Abstract
Incorrectly diagnosing plant diseases can lead to various undesirable outcomes. This includes the potential for the misuse of unsuitable herbicides, resulting in harm to both plants and the environment. Examining plant diseases visually is a complex and challenging procedure that demands considerable time and resources. Moreover, it necessitates keen observational skills from agronomists and plant pathologists. Precise identification of plant diseases is crucial to enhance crop yields, ultimately guaranteeing the quality and quantity of production. The latest progress in deep learning (DL) models has demonstrated encouraging outcomes in the identification and classification of plant diseases. In the context of this study, we introduce a novel hybrid deep learning architecture named “CTPlantNet”. This architecture employs convolutional neural network (CNN) models and a vision transformer model to efficiently classify plant foliar diseases, contributing to the advancement of disease classification methods in the field of plant pathology research. This study utilizes two open-access datasets. The first one is the Plant Pathology 2020-FGVC-7 dataset, comprising a total of 3526 images depicting apple leaves and divided into four distinct classes: healthy, scab, rust, and multiple. The second dataset is Plant Pathology 2021-FGVC-8, containing 18,632 images classified into six categories: healthy, scab, rust, powdery mildew, frog eye spot, and complex. The proposed architecture demonstrated remarkable performance across both datasets, outperforming state-of-the-art models with an accuracy (ACC) of 98.28% for Plant Pathology 2020-FGVC-7 and 95.96% for Plant Pathology 2021-FGVC-8.
1. Introduction
The apple is a largely produced fruit thanks to its exceptional flavor, texture, nutritional benefits, and aesthetic appeal. Apple production is an important economic activity in many countries of the world. As reported by the Food and Agriculture Organization (FAO) of the United Nations, 2022 saw a global apple production exceeding 95.8 million tons. Leading this production was China with 49.7% (47.5 million metric tons), followed by Turkey at 5% (4.82 million tons) and the U.S. close behind at 4.6% (4.43 million tons) [1]. Apples hold significant value in the U.S. horticultural sector. Producing close to 2 billion pounds annually, the apple sector notably contributes to the U.S. economy, generating up to USD 15 billion each year [2,3]. However, apple diseases can significantly reduce yield and fruit quality. Identifying apple diseases at an early stage is crucial for the proficient management of apple orchards. Research indicates that Damage to crops caused by diseases can account for up to 50% of the overall yield, particularly from parasitic and prevalent plant ailments [4]. Scab stands out as a common fungal disease affecting apple trees. It can cause up to a 50% yield loss if not treated promptly [5]. Another widespread fungal ailment in apple trees is apple rust, which can lead to substantial yield reductions if not identified and addressed in a timely manner. According to a study conducted by the Australian government, fungal diseases of apple leaves are the major health risk of fruit trees in Australian apple-producing areas [6]. A good knowledge of the different diseases that affect apple trees and the proper control methods is essential for agronomists to avoid crop losses caused by these diseases.
Traditional optical methods such as multispectral, hyperspectral, and fluorescence imaging provide valuable information on plant health [7]. However, they generally rely on the analysis of grayscale images, which do not always capture the complex color variations associated with disease symptoms [8]. By contrast, AI-based solutions excel at processing colored images, exploiting the rich spectrum of hues and tones to detect subtle changes indicative of disease [9]. By harnessing the power of deep learning algorithms, AI can accurately identify patterns and anomalies in colored images, enabling a more accurate and reliable diagnosis of disease compared to traditional methods. What is more, AI’s ability to learn from large datasets enhances its adaptability to diverse environmental conditions and disease states, further improving its effectiveness in early disease detection and management.
Developing AI-based solutions for swift disease identification in apple leaves is crucial to assist farmers and orchard managers in safeguarding their crops. Advances in DL allowed the development of image-based techniques for the automated identification and classification of various plant ailments. The technology has the potential to provide rapid and accurate identification of plant diseases, which would facilitate farm management decisions. In addition, the use of DL-powered systems for apple foliar disease detection offers continuous crop monitoring, thus enabling early detection of diseases, preventing their spread, and supporting targeted interventions. Ultimately, the implementation of such automated diagnostic systems can significantly contribute to the production of healthy, high-quality fruit.
