Enhancing Cardiovascular Disease Detection Through Exploratory Predictive Modeling Using DenseNet-Based Deep Learning
Abstract
1. Introduction
2. Associate Work
DenseNet in Cardiovasular Disease Detection
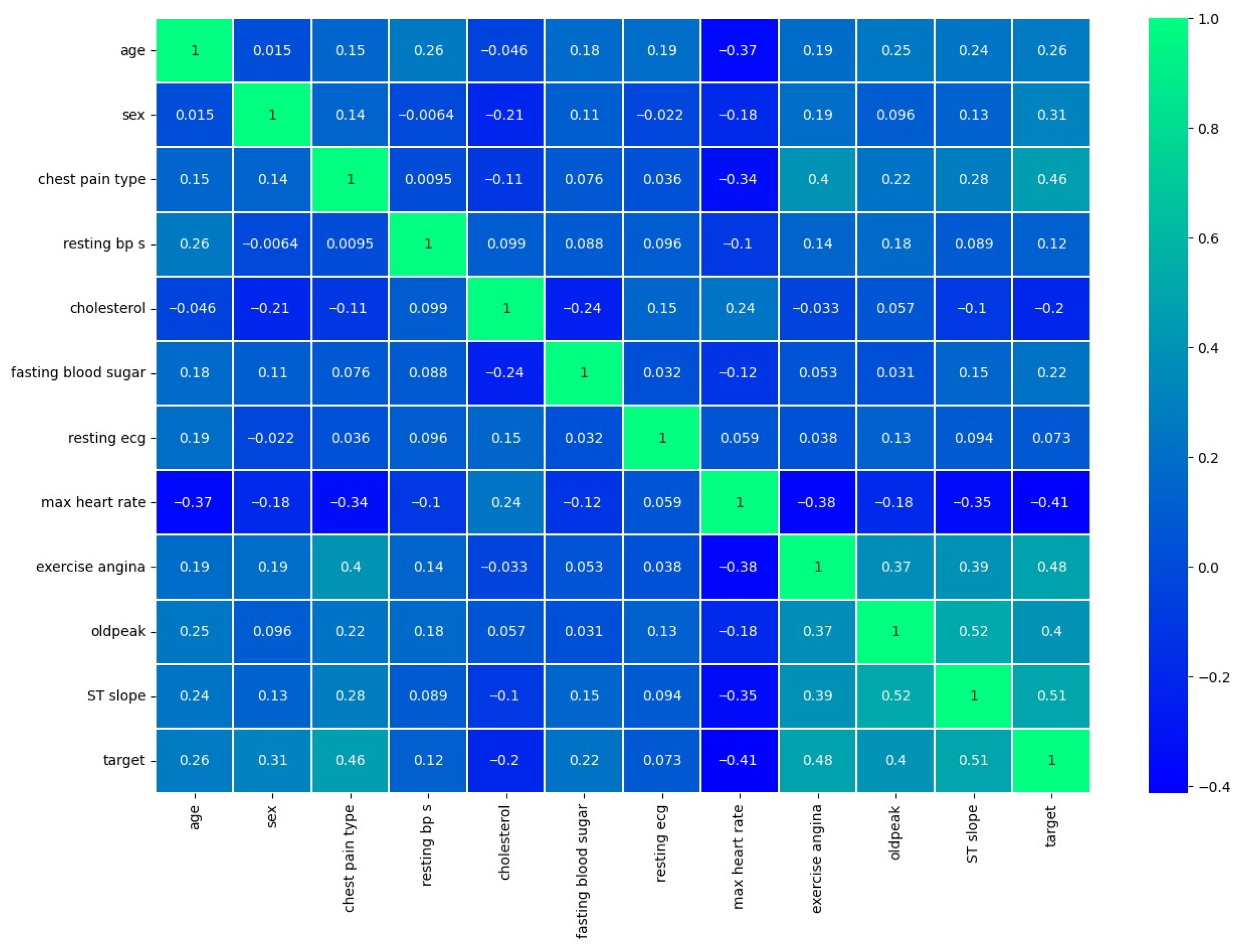
3. Dataset Overview
4. Proposed Methodology
4.1. DenseNet Architecture Design
4.2. Layer-Wise Details
4.2.1. Transition Layers
4.2.2. Overall DenseNet Structure
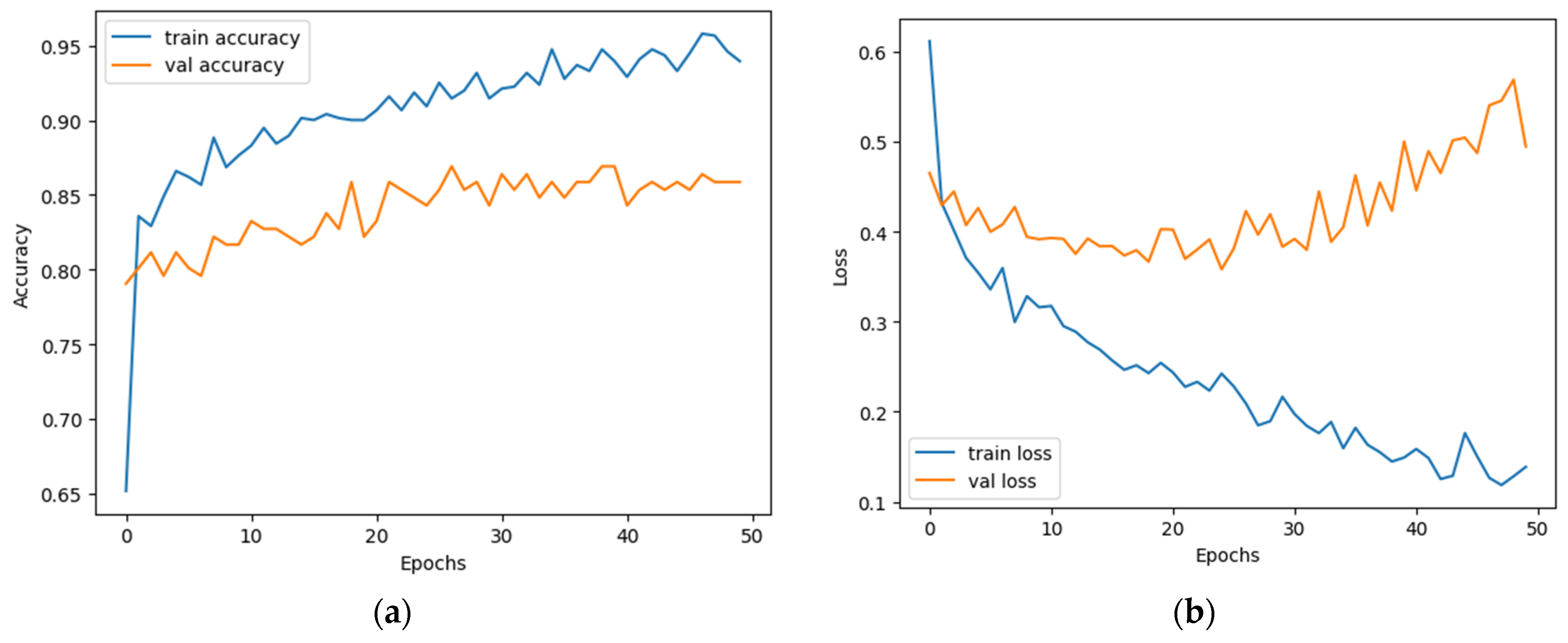
5. Result Analysis
5.1. Experimentation Setup
5.2. Performance Parameters
- N = no of samples
- = true label
- p = predicted class
5.3. Objective Analysis
Performance Parameters
6. Comparative Evaluation
6.1. Comparison with Existing Methods
6.2. Benchmarking Analysis
7. Conclusions
7.1. Limitations
7.2. Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbas, S.; Sampedro, G.A.; Alsubai, S.; Almadhor, A.; Kim, T. An efficient stacked ensemble model for heart disease detection and classification. Comput. Mater. Contin. 2023, 77, 665–680. [Google Scholar] [CrossRef]
- Arvaniti, E.; Claassen, M.; Giamberardino, G. Automated cardiac diagnosis challenge (ACDC) & caudate nucleus segmentation challenge—Preliminary results. In Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges; Springer: Cham, Switzerland, 2017; pp. 196–206. [Google Scholar]
- Asif, D.; Bibi, M.; Arif, M.S.; Mukheimer, A. Enhancing heart disease prediction through ensemble learning techniques with hyperparameter optimization. Algorithms 2023, 16, 308. [Google Scholar] [CrossRef]
- Bhatt, C.M.; Patel, P.; Ghetia, T.; Mazzeo, P.L. Effective heart disease prediction using machine learning techniques. Algorithms 2023, 16, 88. [Google Scholar] [CrossRef]
