Enzymology of Alternative Carbohydrate Catabolic Pathways
Abstract
1. Introduction
2. Important Enzymes of the ED Pathway/Non-Phosphorylative Pathways
2.1. Dehydratases
2.1.1. Dehydratases of the Enolase Superfamily
2.1.2. Dehydratases in Hexuronate Metabolism
2.1.3. Dehydratases of the IlvD/EDD Superfamily
2.2. Aldolases
3. Alternative Metabolism of Sugars Other than Glucose
3.1. D-Xylose Metabolism
3.2. L-Arabinose Metabolism
3.3. L-Rhamnose Metabolism
3.4. L-Fucose Metabolism
3.5. D-Mannose Metabolism
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AldT | aldohexose dehydrogenase |
| CCM | central carbon metabolism |
| DHAD | dihydroxyacid dehydratase |
| DHAP | dihydroacetone phosphate |
| ED | Entner–Doudoroff |
| EDD | 6-phosphogluconate dehydratase |
| EMP | Embden–Meyerhof–Parnas |
| ExuT | hexuronate transporter |
| GAD | gluconate dehydratase |
| GAP | glyceraldehyde-3-phosphate |
| IlvD/EDD | dihydroxyacid dehydratase/EDD |
| KDG | 2-keto-3-deoxygluconate |
| KDGA | 2-keto-3-deoxygluconate aldolase |
| KDGK | 2-keto-3-deoxygluconate kinase |
| KDPG | 2-keto-3-deoxy-6-phosphogluconate |
| KDGal | 2-keto-3-deoxygalactonate |
| KDPGal | 2-keto-3-deoxy-6-phosphogalactonate |
| KDX | 2-keto-3-deoxy-D-xylonate |
| L-KDA | L-2-keto-3-deoxyarabinonate |
| L-KDR | 2-keto-3-deoxy-L-rhamnonate |
| ManD/UxuA | mannonate dehydratase |
| MLE | muconate lactonising enzymes |
| MR | mandelate racemases |
| MR/MLE | mandelate racemase/muconate lactonising enzymes |
| np-ED | non-phosphorylative |
| sp-ED | semi-phosphorylative |
| SCG | spent coffee grounds |
| UxaC | uronate isomerase |
| XAD | xylonate dehydratase |
| XDH | xylose dehydrogenase |
| XI | xylose isomerase |
| X5P | D-xylulose-5-phosphate |
| ZWF | glucose-6-phosphate dehydrogenase |
References
- Kopp, D.; Sunna, A. Alternative carbohydrate pathways—Enzymes, functions and engineering. Crit. Rev. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, U.; Sutter, J.-M.; Reinhardt, A.; Picki, A.; Wang, R.; Xiang, H.; Schönheit, P. D-Ribose catabolism in Archaea: Discovery of a novel oxidative pathway in Haloarcula species. J. Bacteriol. 2020, e00608-19. [Google Scholar] [CrossRef] [PubMed]
- Benisch, F.; Boles, E. The bacterial Entner-Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron-sulfur cluster enzyme 6-phosphogluconate dehydratase. J. Biotechnol. 2014, 171, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Scopes, R.K.; Griffiths-Smith, K. Use of differential dye-ligand chromatography with affinity elution for enzyme purification: 6-phosphogluconate dehydratase from Zymomonas mobilis. Anal. Biochem. 1984, 136, 530–534. [Google Scholar] [CrossRef]
- Gardner, P.R.; Fridovich, I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem. 1991, 266, 1478–1483. [Google Scholar]
- Kornberg, H.L.; Soutar, A.K. Utilization of gluconate by Escherichia coli. Induction of gluconate kinase and 6-phosphogluconate dehydratase activities. Biochem. J. 1973, 134, 489–498. [Google Scholar] [CrossRef]
- Cuskey, S.M.; Wolff, J.A.; Phibbs, P.V., Jr.; Olsen, R.H. Cloning of genes specifying carbohydrate catabolism in Pseudomonas aeruginosa and Pseudomonas putida. J. Bacteriol. 1985, 162, 865–871. [Google Scholar] [CrossRef]
- Kovachevich, R.; Wood, W.A. Carbohydrate metabolism by Pseudomonas fluorescens. III. Purification and properties of a 6-phosphogluconate dehydrase. J. Biol. Chem. 1955, 213, 745–756. [Google Scholar]
- Kim, S.; Lee, S.B. Identification and characterization of the bacterial D-gluconate dehydratase in Achromobacter xylosoxidans. Biotechnol. Bioprocess Eng. 2008, 13, 436–444. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.B. Catalytic promiscuity in dihydroxy-acid dehydratase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Biochem. 2006, 139, 591–596. [Google Scholar] [CrossRef]
- Rahman, M.M.; Andberg, M.; Koivula, A.; Rouvinen, J.; Hakulinen, N. The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family. Sci. Rep. 2018, 8, 865. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Shimada, N.; Tajima, K.; Kodaki, T.; Makino, K. Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism: Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 2006, 281, 33521–33536. [Google Scholar] [CrossRef] [PubMed]