To address the challenges faced by specialized pathologists, we propose an innovative DL solution to identify diseases in apple leaves. Our approach consists of using advanced computer vision and DL techniques to design an advanced architecture, named “CTPlantNet”, for preprocessing and classifying images according to the observed symptoms on apple leaves. More specifically, this paper provides significant contributions to the field by adapting recent DL models (SEResNext, EfficientNet-V2S, and Swin-Large) and proposing a novel hybrid DL architecture (CNN-transformer) for apple disease preprocessing, detection, and classification using apple leaf images. The architecture of CTPlantNet comprises two blocks of models. The initial block comprises two adapted convolutional neural network (CNN) models: SEResNext-50 and EfficientNet-V2S. The subsequent block incorporates an adapted Swin-Large transformer model. To classify multiple apple leaf diseases, the models in CTPlantNet were trained and tested on two public datasets, specifically Plant Pathology 2020-FGVC-7 and Plant Pathology 2021-FGVC-8, using a five-fold cross-validation method. Outputs from the two blocks are merged using an averaging method to derive the ultimate prediction. Additionally, the study details data augmentation approaches utilized to overcome the imbalance in the data. In conclusion, the efficacy of the introduced architecture was assessed against other leading-edge models and recent scholarly contributions.
The main contributions of our paper are as follows:
- A novel DL architecture, called CTPlantNet, which integrates two CNNs as the first block and one vision transformer as the second block, is proposed for the recognition of plant diseases, thus enhancing the performance of deep learning-based plant-anomaly-detection tasks.
- The recent deep CNNs (SEResNext and EfficientNet-V2S) and the Swin-Large vision transformer were adapted for classifying plant disease images, leveraging the advantages of the transfer learning technique.
- The performances of the proposed models were assessed against each other and compared with state-of-the-art models using two open-access plant pathology datasets (Plant Pathology 2020-FGVC-7 and Plant Pathology 2021-FGVC-8), applying a cross-validation strategy.
- The testing results show that the proposed DL models, including the CNNs and the vision transformer, performed better than the state-of-the-art (SOTA) works, where the proposed CTPlantNet architecture slightly increases the performance of the three models (SEResNext, EfficientNet-V2S, and Swin-Large) when trained and tested separately.
- The introduced DL models demonstrate their ability to detect the related challenges faced by the traditional optical inspection methods, such as the similarities between the features of some diseases, thus resulting in an early and swift detection of the pathologies in apple leaves and assisting farmers and orchard managers in safeguarding their crops and producing healthy and high-quality fruits.
This paper is organized as follows: Section 2 provides a review of existing deep learning (DL) models proposed for plant pathology identification. Section 3 describes the materials and methods used, including the datasets utilized, the adapted DL models, the experimental implementation details, and the evaluation metrics employed in this study. Section 4 presents the obtained results. Section 5 interprets and analyzes these results, highlighting the potential of the proposed architecture and suggesting future improvements. Finally, Section 6 provides an overview and conclusion of this work.
2. Related Work
In recent years, DL methods have gained prominence, due to the availability of datasets and the significant progress in the computing power and memory capacity of Graphical Processing Units (GPUs). An automated plant-disease-detection system would be a valuable decision support and assistance tool for agronomists, who tend to accomplish this task through visual observation of infected plants. Consequently, many studies over the past decade have focused on the detection of plant diseases using DL techniques.
Kirola et al. [10] carried out a study contrasting machine learning (ML) and DL methods in terms of performance. For ML, plant leaf image classification was performed using five algorithms: Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Logistic Regression (LR), Random Forest (RF), and Naïve Bayes (NB). For deep learning, a custom CNN model was employed in this experiment. An image dataset consisting of 53,200 samples distributed across three categories (bacterial, viral, and fungal) served as the basis to train and test the models in this study. The proposed CNN model was the best performing with an accuracy (ACC) of 98.43%, while RF emerged as the top machine learning classifier, scoring an ACC of 97.12%. Ahmed et al. [11] used a DL model (AlexNet) to classify plant leaf images into healthy or unhealthy. A large dataset of 87,000 images was used for this experiment. The CNN model proposed in this study obtained an ACC of 96.50%. Kawasaki et al. [12] introduced a CNN-based automated detection system to identify foliar diseases in cucumbers, which achieved an impressive ACC score of 94.90%. Ma et al. [13] presented a symptom-based method using a CNN model to identify four cucumber leaf diseases: Anthracnose, downy mildew, leaf spots, and powdery mildew. To avoid overfitting, they used data augmentation techniques on the set of cucumber data, which generated a total of 14,208 symptom images. Among all the models tested, the AlexNet model with data augmentation techniques performed the best with an ACC of 93.40%. Chen et al. [14] proposed an AlexNet architecture to identify different diseases in tomato leaf images. The model obtained a mean ACC of 98.00%.