- Bilgaiyan, S.; Ayon, T.; Khan, A.; Johora, F.; Parvin, M.; Alam, M. Heart disease prediction using machine learning. In Proceedings of the 2023 International Conference on Computing, Communication, and Information Technology (ICCCI), Coimbatore, India, 23–25 January 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Alsharqi, M.; Edelman, E.R. Artificial Intelligence in Cardiovascular Imaging and Interventional Cardiology: Emerging Trends and Clinical Implications. J. Soc. Cardiovasc. Angiogr. Interv. 2025, 4 Pt B, 102558. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bentley, P.; Mori, K. Deep learning in medical image analysis. In Medical Imaging Technology; Springer: Cham, Switzerland, 2019; pp. 25–46. [Google Scholar]
- Coursera. Deep Learning Specialization by Andrew Ng. Coursera. Available online: https://www.coursera.org/specializations/deep-learning (accessed on 26 July 2025).
- Divya, K.; Sirohi, A.; Pande, S.; Malik, R. An IoMT assisted heart disease diagnostic system using machine learning techniques. In Cognitive Internet of Medical Things for Smart Healthcare; Springer: Berlin/Heidelberg, Germany, 2021; pp. 145–161. [Google Scholar]
- Enad, H.G.; Mohammed, M.A. Cloud computing-based framework for heart disease classification using quantum machine learning approach. J. Intell. Syst. 2024, 33, 20230261. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Duan, W.; Zhou, T.; Jin, X. Automated detection of cardiac abnormalities from echocardiographic images using convolutional neural networks. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; IEEE: Piscataway, NJ, USA, 2018. [Google Scholar]