- Lamble, H.J.; Milburn, C.C.; Taylor, G.L.; Hough, D.W.; Danson, M.J. Gluconate dehydratase from the promiscuous Entner-Doudoroff pathway in Sulfolobus solfataricus. FEBS Lett. 2004, 576, 133–136. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.B. Identification and characterization of Sulfolobus solfataricus D-gluconate dehydratase: A key enzyme in the non-phosphorylated Entner-Doudoroff pathway. Biochem. J. 2005, 387, 271–280. [Google Scholar] [CrossRef]
- Reher, M.; Fuhrer, T.; Bott, M.; Schönheit, P. The nonphosphorylative Entner-Doudoroff pathway in the thermoacidophilic euryarchaeon Picrophilus torridus involves a novel 2-keto-3-deoxygluconate- specific aldolase. J. Bacteriol. 2010, 192, 964–974. [Google Scholar] [CrossRef]
- Matsubara, K.; Köhling, R.; Schönenberger, B.; Kouril, T.; Esser, D.; Bräsen, C.; Siebers, B.; Wohlgemuth, R. One-step synthesis of 2-keto-3-deoxy-D-gluconate by biocatalytic dehydration of D-gluconate. J. Biotechnol. 2014, 191, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.; Gottschalk, G. Purification and properties of D--gluconate dehydratase from Clostridium pasteurianum. Eur. J. Biochem. 1973, 40, 309–321. [Google Scholar] [CrossRef]
- Sutter, J.M.; Tästensen, J.B.; Johnsen, U.; Soppa, J.; Schönheit, P. Key enzymes of the semiphosphorylative Entner-Doudoroff Pathway in the haloarchaeon Haloferax volcanii: Characterization of glucose dehydrogenase, gluconate dehydratase, and 2-keto-3-deoxy-6-phosphogluconate aldolase. J. Bacteriol. 2016, 198, 2251–2262. [Google Scholar] [CrossRef]
- Johnsen, U.; Dambeck, M.; Zaiss, H.; Fuhrer, T.; Soppa, J.; Sauer, U.; Schönheit, P. D-Xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J. Biol. Chem. 2009, 284, 27290–27303. [Google Scholar] [CrossRef]
- Brouns, S.J.J.; Walther, J.; Snijders, A.P.L.; Van De Werken, H.J.G.; Willemen, H.L.D.M.; Worm, P.; de Vos, M.G.J.; Anderson, A.; Lundgren, M.; Mazon, H.F.M.; et al. Identification of the missing links in prokaryotic pentose oxidation pathways: Evidence for enzyme recruitment. J. Biol. Chem. 2006, 281, 27378–27388. [Google Scholar] [CrossRef]
- Rakus, J.F.; Fedorov, A.A.; Fedorov, E.V.; Glasner, M.E.; Hubbard, B.K.; Delli, J.D.; Babbitt, P.C.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase. Biochemistry 2008, 47, 9944–9954. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Watanabe, S.; Saimura, M.; Makino, K. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J. Biol. Chem. 2008, 283, 20372–20382. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Makino, K. Novel modified version of nonphosphorylated sugar metabolism—An alternative L-rhamnose pathway of Sphingomonas sp. FEBS J. 2009, 276, 1554–1567. [Google Scholar] [CrossRef] [PubMed]
- Wichelecki, D.J.; Alyxa, J.; Vendiola, F.; Jones, A.M.; Al-obaidi, N.; Almo, S.C.; Gerlt, J.A. Investigating the physiological roles of low-efficiency D--mannonate and D--gluconate dehydratases in the enolase superfamily: Pathways for the catabolism of L--gulonate and L--idonate. Biochemistry 2014, 53, 5692–5699. [Google Scholar] [CrossRef]
- Wichelecki, D.J.; Balthazor, B.M.; Chau, A.C.; Vetting, M.W.; Fedorov, A.A.; Fedorov, E.V.; Lukk, T.; Patskovsky, Y.V.; Stead, M.B.; Hillerich, B.S.; et al. Discovery of function in the enolase superfamily: D-mannonate and D-gluconate dehydratases in the D-mannonate dehydratase subgroup. Biochemistry 2014, 53, 2722–2731. [Google Scholar] [CrossRef]
- Rakus, J.F.; Fedorov, A.A.; Fedorov, E.V.; Glasner, M.E.; Vick, J.E.; Babbitt, P.C.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: D-mannonate dehydratase from Novosphingobium aromaticivorans. Biochemistry 2007, 46, 12896–12908. [Google Scholar] [CrossRef]
- Yew, W.S.; Fedorov, A.A.; Fedorov, E.V.; Rakus, J.F.; Pierce, R.W.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry 2006, 45, 14582–14597. [Google Scholar] [CrossRef]
- Zhang, Q.; Gao, F.; Peng, H.; Cheng, H.; Liu, Y.; Tang, J.; Thompson, J.; Wei, G.; Zhang, J.; Du, Y.; et al. Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: Genetic and biochemical evidence for a catalytic mechanism. J. Bacteriol. 2009, 191, 5832–5837. [Google Scholar] [CrossRef]
- Kopp, D.; Willows, R.; Sunna, A. Characterisation of the first archaeal mannonate dehydratase from Thermoplasma acidophilum and its potential in the catabolism of D-mannose. Catalysts 2019, 9, 234. [Google Scholar] [CrossRef]
- Ahmed, H.; Ettema, T.J.G.; Tjaden, B.; Geerling, A.C.M.; van der Oost, J.; Siebers, B. The semi-phosphorylative Entner-Doudoroff pathway in hyperthermophilic archaea: A re-evaluation. Biochem. J. 2005, 390, 529–540. [Google Scholar] [CrossRef]