For apple diseases, Sulistyowati et al. [15] employed the VGG-16 model to automatically categorize apple leaf images into four groups: healthy, apple scab, apple rust, and multiple diseases. The model achieved the best performance when used with the Synthetic Minority Over-Sampling (SMOTE) technique. The Plant Pathology 2020-FGVC-7 dataset was used in this study, and the highest ACC obtained was 92.94%. Yadav et al. [16] developed a DL model called AFD-Net for the multi-classification of apple leaves. AFD-Net comprises an ensemble learning architecture of two EfficientNet models (EfficientNet-B3 and EfficientNet-B4). Two Kaggle datasets were used in this study (Plant Pathology 2020-FGVC-7 and Plant Pathology 2021-FGVC-8). The AFD-Net model surpassed several models, including Inception, ResNet-50, ResNet-101, and VGG-16, achieving 98.7% and 92.6% in terms of the ACC consecutively for the Plant Pathology 2020-FGVC-7 and Plant Pathology 2021-FGVC-8 datasets. Alsayed et al. [17] Proposed a deep learning methodology for identifying leaf diseases in apple trees. This approach employed CNN models, including MobileNet-V2, Inception-V3, VGG-16, and ResNet-V2, on the Plant Pathology 2020-FGVC-7 dataset to categorize images into four categories: healthy, scab, rust, and multiple diseases. When employed with the Adam optimizer, the ResNet-V2 model yielded the highest performance, achieving an ACC of 94.00%. Bansal et al. [18] proposed an ensemble learning architecture that incorporates three CNN models: DenseNet-121, EfficientNet-B7, and an EfficientNet model pre-trained on the NoisyStudent dataset. Data augmentation methods were implemented to expand the dataset (Plant Pathology 2020-FGVC-7) and to tackle the challenge of imbalanced data. The proposed architecture achieved remarkable results with an ACC of 96.25%. Subetha et al. [19] presented an automatic DL model for the classification of apple leaf images. The Plant Pathology 2020-FGVC-7 dataset was used for training, validating, and evaluating two CNN models: VGG-19 and ResNet-50. Among the two, the ResNet-50 architecture demonstrated superior performance, outperforming VGG-19 with an average ACC of 87.70%. Kejriwal et al. [20] presented an ensemble learning-based method consisting of three CNN models, namely InceptionResNetV2, ResNet101V2, and Xception. The proposed method was trained and validated using the Plant Pathology 2021-FGVC8 dataset. The ensemble learning architecture achieved a precision of 97.43%.
3. Materials and Methods
In this section, we present the two publicly available datasets used in our experiment. We introduce the proposed DL approach, which performs multi-classification of apple leaf diseases based on symptoms identified using images. Additionally, we discuss the DL models implemented in our architecture. Finally, we demonstrate how the proposed approach was implemented, as well as the techniques used to increase its performance.
3.1. Datasets
In this study, we used two editions of the Plant Pathology dataset to train and evaluate the potential of the introduced DL architecture in classifying various apple leaf diseases, relying on the visual indications present in the images. The datasets used are described as follows:
- Plant Pathology 2020-FGVC-7 [2,21] is a publicly available collection of apple leaf images with different angles, illumination, noise, and backgrounds captured and labeled manually by specialists. The main objective of releasing this dataset was to provide a resource for researchers and developers to create models for automatic image classification in the field of apple leaf pathology. This dataset comprises 3651 apple leaf images distributed across four categories: 865 healthy leaf images, 1200 with scab, 1399 with rust, and 187 showcasing multiple diseases. While each image predominantly displays a singular disease, the “multiple disease” category includes images where each leaf bears more than one ailment. All images are rendered in high resolution and RGB color mode.
