase, from Arthrobacter sp. KNK168 (FERM BP-5228): Purification, characterization, and gene cloning. Appl. Microbiol. Biotechnol. 2012, 93, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Bezsudnova, E.Y.; Popov, V.O.; Boyko, K.M. Structural insight into the substrate specificity of PLP fold type IV transaminases. Appl. Microbiol. Biotechnol. 2020, 104, 2343–2357. [Google Scholar] [CrossRef]
- Mathew, S.; Nadarajan, S.P.; Chung, T.; Park, H.H.; Yun, H. Biochemical characterization of thermostable ω-transaminase from Sphaerobacter thermophilus and its application for producing aromatic β- and γ-amino acids. Enzym. Microb. Technol. 2016, 87–88, 52–60. [Google Scholar] [CrossRef]
- Kim, G.H.; Jeon, H.; Khobragade, T.P.; Patil, M.D.; Sung, S.; Yoon, S.; Won, Y.; Choi, I.S.; Yun, H. Enzymatic synthesis of sitagliptin intermediate using a novel ω-transaminase. Enzym. Microb. Technol. 2019, 120, 52–60. [Google Scholar] [CrossRef]
- Han, S.W.; Park, E.S.; Dong, J.Y.; Shin, J.S. Mechanism-guided engineering of ω-transaminase to accelerate reductive amination of ketones. Adv. Synth. Catal. 2015, 357, 1732–1740. [Google Scholar] [CrossRef]
- Scheidt, T.; Land, H.; Anderson, M.; Chen, Y.; Berglund, P.; Yi, D.; Fessner, W.D. Fluorescence-based kinetic assay for high-throughput discovery and engineering of stereoselective ω-transaminases. Adv. Synth. Catal. 2015, 357, 1721–1731. [Google Scholar] [CrossRef]
- Hohne, M.; Schatzle, S.; Jochens, H.; Robins, K.; Bornscheuer, U.T. Rational assignment of key motifs for function guides in silico enzyme identification. Nat. Chem. Biol. 2010, 6, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chen, X.; Zhang, D.; Wu, Q.; Zhu, D. Characterization of (R)-selective amine transaminases identified by in silico motif sequence blast. Appl. Microbiol. Biotechnol. 2015, 99, 2613–2621. [Google Scholar] [CrossRef] [PubMed]
- Sayer, C.; Martinez-Torres, R.J.; Richter, N.; Isupov, M.N.; Hailes, H.C.; Littlechild, J.A.; Ward, J.M. The substrate specificity, enantioselectivity and structure of the (R)-selective amine: Pyruvate transaminase from Nectria haematococca. FEBS J. 2014, 281, 2240–2253. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Ren, R.; Meng, D.; Tian, Q.; Guan, Z.; Cai, Y.; Liao, X. Comparison of aminotransferases of three Bacillus strains Bacillus altitudinis W3, Bacillus velezensis SYBC H47, and Bacillus amyloliquefaciens YP6 via genome analysis and bioinformatics. J. Appl. Genet. 2019, 60, 427–430. [Google Scholar] [CrossRef]
- Zhai, L.; Yang, S.; Lai, Y.; Meng, D.; Tian, Q.; Guan, Z.; Cai, Y.; Liao, X. Mining of aminotransferase gene ota3 from Bacillus pumilus W3 via genome analysis, gene cloning and expressing for compound bioamination. Gene 2019, 686, 21–28. [Google Scholar] [CrossRef]
- Gao, S.; Su, Y.; Zhao, L.; Li, G.; Zheng, G. Characterization of a (R)-selective amine transaminase from Fusarium oxysporum. Process Biochem. 2017, 63, 130–136. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Dibrova, D.V.; Nikolaeva, A.Y.; Rakitina, T.V.; Popov, V.O. Identification of branched-chain amino acid aminotransferases active towards (R)-(+)-1-phenylethylamine among PLP fold type IV transaminases. J. Biotechnol. 2018, 271, 26–28. [Google Scholar] [CrossRef]
- Cerioli, L.; Planchestainer, M.; Cassidy, J.; Tessaro, D.; Paradisi, F. Characterization of a novel amine transaminase from Halomonas Elongate. J. Mol. Catal. B Enzym. 2015, 120, 141–150. [Google Scholar] [CrossRef]
- Kaulmann, U.; Smithies, K.; Smith, M.E.; Hailes, H.C.; Ward, J.M. Substrate spectrum of ω-transaminase from Chromobacterium violaceum DSM30191 and its potential for biocatalysis. Enzyme Microb. Technol. 2007, 41, 628–637. [Google Scholar] [CrossRef]
- Shin, J.S.; Yun, H.; Jang, J.W.; Park, I.; Kim, B.G. Purification, characterization, and molecular cloning of a novel amine: Pyruvate transaminase from Vibrio fluvialis JS17. Appl. Microbiol. Biotechnol. 2003, 61, 463–471. [Google Scholar] [CrossRef]
- Guan, Z.; Cai, Y.; Zhang, Y.; Zhao, H.; Liao, X. Complete genome sequence of Bacillus pumilus W3: A strain exhibiting high laccase activity. J. Biotechnol. 2015, 207, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Meng, D.; Tian, Q.P.; Yang, S.L.; Deng, H.X.; Guan, Z.B.; Cai, Y.J.; Liao, X.R. Characterization of a novel carboxylesterase from Bacillus velezensis SYBC H47 and its application in degradation of phthalate esters. J. Biosci. Bioeng. 2020, 129, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Wallace, B.A. CDtoolX, a Downloadable Software Package for Processing and Analyses of Circular Dichroism Spectroscopic Data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein Secondary Structure Analyses from Circular Dichroism Spectroscopy: Methods and Reference Databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, 668–673. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).