- Lamble, H.J.; Heyer, N.I.; Bull, S.D.; Hough, D.W.; Danson, M.J. Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J. Biol. Chem. 2003, 278, 34066–34072. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, U.; Schönheit, P. Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 2004, 186, 6198–6207. [Google Scholar] [CrossRef] [PubMed]
- Yew, W.S.; Fedorov, A.A.; Fedorov, E.V.; Almo, S.C.; Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: L-talarate/galactarate dehydratase from Salmonella typhimurium LT2. Biochemistry 2007, 46, 9564–9577. [Google Scholar] [CrossRef] [PubMed]
- Babbitt, P.C.; Gerlt, J.A. Understanding enzyme superfamilies—Chemistry as the fundamental determinant in the evolution of new catalytic activities. J. Biol. Chem. 1997, 272, 30591–30594. [Google Scholar] [CrossRef] [PubMed]
- Gerlt, J.A.; Babbitt, P.C.; Rayment, I. Divergent evolution in the enolase superfamily: The interplay of mechanism and specificity. Arch. Biochem. Biophys. 2005, 433, 59–70. [Google Scholar] [CrossRef]
- Gulick, A.M.; Hubbard, B.K.; Gerlt, J.A.; Rayment, I. Evolution of enzymatic activities in the enolase superfamily: Identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli. Biochemistry 2001, 40, 10054–10062. [Google Scholar] [CrossRef]
- Rothe, M.; Alpert, C.; Loh, G.; Blaut, M. Novel insights into E. coli’s hexuronate metabolism: KduI facilitates the conversion of galacturonate and glucuronate under osmotic stress conditions. PLoS ONE 2013, 8, e56906. [Google Scholar] [CrossRef]
- Seibert, C.M.; Raushel, F.M. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef]
- Portalier, R.; Robert-Baudouy, J.; Stoeber, F. Regulation of Escherichia coli K-12 hexuronate system genes: Exu regulon. J. Bacteriol. 1980, 143, 1095–1107. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N.; Robert-Baudouy, J. Hexuronate catabolism in Erwinia Chrysanthemi. J. Bacteriol. 1987, 169, 1223–1231. [Google Scholar] [CrossRef]
- Mekjian, K.R.; Bryan, E.M.; Beall, B.W.; Moran, C.P. Regulation of hexuronate utilization in Bacillus subtilis. J. Bacteriol. 1999, 181, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Shulami, S.; Gat, O.; Sonenshein, A.L.; Shoham, Y. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 1999, 181, 3695–3704. [Google Scholar] [CrossRef] [PubMed]
- Kuivanen, J.; Sugai-Guérios, M.H.; Arvas, M.; Richard, P. A novel pathway for fungal D-glucuronate catabolism contains an L-idonate forming 2-keto-L-gulonate reductase. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Reis, D.; Vian, B.; Roland, J.C. Cellulose-glucuronoxylans and plant cell wall structure. Micron 1994, 25, 171–187. [Google Scholar] [CrossRef]
- Chang, D.-E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, W.E.; Stevenson, S.J.; Anderson, A.B.; Grissom, J.E.; Laux, D.C.; Cohen, P.D.; et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef] [PubMed]
- Fabich, A.J.; Jones, S.A.; Chowdhury, F.Z.; Cernosek, A.; Anderson, A.; Smalley, D.; McHargue, J.W.; Hightower, G.A.; Smith, J.T.; Autieri, S.M.; et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 2008, 76, 1143–1152. [Google Scholar] [CrossRef]
- Peekhaus, N.; Conway, T. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 1998, 180, 3495–3502. [Google Scholar] [CrossRef]
- Egan, S.E.; Fliege, R.; Tong, S.; Shibata, A.; Wolf, R.E., Jr.; Conway, T. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: Sequence analysis and localization of promoters for the edd-eda operon. J. Bacteriol. 1992, 174, 4638–4646. [Google Scholar] [CrossRef]
- Flint, D.H.; Emptage, M.H. Dihydroxy acid dehydratase from spinach contains a [2Fe-2S] cluster. J. Biol. Chem. 1988, 263, 3558–3564. [Google Scholar]
- Rahman, M.M.; Andberg, M.; Thangaraj, S.K.; Parkkinen, T.; Penttilä, M.; Jänis, J.; Koivula, A.; Rouvinen, J.; Hakulinen, N. The crystal structure of a bacterial L-arabinonate dehydratase contains a [2Fe-2S] cluster. ACS Chem. Biol. 2017, 12, 1919–1927. [Google Scholar] [CrossRef]
- Flint, D.H.; Emptage, M.H.; Finnegan, M.G.; Fu, W.; Johnson, M.K. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J. Biol. Chem. 1993, 268, 14732–14742. [Google Scholar] [PubMed]
- Rodriguez, M.; Wedd, A.G.; Scopes, R.K. 6-phosphogluconate dehydratase from Zymomonas mobilis: An iron-sulfur-manganese enzyme. Biochem. Mol. Biol. Int. 1996, 38, 783–789. [Google Scholar] [PubMed]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef] [PubMed]