- Liang, G.; Zheng, L.; Zhang, L.; Lin, L.; Xiang, D. Deep learning-based cardiovascular disease diagnosis and monitoring: A review. Front. Cardiovasc. Med. 2020, 7, 169. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Arnaout, R.; Mofrad, M.; Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit. Med. 2018, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.B.; Gupta, S.; Singh, D.; Acharya, B.; Gerogiannis, V.C.; Kanavos, A.; Pintelas, P. Heart disease prediction using concatenated hybrid ensemble classifiers. Algorithms 2023, 16, 538. [Google Scholar] [CrossRef]
- Melendez, J.; Sánchez, C.I. Deep learning in medical image analysis: A comprehensive review. Biomed. Signal Process. Control 2020, 60, 101986. [Google Scholar]
- Mohan, S.; Thirumalai, C.; Srivastava, G. Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 2019, 7, 81542–81554. [Google Scholar] [CrossRef]
- Official DenseNet GitHub Repository. GitHub. Available online: https://github.com/liuzhuang13/DenseNet (accessed on 26 July 2025).
- Kabir, P.B.; Akter, S. Emphasised research on heart disease divination applying tree-based algorithms and feature selection. In Proceedings of the 2021 International Conference on Innovative Computing, Intelligent Communication and Smart Electrical Systems (ICSES), Chennai, India, 24–25 September 2021; pp. 1–6. [Google Scholar] [CrossRef]
- PyTorch documentation on DenseNet. PyTorch. Available online: https://pytorch.org/hub/pytorch_vision_densenet/ (accessed on 26 July 2025).