- Plant Pathology 2021-FGVC-8 [22] is a large publicly available collection of images of plant leaves, which are labeled with different disease categories. The dataset was provided in the 2021 Fine-Grained Visual Categorization (FGVC) challenge, specifically for plant pathology detection. This is an extended version of Plant Pathology 2020-FGVC-7 with more images and more diseases. This dataset contains a total of 18,632 images classified into six different classes (6225 healthy images, 4826 images with scab, 3434 images with rust, 3181 images with powdery mildew, 2010 images with frog eye spot, and 956 images with multiple diseases).
The collection of the two datasets was sponsored by the Cornell Initiative for Digital Agriculture (CIDA) and released as a Kaggle competition. Plant pathology datasets provide a rich source of data for the development and testing of automated algorithms for plant abnormalities’ recognition and classification. Samples of plant foliar images from the Plant Pathology 2020-FGVC-7 and Plant Pathology 2021-FGVC-8 datasets are shown in Figure 1.
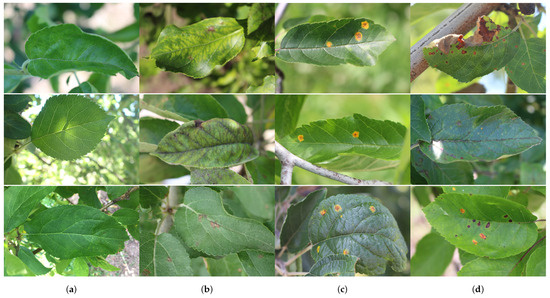
Figure 1.
Sample images of apple leaves from Plant Pathology 2020-FGVC-7 and 2021-FGVC-8 Datasets [23]: (a) healthy, (b) scab, (c) rust, and (d) multiple diseases.
3.2. Proposed Approach
To accurately classify apple leaf diseases, we introduce a hybrid DL architecture known as “CTPlantNet”. It uses pre-processing and data augmentation techniques to achieve the highest performance of the models used in this architecture. Three models are implemented in CTPlantNet as model blocks. The first block incorporates two modified convolution-based models (SEResNext-50 and EfficientNet-V2S). The second block consists of a modified Swin-Large transformer model. These two blocks were used as CNN-transformer backbones to extract the features of apple foliar diseases. After training the models separately, an averaging technique was used to combine their predictions. Additionally, training and validating the performance of these models involved a 5-fold cross-validation strategy coupled with a bagging technique. As a result, a diverse range of models, each trained on different sub-datasets, was generated (five SEResNext-50 models, five EfficientNet-V2S models, and five Swin-Large models). The proposed architecture, named CTPlantNet, is illustrated in Figure 2.
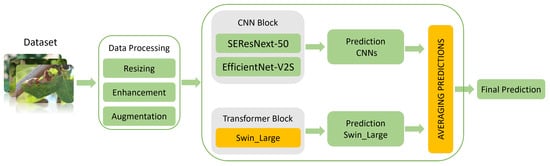
Figure 2.
Architecture of CTPlantNet for classifying apple foliar diseases, adapted from [23].
Our research involved the training and evaluation of three distinct models across two blocks, specifically a CNN block and a transformer block. Employing various DL models as backbones, combined with a 5-fold cross-validation method, enables the extraction of diverse features. Moreover, leveraging the error-correction mechanism between models ultimately allows the optimization of our hybrid architecture, thus enhancing its overall performance. To determine the best models for our architecture, we conducted several experiments, evaluating their performance on similar classification tasks. After a thorough analysis of the results, we selected the best performing models, namely SEResNext-50, EfficientNet-V2S, and Swin-Large. Following this, the models were effectively incorporated into the CTPlantNet architecture, leading to enhanced accuracy.
3.2.1. Convolutional Neural Network Models
A CNN is a type of DL model widely used for image segmentation [24], image classification [25,26], and object detection [27], where the connection between neurons is inspired by the visual cortex of animals [28]. In a CNN’s architecture, multiple layers are present, encompassing convolutional, pooling, and fully connected layers. Convolutional layers perform convolution, a mathematical operation, to extract features from the input image. Pooling layers are used in CNNs to decrease the spatial dimension of the feature maps, which reduces the computational complexity of the network. Finally, fully connected layers, by connecting all neurons in one layer to all neurons in the subsequent layer, enable the generation of predictions. CNNs use the backpropagation technique to train the network, where neuron weights are updated based on the error between the predicted and the actual values. Figure 3 depicts an example of a standard architecture for a CNN. The CNN block within CTPlantNet’s architecture is composed of two distinct models: SEResNext-50 and EfficientNet-V2S. The details of these models are provided below:

Figure 3.