- Carsten, J.M.; Schmidt, A.; Sieber, V. Characterization of recombinantly expressed dihydroxy-acid dehydratase from Sulfobus solfataricus—A key enzyme for the conversion of carbohydrates into chemicals. J. Biotechnol. 2015, 211, 31–41. [Google Scholar] [CrossRef]
- Guterl, J.K.; Garbe, D.; Carsten, J.; Steffler, F.; Sommer, B.; Reiße, S.; Philipp, A.; Haack, M.; Rühmann, B.; Koltermann, A.; et al. Cell-free metabolic engineering: Production of chemicals by minimized reaction cascades. Chemsuschem 2012, 5, 2165–2172. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Li, Z.; Zhang, L.; Wang, C.; Li, K.; Ma, C.; Xu, P. An artificial enzymatic reaction cascade for a cell-free bio-system based on glycerol. Green Chem. 2015, 17, 804–807. [Google Scholar] [CrossRef]
- Xie, L.; Wei, X.; Zhou, X.; Meng, D.; Zhou, R.; Zhang, Y.-H.P.J.; Xu, S.; You, C. Conversion of D-glucose to L-lactate via pyruvate by an optimized cell-free enzymatic biosystem containing minimized reactions. Synth. Syst. Biotechnol. 2018, 3, 204–210. [Google Scholar] [CrossRef]
- Wong, C.; Whitesides, M. Chemical and enzymatic syntheses of 6-deoxyhexoses. Conversion to 2,5-dimethyl-4-hydroxy-2,3-dihydrofuran-3-one (Furaneol) and analogues. J. Org. Chem. 1983, 48, 3493–3497. [Google Scholar] [CrossRef]
- Hecquet, L.; Hélaine, V.; Charmantray, F.; Lemaire, M. Enzymes catalyzing C-C bond formation for the synthesis of monosaccharide analogs. In Modern Biocatalysis: Stereoselective and Environmentally Friendly Reactions; Fessner, W.-D., Anthonsen, T., Eds.; Wiley-VCH: Weinheim, Germany, 2008; pp. 287–298. [Google Scholar]
- Clapés, P.; Fessner, W.D.; Sprenger, G.A.; Samland, A.K. Recent progress in stereoselective synthesis with aldolases. Curr. Opin. Chem. Biol. 2010, 14, 154–167. [Google Scholar] [CrossRef]
- Theodossis, A.; Walden, H.; Westwick, E.J.; Connaris, H.; Lamble, H.J.; Hough, D.W.; Danson, M.J.; Taylor, G.L. The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus Solfataricus. J. Biol. Chem. 2004, 279, 43886–44892. [Google Scholar] [CrossRef]
- Wolf, J.; Stark, H.; Fafenrot, K.; Albersmeier, A.; Pham, T.K.; Müller, K.B.; Meyer, B.H.; Hoffmann, L.; Shen, L.; Albaum, S.P.; et al. A systems biology approach reveals major metabolic changes in the thermoacidophilic archaeon Sulfolobus solfataricus in response to the carbon source L-fucose versus D-glucose. Mol. Microbiol. 2016, 102, 882–908. [Google Scholar] [CrossRef] [PubMed]
- Siebers, B.; Tjaden, B.; Michalke, K.; Do, C.; Ahmed, H.; Zaparty, M.; Gordon, P.; Sensen, C.W.; Zibat, A.; Klenk, H.-P.; et al. Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J. Bacteriol. 2004, 186, 2179–2194. [Google Scholar] [CrossRef] [PubMed]
- Wolterink-van Loo, S.; van Eerde, A.; Siemerink, M.A.J.; Akerboom, J.; Dijkstra, B.W.; van der Oost, J. Biochemical and structural exploration of the catalytic capacity of Sulfolobus KDG aldolases. Biochem. J. 2007, 403, 421–430. [Google Scholar] [CrossRef]
- Pauluhn, A.; Ahmed, H.; Lorentzen, E.; Buchinger, S.; Schomburg, D.; Siebers, B.; Pohl, E. Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax. Proteins 2008, 72, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Roodveldt, C.; Tawfik, D.S. Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006, 10, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Van der Oost, J.; Wolterink-Van Loo, S.; Siemerink, M.A.J.; Perrakis, G.; Kaper, T.; Kengen, S.W.M. Improving low-temperature activity of Sulfolobus acidocaldarius 2-keto-3-deoxygluconate aldolase. Archaea 2009, 2, 233–239. [Google Scholar] [CrossRef]
- Royer, S.F.; Haslett, L.; Crennell, S.J.; Hough, D.W.; Danson, M.J.; Bull, S.D. Structurally informed site-directed mutagenesis of a stereochemically promiscuous aldolase to afford stereochemically complementary biocatalysts. J. Am. Chem. Soc. 2010, 132, 11753–11758. [Google Scholar] [CrossRef]
- Tästensen, J.-B.; Johnsen, U.; Reinhartdt, A.; Ortjohjann, M.; Schönheit, P. D-Galactose catabolism in archaea: Operation of the DeLey-Doudoroff pathway in Haloferax volcanii. FEMS Microbiol. Lett. 2020, 367, fnaa029. [Google Scholar] [CrossRef]
- Kenney, K.L.; Smith, W.A.; Gresham, G.L.; Westover, T.L. Understanding biomass feedstock variability. Biofuels 2013, 4, 111–127. [Google Scholar] [CrossRef]
- Aden, A.; Ruth, M.; Ibsen, K.; Jechura, J.; Neeves, K.; Sheehan, J.; Wallace, B. Lignocellulosic Biomass to Ethanol Process Design and Economics Utilizing Co-Current Dilute Acid Prehydrolysis and Enzymatic Hydrolysis for Corn Stover; National Renewable Energy Lab.: Golden, CO, USA, 2002. [Google Scholar]