- Quang, N.N.; Gatt, A. A survey of deep learning techniques for medical image classification. In Proceedings of the 3rd International Conference on Image and Graphics Processing, Jeju Island, Republic of Korea, 24–26 February 2019; pp. 87–93. [Google Scholar]
- Samad, M.D.; Ulloa, A.; Wehner, G.J.; Jing, L.; Hartzel, D.N.; Good, C.W.; Williams, B.A.; Haggerty, C.M.; Fornwalt, B.K. Predicting survival from large echocardiography and electronic health record datasets: Optimization with machine learning. JACC Cardiovasc. Imaging 2019, 12, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Maneesha, S.; Mohanty, S.; Patra, P.K.; Bhoi, S.K.; Sahoo, K.S.; Gandomi, A.H. A stacking classifiers model for detecting heart irregularities and predicting cardiovascular disease. Healthc. Anal. 2023, 3, 100133. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Zhou, Z. DenseNet-based deep learning for coronary artery disease detection from angiograms. Med. Image Anal. 2019, 57, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Chen, Y.; Zhang, Y. Application of DenseNet architecture in cardiac magnetic resonance image analysis for myocardial infarction detection. IEEE Trans. Med. Imaging 2020, 39, 2986–2996. [Google Scholar]
- Yao, J.; Burns, J.E.; Munoz, H.E.; Summers, R.M.; Yao, J. Dense-inception network for abdominal multi-organ segmentation. In International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis; Springer: Cham, Switzerland, 2017; pp. 164–172. [Google Scholar]
- Available online: https://www.kaggle.com/datasets/sid321axn/heart-statlog-cleveland-hungary-final (accessed on 24 February 2024).
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Almulihi, A.; Saleh, H.; Hussien, A.M.; Mostafa, S.; El-Sappagh, S.; Alnowaiser, K.; Ali, A.A.; Refaat Hassan, M. Ensemble Learning Based on Hybrid Deep Learning Model for Heart Disease Early Prediction. Diagnostics 2022, 12, 3215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Chen, J.; Zhang, X.; He, W.; Tong, L. Multi-class classification of skin cancer images with deep learning techniques. IEEE Access 2019, 7, 11580–11587. [Google Scholar]
- Zhuang, X.; Wei, Y.; Li, Y.; Jia, Y.; Shi, J.; Lin, J. Attend in, localize and segment (AILS): Weakly supervised learning of a dense event captioning model. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 10419–10428. [Google Scholar]
- Jaware, T.; Khanchandani, K.; Badgujar, R. A novel hybrid atlas-free hierarchical graph-based segmentation of newborn brain MRI using wavelet filter banks. Int. J. Neurosci. 2019, 130, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Dritsas, E.; Trigka, M. Application of deep learning for heart attack prediction with explainable artificial intelligence. Computers 2024, 13, 244. [Google Scholar] [CrossRef]









| Parameters | Training | Validation | Testing |
|---|---|---|---|
| Accuracy | 0.964 | 0.910 | 0.924 |
| Precision | 0.962 | 0.905 | 0.909 |
| F1-score | 0.963 | 0.915 | 0.930 |
| Log Loss | 0.068 | 0.315 | 0.275 |
| Sensitivity | 0.969 | 0.927 | 0.952 |
| Specificity | 0.875 | 0.875 | 0.893 |
| Matthews Correlation Coefficient | 0.772 | 0.772 | 0.849 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| FT-DNN [10] | 80.19 | 77.03 | 86.77 | 69.43 |