Example of a standard architecture of a convolutional neural network.
- SEResNeXt-50 is a CNN model that incorporates a Squeeze-and-Excitation (SE) block with the ResNeXt architecture. The SE block in the SEResNeXt model employs a channelwise mechanism to capture channel dependencies, allowing for the adaptive recalibration of feature maps. This mechanism utilizes global average pooling to extract spatial information from the feature maps, followed by the application of a fully connected layer to recalibrate the feature maps based on the learned importance of each channel. SEResNext-50 effectively enhances the feature representation by leveraging the SE block. In addition to the SE block, SEResNext-50 takes advantage of the ResNeXt architecture [29], which is an extension of the widely used ResNet architecture. ResNeXt introduces the concept of “cardinality” as a new dimension with depth and width, allowing the network to better exploit the potential of parallel paths. Combining SE blocks with the ResNeXt architecture systematically enhances performance over different depths at an extremely minimal rise in computing complexity [30]. The SEResNeXt-50 model is capable of providing highly accurate predictions in a wide range of classification tasks, including image recognition and object detection.
- EfficientNet-V2S is a cutting-edge CNN model developed by Google [31]. This model is a member of the EfficientNet family, which is widely known for its superior efficiency and performance. The EfficientNet-V2S model is particularly noteworthy for its ability to achieve high accuracy with relatively few parameters. This is achieved through the use of novel techniques, such as compound scaling, which optimizes the architecture and scalability coefficients of the network, in addition to the Fused-MBConv blocks, which combine and employ two types of convolutions (pointwise and depthwise) into a single layer, resulting in lower computational complexity and speeding up the training of the EfficientNet-V2S model. As a result of the incorporated techniques in EfficientNet-V2S, the model shows strong potential in image classification tasks, achieving high performance while maintaining a relatively reduced size [31].
3.2.2. Vision Transformer Model
Vision transformers are a recently proposed type of DL model applied to computer vision tasks. These models employ the transformer architecture, which was originally developed for tasks in natural language processing [32]. Unlike traditional convolutional neural networks, vision transformers process visual information in parallel, without requiring convolutional operations. This allows the transformer model to process the entire image at once, capturing both local and global information from the image. In addition, vision transformers have a large number of parameters, which makes them well suited for large-scale image classification tasks. Vision transformer models adopt the self-attention mechanism, which allows for evaluating the importance of each part of the image and making more informed decisions on which features to extract and use for the classification task. Figure 4 presents an illustration of a typical vision transformer model architecture. In this work, we used the Swin-Large transformer described in the following:
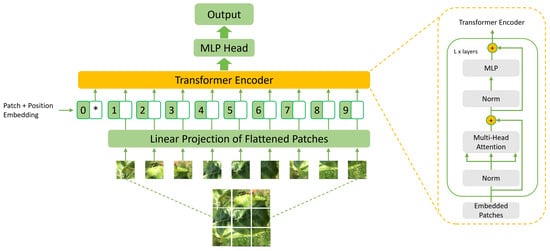
Figure 4.
Example of a standard architecture of a vision transformer model.
- Swin-Large is a transformer-based DL model consisting of 197 million trainable parameters trained on the ImageNet22k dataset for image classification tasks. Its name refers to the “shifted window” method employed to calculate the hierarchical representation of the model. This method enhances the model’s efficiency by restricting the computation of self-attention to non-overlapping local windows, simultaneously permitting connections across these windows. This approach enables the Swin-Large model to capture both local and global contextual information in images while keeping computational complexity manageable. In addition, the Swin transformer architecture incorporates a learnable relative position bias to account for the spatial relationships between image patches, further improving its performance in computer vision tasks [33].