- Kobayashi, H.; Fukuoka, A. Synthesis and utilisation of sugar compounds derived from lignocellulosic biomass. Green Chem. 2013, 15, 1740–1763. [Google Scholar] [CrossRef]
- McMillan, J.D. Xylose Fermentation to Ethanol: A Review; National Renewable Energy Lab.: Golden, CO, USA, 1993. [Google Scholar]
- Moysés, D.N.; Reis, V.C.B.; de Almeida, J.R.M.; de Moraes, L.M.P.; Torres, F.A.G. Xylose fermentation by Saccharomyces cerevisiae: Challenges and prospects. Int. J. Mol. Sci. 2016, 17, 207. [Google Scholar] [CrossRef] [PubMed]
- Oreb, M.; Dietz, H.; Farwick, A.; Boles, E. Novel strategies to improve co-fermentation of pentoses with D-glucose by recombinant yeast strains in lignocellulosic hydrolysates. Bioengineering 2012, 3, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Weimberg, R. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 1961, 236, 629–635. [Google Scholar] [PubMed]
- Dahms, S.A. 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation. Biochem. Biophys. Res. Commun. 1974, 60, 1433–1439. [Google Scholar] [CrossRef]
- Watanabe, S.; Kodak, T.; Makino, K. Cloning, expression, and characterization of bacterial L-arabinose 1-dehydrogenase involved in an alternative pathway of L-arabinose metabolism. J. Biol. Chem. 2006, 281, 2612–2623. [Google Scholar] [CrossRef]
- Radek, A.; Krumbach, K.; Gätgens, J.; Wendisch, V.F.; Wiechert, W.; Bott, M.; Noack, S.; Marienhagen, J. Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J. Biotechnol. 2014, 192, 156–160. [Google Scholar] [CrossRef]
- Salusjärvi, L.; Toivari, M.; Vehkomäki, M.L.; Koivistoinen, O.; Mojzita, D.; Niemelä, K.; Penttilä, M.; Ruohonen, L. Production of ethylene glycol or glycolic acid from D-xylose in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 8151–8163. [Google Scholar] [CrossRef]
- Wasserstrom, L.; Portugal-Nunes, D.; Almqvist, H.; Sandström, A.G.; Lidén, G.; Gorwa-Grauslund, M.F. Exploring D -xylose oxidation in Saccharomyces cerevisiae through the Weimberg pathway. AMB Express 2018, 8, 33. [Google Scholar] [CrossRef]
- Toivari, M.; Nygård, Y.; Kumpula, E.P.; Vehkomäki, M.L.; Benčina, M.; Valkonen, M.; Maaheimo, H.; Andberg, M.; Koivula, A.; Ruohonen, L.; et al. Metabolic engineering of Saccharomyces cerevisiae for bioconversion of D-xylose to D-xylonate. Metab. Eng. 2012, 14, 427–436. [Google Scholar] [CrossRef]
- Nygård, Y.; Maaheimo, H.; Mojzita, D.; Toivari, M.; Wiebe, M.; Resnekov, O.; Pesce, C.G.; Ruohonen, L.; Penttilä, M. Single cell and in vivo analyses elucidate the effect of xylC lactonase during production of D-xylonate in Saccharomyces cerevisiae. Metab. Eng. 2014, 25, 238–247. [Google Scholar] [CrossRef]
- Choi, S.Y.; Kim, W.J.; Yu, S.J.; Park, S.J.; Im, S.G.; Lee, S.Y. Engineering the xylose-catabolizing Dahms pathway for production of poly(D-lactate-co-glycolate) and poly(D-2-hydroxybutyrate) in Escherichia coli. Microb. Biotechnol. 2017, 10, 1353–1364. [Google Scholar] [CrossRef]
- Boer, H.; Andberg, M.; Pylkkänen, R.; Maaheimo, H.; Kolvula, A. In vitro reconstitution and characterisation of the oxidative D-xylose pathway for production of organic acids and alcohols. AMB Express 2019, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Borgström, C.; Wasserstrom, L.; Almqvist, H.; Broberg, K.; Klein, B.; Noack, S.; Lidén, G.; Gorwa-Grausland, M.F. Identification of modifications procuring growth on xylose of recombinant Saccharomyces cerevisae strains carrying the Weimberg pathway. Metab. Eng. 2019, 55, 1–11. [Google Scholar] [CrossRef]
- Halmschlag, B.; Hoffmann, K.; Hanke, R.; Puytri, S.; Fukusaki, E.; Büchs, J.; Blank, L. Comparison of isomerase and Weimberg pathway for γ-PGA production from xylose by engineered Bacillus Subtilus. Front. Bioeng. Biotechnol. 2020, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Meijnen, J.-P.; de Winde, J.H.; Ruijssenaars, H.J. Establishment of oxidative D-xylose metabolism in Pseudomonas putida S12. Appl. Environ. Microbiol. 2009, 75, 2784–2791. [Google Scholar] [CrossRef]
- Bator, I.; Wittgens, A.; Rosenau, F.; Tiso, T.; Blank, L.M. Comparison of three xylose pathways in Pseudomonas putida KT2440 for the synthesis of valuable products. Front. Bioeng. Biotechnol. 2020, 7, 480. [Google Scholar] [CrossRef]
- Van de Werken, H.J.G.; Brouns, S.J.J.; van der Oost, J. Pentose metabolism in archaea. In Archaea: New Models for Prokaryotic Biology; Blum, P., Ed.; Caister Academic Press: Norfolk, UK, 2008; pp. 71–94. [Google Scholar]