| DNN [10] | 76.73 | 72.85 | 86.19 | 67.32 |
| AdaBoost [13] | 80.33 | 88.00 | 72.00 | 79.00 |
| RF [13] | 83.61 | 89.00 | 78.00 | 83.00 |
| KNN [13] | 81.79 | 89.00 | 72.00 | 81.00 |
| XGBoost [13] | 80.33 | 86.00 | 75.00 | 80.00 |
| LR [13] | 81.97 | 84.00 | 81.00 | 83.00 |
| Concatenated Hybrid Ensemble Classifiers [15] | 86.89 | 81.8 | 86.9 | 84.3 |
| QNN [10] | 77.00 | 76.00 | 73.00 | 75.00 |
| QSVM [10] | 85.00 | 79.00 | 90.00 | 84.00 |
| MLP [35] | 85.00 | 83.00 | 84.00 | 84.00 |
| RNN [35] | 84.00 | 82.00 | 83.00 | 82.00 |
| GRU [35] | 89.00 | 87.00 | 88.00 | 87.00 |
| LSTM [35] | 88.00 | 86.00 | 87.00 | 87.00 |
| CNN [35] | 87.00 | 85.00 | 86.00 | 85.00 |
| XAI [35] | 90.00 | 89.00 | 90.00 | 89.00 |
| Proposed (DenseNet) | 92.44 | 90.91 | 95.24 | 93.02 |
| Models | Training | Validation | ||
|---|---|---|---|---|
| Accuracy | Loss | Accuracy | Loss | |
| ResNet50 | 0.960 | 0.093 | 0.796 | 0.852 |
| ResNet101 | 0.958 | 0.002 | 0.863 | 1.799 |
| VGG-16 | 0.952 | 0.120 | 0.874 | 0.523 |
| VGG-19 | 0.942 | 0.161 | 0873 | 0.566 |
| DenseNet (proposed) | 0.964 | 0.067 | 0.903 | 0.314 |
| Parameters | VGG 16 | VGG 19 | ResNet 50 | ResNet 101 | DenseNet |
|---|---|---|---|---|---|
| Accuracy | 0.874 | 0.873 | 0.796 | 0.863 | 0.903 |
| Recall (Sensitivity) | 0.900 | 0.900 | 0.860 | 0.900 | 0.927 |
| Precision | 0.893 | 0.917 | 0.810 | 0.893 | 0.905 |
| F1 Score | 0.897 | 0.914 | 0.880 | 0.897 | 0.915 |
| Specificity | 0.847 | 0.897 | 0.906 | 0.906 | 0.875 |
| MCC | 0.778 | 0.907 | 0.887 | 0.907 | 0.772 |
| Parameter | VGG16 | VGG19 | ResNet50 | ResNet101 | DenseNet |
|---|---|---|---|---|---|
| Accuracy | 0.8739 | 0.8824 | 0.8908 | 0.9160 | 0.9244 |
| Precision | 0.8871 | 0.8769 | 0.9032 | 0.9206 | 0.9091 |
| Recall (Sensitivity) | 0.8730 | 0.9048 | 0.8889 | 0.9206 | 0.9524 |
| Specificity | 0.8750 | 0.8571 | 0.8929 | 0.9107 | 0.8929 |
| F1 Score | 0.8800 | 0.8906 | 0.8960 | 0.9206 | 0.9302 |
| MCC | 0.7474 | 0.7639 | 0.7811 | 0.8314 | 0.8489 |
| Test Loss | 0.5243 | 0.5023 | 0.4758 | 0.4836 | 0.2748 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadi, W.; Jaware, T.; Khalifa, T.; Aburub, F.; Ali, N.; Saini, R. Enhancing Cardiovascular Disease Detection Through Exploratory Predictive Modeling Using DenseNet-Based Deep Learning. Computers 2025, 14, 330. https://doi.org/10.3390/computers14080330
Hadi W, Jaware T, Khalifa T, Aburub F, Ali N, Saini R. Enhancing Cardiovascular Disease Detection Through Exploratory Predictive Modeling Using DenseNet-Based Deep Learning. Computers. 2025; 14(8):330. https://doi.org/10.3390/computers14080330
Chicago/Turabian StyleHadi, Wael, Tushar Jaware, Tarek Khalifa, Faisal Aburub, Nawaf Ali, and Rashmi Saini. 2025. "Enhancing Cardiovascular Disease Detection Through Exploratory Predictive Modeling Using DenseNet-Based Deep Learning" Computers 14, no. 8: 330. https://doi.org/10.3390/computers14080330
APA StyleHadi, W., Jaware, T., Khalifa, T., Aburub, F., Ali, N., & Saini, R. (2025). Enhancing Cardiovascular Disease Detection Through Exploratory Predictive Modeling Using DenseNet-Based Deep Learning. Computers, 14(8), 330. https://doi.org/10.3390/computers14080330