3.3. Implementation
The implementation of the proposed approach includes incorporating three separate pre-trained models, where two, specifically SEResNext-50 and EfficientNet-V2S, are CNN-based and were pre-trained using the ImageNet dataset [34]. The third model (Swin-Large) was pre-trained on ImageNet22k [33]. To enhance the performance of both CNN models, we added a set of layers, starting with a global average pooling (GAP) layer. The layer computes the average of patch values in each feature map, producing a vector that was then fed into two fully connected layers (FCLs) with dimensions of 512 and 256. A 30% Dropout (DP) layer was incorporated following the first FCL to mitigate the risk of overfitting. Additionally, to address the ReLU dying problem [35], we employed a LeakyReLU (LRU) activation function after the second FCL. To improve the accuracy and performance of our models while reducing the training time, we integrated Gaussian Dropout (GD) at a rate of 40% [36]. More details about the added layers are depicted in Figure 5. The Swin-Large model in our architecture was fine-tuned by incorporating an FCL and a softmax activation function.
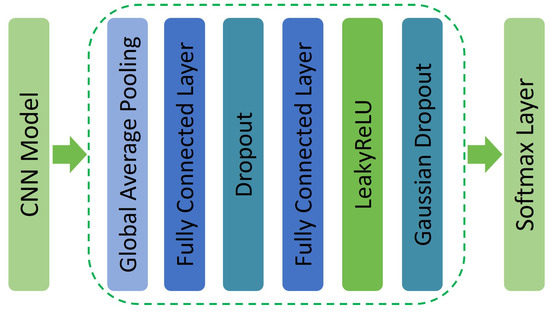
Figure 5.
Overview of the integrated layers in the CNN models, adapted from [23].
Our models were trained using two public datasets, Plant Pathology 2020 FGVC-7 and Plant Pathology 2021 FGVC-8, resizing the images to 384 × 384 pixels. We used TensorFlow version 2.14.0 [37] as a framework and ran our algorithms on a machine equipped with a Tesla P100 GPU and 16 GB of memory. To increase the number of samples and address the problem of imbalanced data, we applied data augmentation techniques, including image rotation within a range of −20 to 20 degrees, random cropping, vertical flipping, horizontal flipping, and random shear. Additionally, we employed enhancement techniques to highlight disease features in the images by adjusting the brightness and contrast. This resulted in a total of 25,557 images for FGVC-7 and 130,424 images for FGVC-8. We selected RectifiedAdam as the optimizer and used a batch size of 64 to train our models. We opted for Stochastic Gradient Descent (SGDR) with warm-up restarts, rather than traditional learning rate (LR) annealing. With each restart, the LR was decreased in accordance with the cosine function [38]. The SGDR formula is presented as follows:
The LR was determined using the SGDR formula, which incorporates the LR ranges and , along with the number of epochs since the last restart () and the epoch for the upcoming restart (). In this experiment, we employed an of 0.0007, an of 0, and a set to 10, triggering a new warm restart at every 10-epoch interval.
3.4. Evaluation Metrics
To assess the performance of the introduced architecture (CTPlantNet), we employed three different metrics commonly used in classification problems. The metrics are detailed as follows:
- Accuracy (ACC): describes the number of correct predictions over all predictions considering the true/false negative/positive predictions (TP, TN, FP, FN).
- Precision (PRE): quantifies the ratio of accurately classified positive instances among all instances classified as positive.
- F1-score (F1): quantifies the harmonic mean of precision and recall, providing a balance between the two metrics.
- Area the under curve (AUC): widely used in classification problems. This metric measures the performance of the proposed pipeline by highlighting the difference between its good and bad predictions. It is computed by measuring the area under the Receiver Operating Characteristic (ROC) curve [39].
4. Experimental Results
After pre-processing the images from the Plant Pathology 2020-FGVC-7 dataset and applying the data augmentation techniques, different DL models were trained and tested using a cross-validation method. The most promising models were then implemented in our architecture to perform an accurate classification of apple foliar diseases. The performance of the introduced architecture (CTPlantNet) was compared against that of every single model within the architecture, trained and tested separately. The obtained results were also compared against those obtained by Sultistowati et al. [15], Bansal et al. [18], Alsayed et al. [17], Yadav et al. [16], and Subetha et al. [19]. The analysis of the results presented in Table 1 indicates that our architecture demonstrated high performance, achieving an average AUC of 99.82% and an average ACC of 98.20%, outperforming previous state-of-the-art models.

Table 1.
Comparative overview of CTPlantNet’s results against separate models and state-of-the-art approaches using the Plant Pathology 2020-FGVC-7 dataset.