- Nunn, C.E.M.; Johnsen, U.; Schönheit, P.; Fuhrer, T.; Sauer, U.; Hough, D.W.; Danson, M.J. Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 2010, 285, 33701–33709. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, U.; Sutter, J.M.; Zaiß, H.; Schönheit, P. L-Arabinose degradation pathway in the haloarchaeon Haloferax volcanii involves a novel type of L-arabinose dehydrogenase. Extremophiles 2013, 17, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Kodaki, T.; Makino, K. A novel α-ketoglutaric semialdehyde dehydrogenase: Evolutionary insight into an alternative pathway of bacterial L-arabinose metabolism. J. Biol. Chem. 2006, 281, 28876–28888. [Google Scholar] [CrossRef]
- Watanabe, S.; Fukumori, F.; Watanabe, Y. Substrate and metabolic promiscuities of D-altronate dehydratase family proteins involved in non-phosphorylative D-arabinose, sugar acid, L-galactose and L-fucose pathways from bacteria. Mol. Microbiol. 2019, 112, 147–165. [Google Scholar] [CrossRef]
- Watanabe, S.; Fukumori, F.; Nishiwaki, H.; Sakurai, Y.; Tajima, K.; Watanabe, Y. Novel non-phosphorylative pathway of pentose metabolism from bacteria. Sci. Rep. 2019, 9, 155. [Google Scholar] [CrossRef]
- Lachaux, C.; Frazao, C.J.R.; Kraußer, F.; Morin, N.; Walther, T.; François, J.M. A new synthetic pathway for the bioproduction of glycolic acid from lignocellulosic sugars aimed at maximal carbon conservation. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef]
- Kim, S.M.; Paek, K.H.; Lee, S.B. Characterization of NADP+–specific L-rhamnose dehydrogenase from the thermoacidophilic Archaeon Thermoplasma acidophilum. Extremophiles 2012, 16, 447–454. [Google Scholar] [CrossRef]
- Baldomà, L.; Aguilar, J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: Aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J. Bacteriol. 1988, 170, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Twerdochlib, A.L.; Pedrosa, F.O.; Funayama, S.; Rigo, L.U. L-Rhamnose metabolism in Pichia stipitis and Debaryomyces polymorphus. Can. J. Microbiol. 1994, 40, 896–902. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Zhang, X.; Yan, C.; Zhang, Y.; Xu, X.; Zhang, W. Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci. Rep. 2016, 6, 27352. [Google Scholar] [CrossRef]
- Khosravi, C.; Kun, R.S.; Visser, J.; Aguilar-Pontes, M.V.; de Vries, R.P.; Battaglia, E. In vivo functional analysis of L-rhamnose metabolic pathway in Aspergillus niger: A tool to identify the potential inducer of RhaR. BMC Microbiol. 2017, 17, 214. [Google Scholar] [CrossRef]
- Boronat, A.; Aguilar, J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: Differences in induction of propanediol oxidoreductase. J. Bacteriol. 1981, 147, 181–185. [Google Scholar] [CrossRef]
- Rigo, L.U.; Marechal, L.R.; Vieira, M.M.; Viega, L.A. Oxidative pathway for L-rhamnose degradation in Pullularia pullulans. Can. J. Microbiol. 1985, 31, 817–822. [Google Scholar] [CrossRef]
- Reinhardt, A.; Fohnen, U.; Schönheit, P. L-Rhamnose catabolism in archaea. Mol. Microbiol. 2019, 111, 1093–1108. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, W.; Zhang, W.; Zhang, T.; Jiang, B.; Mu, W. Characterization of a thermostable recombinant L-rhamnose isomerase from Caldicellulosiruptor obsdiansis OB47 and its application for the production of L-fructose and L-rhamnulose. J. Sci. Food Agric. 2018, 98, 2184–2193. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, B.A.; Rejezk, M.; Kuhaudomlarp, S.; Hill, L.; Mascia, I.; Nepogodiev, S.A.; Dorfmueller, H.C.; Field, R.A. Discovery of an RmlC/Dfusion protein in the microalga Pyrmnesium parvum and its implications for NDP-β-1-rhamnose biosynthesis in microalgae. J. Biol. Chem. 2019, 294, 9172–9185. [Google Scholar] [CrossRef] [PubMed]
- Koivistoinen, O.M.; Arvas, M.; Headman, J.R.; Andberg, M.; Penttilä, M.; Jeffries, T.W.; Richard, P. Characterisation of the gene cluster for L-rhamnose catabolism in the yeast Scheffersomyces (Pichia) stipites. Gene 2012, 492, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.R.; Munera, D.; Waldor, M.K.; Sperandio, V.; Ritchie, J.M. Fucose sensing regulates bacterial intestinal colonization. Nature 2012, 492, 113–117. [Google Scholar] [CrossRef]
- Vanhooren, P.T.; Vandamme, E.J. L-Fucose: Occurrence, physiological role, chemical, enzymatic and microbial synthesis. J. Chem. Technol. Biotechnol. 1999, 74, 479–497. [Google Scholar] [CrossRef]
- Hugdahl, M.B.; Beery, J.T.; Doyle, M.P. Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 1998, 56, 1560–1566. [Google Scholar] [CrossRef]
- Hooper, L.V.; Xu, J.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 1999, 96, 9833–9838. [Google Scholar] [CrossRef]
- Cocks, G.T.; Aguilar, T.; Lin, E.C.C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J. Bacteriol. 1974, 118, 83–88. [Google Scholar] [CrossRef]
- Liu, J.-J.; Lee, J.W.; Yun, E.J.; Jung, S.-M.; Seo, J.-H.; Jin, Y.-S. L-Fucose production by engineered Escherichia coli. Biotechnol. Bioeng. 2019, 116, 904–911. [Google Scholar] [CrossRef]
- Stahl, M.; Friis, L.M.; Nothaft, H.; Liu, X.; Li, J.; Szymanski, C.M.; Stintzi, A. L-Fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. USA 2011, 108, 7194–7199. [Google Scholar] [CrossRef]
- Watanabe, S. Characterization of L-2-keto-3-deoxyfuconate aldolases in a nonphosphorylating L-fucose metabolism pathway in anaerobic bacteria. J. Biol. Chem. 2020, 295, 1338–1349. [Google Scholar] [CrossRef]
- Sharma, V.; Ichikawa, M.; Freeze, H.H. Mannose metabolism: More than meets the eye. Biochem. Biophys. Res. Commun. 2014, 453, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Nakajima, T.; Ichishima, E. A 1,2-α-D-mannosidase from a Bacillus sp.: Purification, characterization, and mode of action. Carbohydr. Res. 1994, 251, 89–98. [Google Scholar] [CrossRef]
- Duffaud, G.D.; McCutchen, C.M.; Leduc, P.; Parker, K.N.; Kelly, R.M. Purification and characterization of extremely thermostable β-mannanase, β-mannosidase, and α-galactosidase from the hyperthermophilic eubacterium Thermotoga neapolitana 5068. Appl. Environ. Microbiol. 1997, 63, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Fushinobu, S.; Imamura, H.; Shoun, H.; Wakagi, T. Crystallization and preliminary X-ray analysis of cytosolic α-mannosidase from Thermotoga maritima. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 104–105. [Google Scholar] [CrossRef] [PubMed]
- Angelov, A.; Putyrski, M.; Liebl, W. Molecular and biochemical characterization of α-glucosidase and α-mannosidase and their clustered genes from the thermoacidophilic archaeon Picrophilus Torridus. J. Bacteriol. 2006, 188, 7123–7131. [Google Scholar] [CrossRef] [PubMed]
- Numao, S.; He, S.; Evjen, G.; Howard, S.; Tollersrud, O.K.; Withers, S.G. Identification of Asp197 as the catalytic nucleophile in the family 38 α-mannosidase from bovine kidney lysosomes. FEBS Lett. 2000, 484, 175–178. [Google Scholar] [CrossRef]
- Gibbs, M.D.; Elinder, A.U.; Reeves, R.A.; Bergquist, P.L. Sequencing, cloning and expression of a β-1,4-mannanase gene, manA, from the extremely thermophilic anaerobic bacterium, Caldicellulosiruptor Rt8B.4. FEMS Microbiol. Lett. 1996, 141, 37–43. [Google Scholar] [CrossRef]
- Sunna, A.; Gibbs, M.D.; Chin, C.W.J.; Nelson, P.J.; Bergquist, P.L. A gene encoding a novel multidomain β-1,4-mannanase from Caldibacillus cellulovorans and action of the recombinant enzyme on kraft pulp. Appl. Environ. Microbiol. 2000, 66, 664–670. [Google Scholar] [CrossRef][Green Version]
- Sunna, A. Modular organisation and functional analysis of dissected modular β-mannanase CsMan26 from Caldicellulosiruptor Rt8B.4. Appl. Microbiol. Biotechnol. 2010, 86, 189–200. [Google Scholar] [CrossRef]
- Szymona, M.; Doudoroff, M. Carbohydrate metabolism in Rhodopseudomonas Spheroides. J. Gen. Microbiol. 1960, 22, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.A.; Cho, E.; Trinh, L.T.P.; Jeong, J.-S.; Bae, H.-J. Development of an integrated process to produce D-mannose and bioethanol from coffee residue waste. Bioresour. Technol. 2017, 244, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Saburi, W.; Sato, S.; Hashiguchi, S.; Muto, H.; Lizuka, T.; Mori, H. Enzymatic characteristics of D-mannose 2-epimerase, a new member of the acylglucosamine 2-epimerase superfamily. Appl. Microbiol. Biotechnol. 2019, 103, 6559–6570. [Google Scholar] [CrossRef] [PubMed]
- Nishiya, Y.; Tamura, N.; Tamura, T. Analysis of bacterial glucose dehydrogenase homologs from thermoacidophilic archaeon Thermoplasma acidophilum: Finding and characterization of aldohexose dehydrogenase. Biosci. Biotechnol. Biochem. 2004, 68, 2451–2456. [Google Scholar] [CrossRef]
- Yasutake, Y.; Nishiya, Y.; Tamura, N.; Tamura, T. Structural Insights into unique substrate selectivity of Thermoplasma acidophilum D-aldohexose dehydrogenase. J. Mol. Biol. 2007, 367, 1034–1046. [Google Scholar] [CrossRef]
- Kopp, D.; Willows, R.D.; Sunna, A. Cell-free enzymatic conversion of spent coffee grounds into the platform chemical lactic acid. Front. Bioeng. Biotechnol. 2019, 7, 389. [Google Scholar] [CrossRef]
- Krause-Jensen, D.; Duarte, C. Substantial role of macroalgae in marine carbon sequestration. Nat. Geosci. 2016, 9, 737–742. [Google Scholar] [CrossRef]
- Kamke, J.; Sczyrba, A.; Ivanova, N.; Schwientek, P.; Rinke, C.; Mavromatis, K.; Woyke, T.; Hentschel, U. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J. 2013, 7, 2287–2300. [Google Scholar] [CrossRef]
- Reisky, L.; Préchoux, A.; Zühlke, M.-K.; Bäumgen, M.; Robb, C.S.; Gerlach, N.; Roret, T.; Stanetty, C.; Larocque, R.; Michel, G.; et al. A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan. Nat. Chem. Biol. 2019, 15, 803–812. [Google Scholar] [CrossRef]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F.; et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020, 5, 1026–1039. [Google Scholar] [CrossRef]
- Cuskin, F.; Lowe, E.C. Glyan degradation writ large in the ocean. Nat. Microbiol. 2020, 5, 980–981. [Google Scholar] [CrossRef]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Baslé, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef] [PubMed]
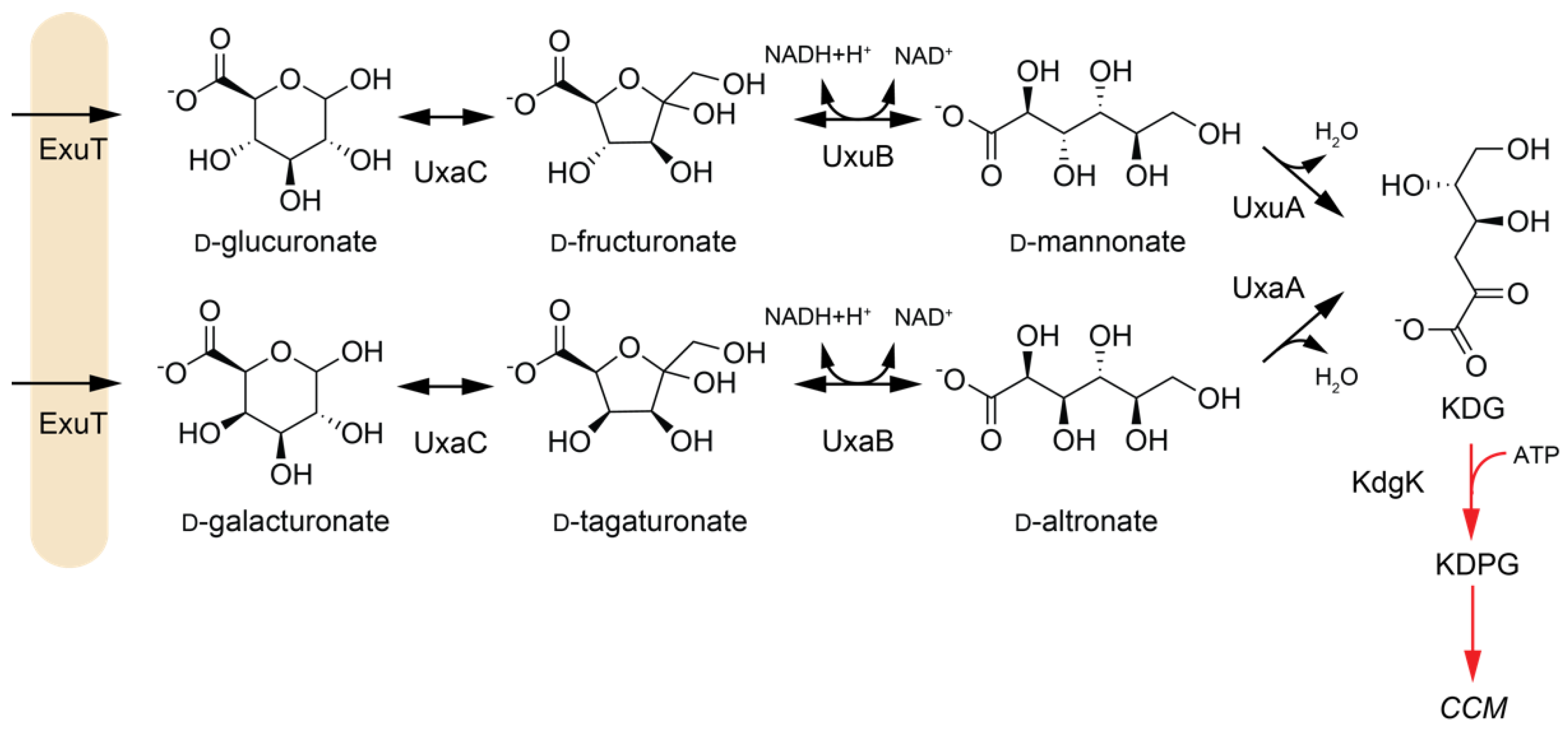
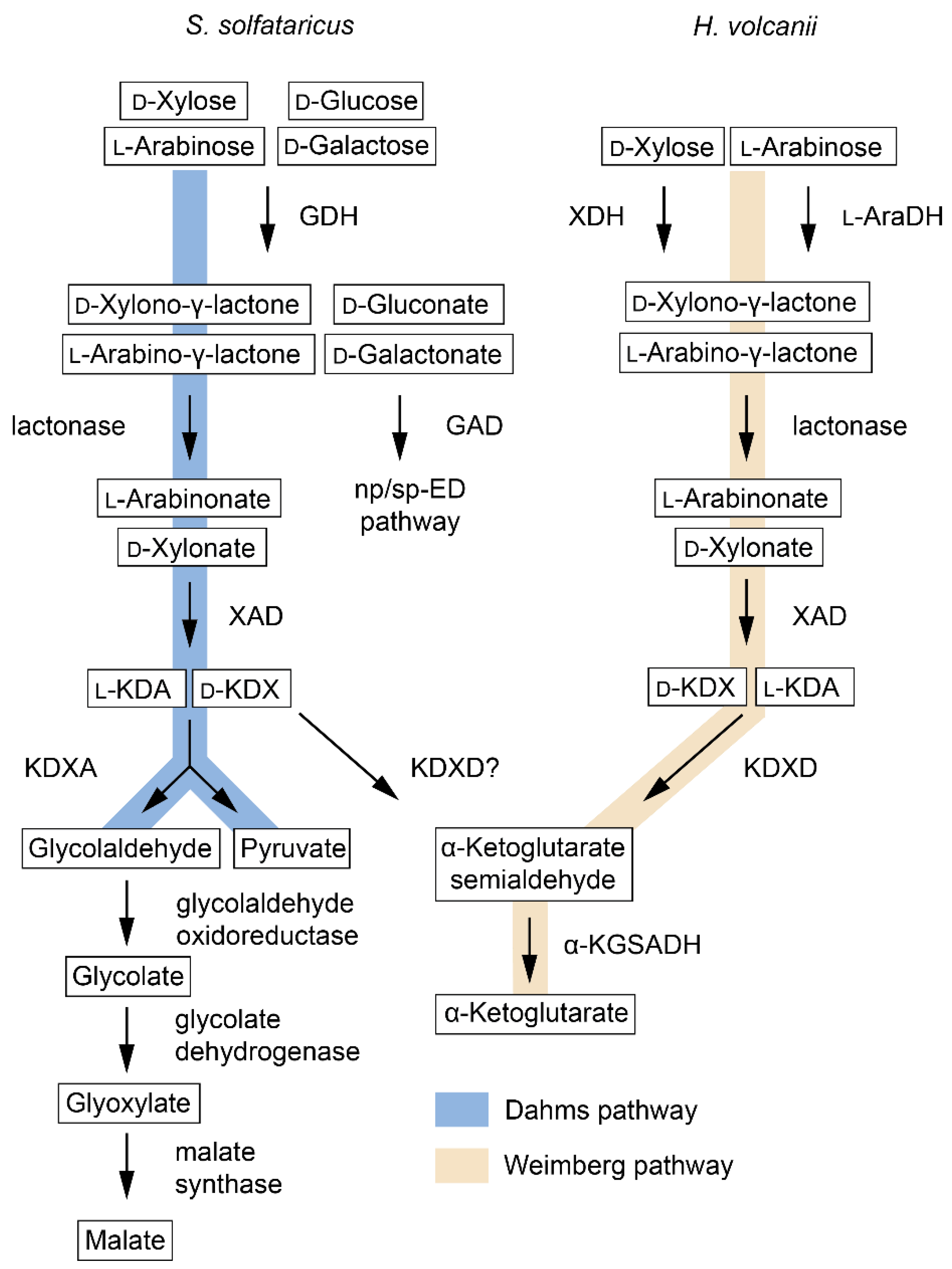
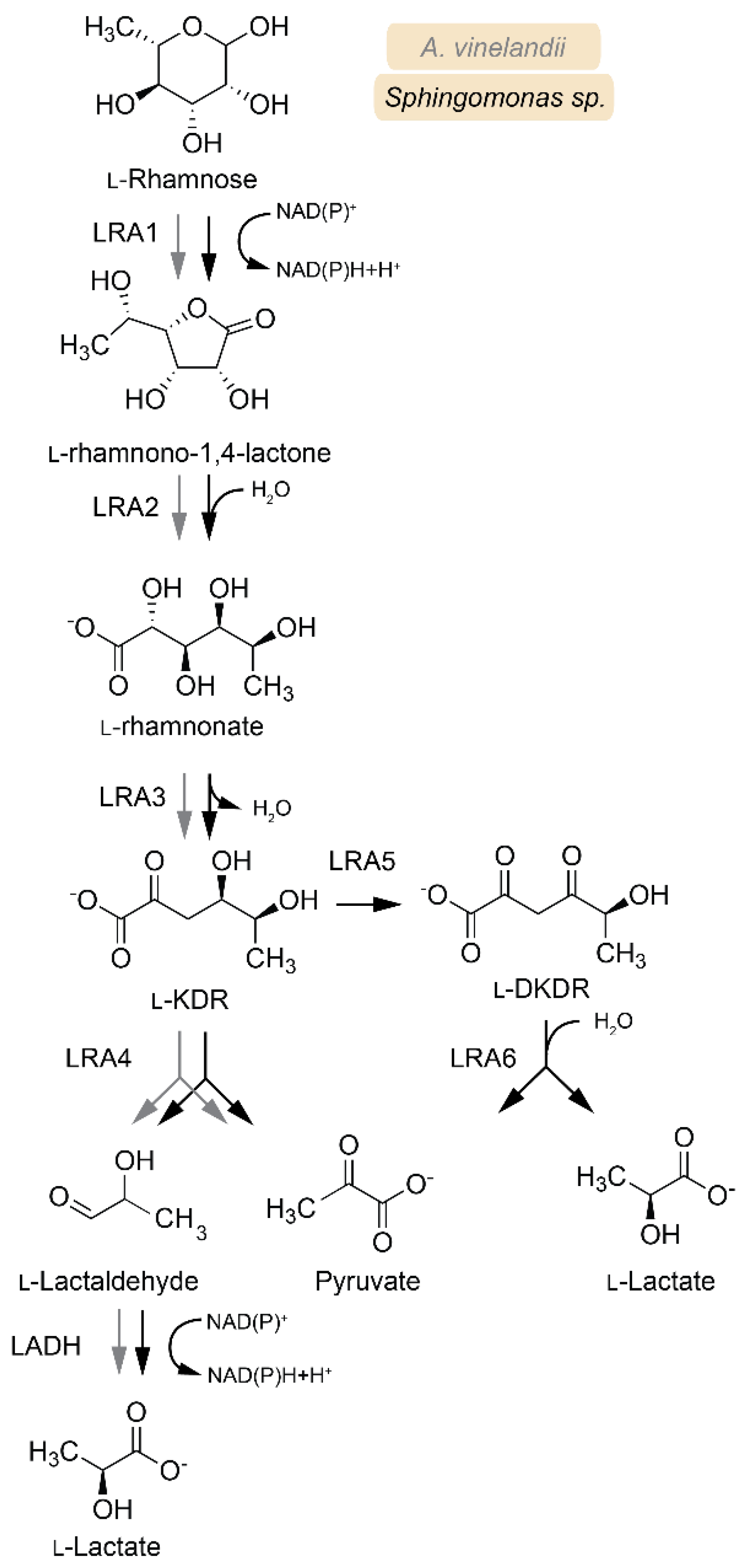
| Enzyme | EC | Abbreviation | Pathway | Species 1 | Active Substrates 2 | Activating Cofactors/Additives |
|---|---|---|---|---|---|---|
| IlvD/EDD superfamily | ||||||
| 6-phospho-gluconate dehydratase | 4.2.13 | EDD | classical ED | E. coli [5,6] Z. mobilis [4] P. aeruginosa [7] P. flourescens [8] P. putida [7] | 6-phosphogluconate | Fe2+, glycerol-3-phosphate, Fe2+, Mn2+, glycerol, gluconate, ascorbate Fe2+, Mg2+, Mn2+ Fe2+, Mg2+, Mn2+, glutathione, cysteine Fe2+, Mg2+, Mn2+ |
| gluconate dehydratase | 4.2.1.39 | GAD | np-ED/sp-ED | A. xylosoxydans [9] | D-Glc, D-Xyl, L-Fuc, D-Gal, D-Man, L-Man, D-Rib, Iso | Mg2+, Mn2+ |
| dihydroxyacid dehydratase | DHAD | valine/isoleucine synthesis | S.solfataricus [10] | Dh-iso, glc, D-Xyl, D-Ara, GlcA, GalA, D-Fuc, L-Thr | Mg2+, Mn2+, Ba2+, NaCl | |
| xylonate deydratase | 4.2.1.82 | XAD | pentose oxidation | C. crescentus [11] | D-Xyl | Fe2+, Mg2+ |
| arabinoate dehydratase | 4.2.1.25 | AraC | pentose oxidation | A. brasiliense [12] | L-Ara, D-Xyl | Fe2+, Mg2+ |
| Enolase superfamily | ||||||
| gluconate/galactonate dehydratase | 4.2.1.140 | GAD | np-ED/sp-ED | S. solfataricus [13,14] P. torridus [15] | D-Glc, D-Gal, D-Glc, D-Gal | Mg2+, Mn2+, Ni2+, Co2+ Mg2+ |
| gluconate dehydratase | 4.2.1.39 | GAD | np-ED/sp-ED | T. tenax [16] C. pasteurianum 3 [17] H. volcanii [18] | D-Glc D-Glc D-Glc | Co2+, Mg2+, Ni2+, Cd2+ (Fe2+ inhibits) Fe2+, Mg2+, Mn2+, Co2+, reducing agents Mg2+, Mn2+ |
| xylonate dehydratase | 4.2.1.82 | XAD | pentose oxidation | H. volcanii [19] | D-Xyl, D-Glc | Mg2+ |
| arabinoate dehydratase | 4.2.1.5 | AraD | pentose oxidation | S. solfataricus [20] | D-Ara | |
| rhamnonate dehydratase | 4.2.1.90 | RhamD | pentose oxidation | E. coli [21] S. typhimurium [21] A. vinelandii [22] Sphingomonas [23] | L-Rha, L-Lyx L-Rha, L-Lyx L-Rha, L-Lyx, L-Man L-Rha, L-Lyx, L-Man | Mg2+ Mg2+ |
| mannonate dehydratases | 4.2.1.8 | ManD | diverse | E. coli [24,25] C. salexigens [24] S. enterica [24,25] N. aromaticivorans [26] many more [25] | D-Man, L-Gul D-Man, L-Gul D-Man, L-Ido D-Man | Mg2+ Mg2+ Mg2+ Mg2+ |
| fuconate dehydratase | 4.2.1.68 | FucD | L-fucose pathway | X. campestris [27] | L-Fuc, D-Ara | Mg2+ |
| Xylose isomerase-like superfamily | ||||||
| mannonate dehydratase | 4.2.1.8 | ManD (uxuA) | hexuronate metabolism | S. suis [28] T. acidophilum [29] | D-Man D-Man | Mg2+ Mg2+, Mn2+, Ni2+, Co2+, Ca2+, ß-mercaptoethanol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopp, D.; Bergquist, P.L.; Sunna, A. Enzymology of Alternative Carbohydrate Catabolic Pathways. Catalysts 2020, 10, 1231. https://doi.org/10.3390/catal10111231
Kopp D, Bergquist PL, Sunna A. Enzymology of Alternative Carbohydrate Catabolic Pathways. Catalysts. 2020; 10(11):1231. https://doi.org/10.3390/catal10111231
Chicago/Turabian StyleKopp, Dominik, Peter L. Bergquist, and Anwar Sunna. 2020. "Enzymology of Alternative Carbohydrate Catabolic Pathways" Catalysts 10, no. 11: 1231. https://doi.org/10.3390/catal10111231
APA StyleKopp, D., Bergquist, P. L., & Sunna, A. (2020). Enzymology of Alternative Carbohydrate Catabolic Pathways. Catalysts, 10(11), 1231. https://doi.org/10.3390/catal10111231