To further evaluate the potential of CTPlantNet, we tested it on a larger and newer dataset, Plant Pathology 2021-FGVC-8. After training the model on this dataset, we achieved impressive results that outperformed Yadav et al. [16], who also examined the potential of their proposed model (AFD-Net) on the same dataset. Moreover, our architecture also outperformed an ensemble learning pipeline proposed by Kejriwal et al. [20] to classify apple foliar disease images from Plant Pathology 2021-FGVC-8. Table 2 provides an overview of the results obtained by CTPlantNet, along with the results obtained by state-of-the-art works. The comparison indicates that our architecture achieved significantly better results than existing models, further highlighting the effectiveness of our approach in detecting apple foliar diseases.

Table 2.
Comparative overview of the results achieved by our proposed approach versus state-of-the-art methods using the Plant Pathology 2021-FGVC-8 dataset.
CTPlantNet achieved a higher accuracy (ACC) of 95.60%, surpassing the model proposed by Yadav et al. [16] by 3.36%, and showed a slight improvement of 0.12% in terms of precision and a notable improvement of 1.32% in terms of the F1-score compared to the model proposed by Kejriwal et al. [20].
5. Discussion
Our architecture, CTPlantNet, has demonstrated high potential in extracting a wide variety of features, which is crucial for discerning subtle distinctions between various diseases. This is particularly significant, as traditional visual inspections often struggle to accurately identify diseases, especially in cases where symptoms are similar or co-occurring. Moreover, the application of data augmentation techniques proved highly effective in addressing the issue of imbalanced data, a common challenge in plant disease datasets, which improved the overall performance of the proposed architecture. Combining and averaging the predictions of the models in our architecture resulted in a further boost in performance across all classes in the dataset, showcasing the robustness and reliability of our approach. Notably, our architecture offers a solution to the limitations of traditional visual inspections, which can be time consuming, labor intensive, and prone to human error. By leveraging deep learning, we can provide farmers with a more accurate, efficient, and scalable tool for disease diagnosis, enabling them to take prompt action and reduce crop losses. This is especially important for small-scale farmers, who often lack the resources and expertise to implement complex disease management strategies. Our architecture can help bridge this gap, providing an accessible and user-friendly tool for disease diagnosis and management. However, the prediction accuracy for the “multiple disease” class was less reliable, potentially attributed to the limited number of samples available in this category, highlighting the need for further data collection and research in this area.
In future studies, we will aim to evaluate the feasibility and effectiveness of our architecture in analyzing apple canopy images, which will enable us to assess its potential for real-world applications. We also will aim to enhance the precision of CTPlantNet and evaluate its performance on diverse datasets encompassing various plant species. Additionally, we will explore methods to enhance its interpretability, as it is crucial to understand the features and characteristics of the data that the models incorporated in CTPlantNet are particularly good at extracting. This will involve implementing techniques such as feature importance analysis, saliency mapping, and visualizations to provide insights into the decision-making process of our models. By highlighting the strengths and weaknesses of our architecture, we can further refine and improve its performance, ultimately leading to more accurate and reliable disease diagnosis and management tools for farmers and agricultural practitioners.
6. Conclusions
Timely and accurate diagnosis of plant diseases can help farmers make informed decisions about crop management, such as choosing the right fungicides or pesticides, selecting resistant cultivars, and adjusting irrigation and fertilization schedules. In this work, we introduced a hybrid deep learning (DL) architecture consisting of two blocks: a convolutional neural network (CNN) block composed of two CNN models (SEResNext-50 and EfficientNet-V2S) and a vision transformer block based on the Swin-Large model. The models were trained separately on the Plant Pathology 2020-FGVC-7 dataset, which comprises 3651 images classified into four classes (healthy, scab, rust, and multiple diseases). A 5-fold cross-validation strategy was employed for the training, which helped the models learn a wide range of patterns from the images. The predictions of the three models were combined using an averaging technique to provide the final output. Data augmentation and preprocessing techniques were utilized to increase the size of the dataset, overcome the imbalanced data issue, and enhance the performance of the models. CTPlantNet, our proposed architecture, outperformed state-of-the-art models and recently published works on the same dataset, achieving a remarkable accuracy (ACC) of 98.28% and an area under the curve (AUC) of 99.82%. To examine the potential of CTPlantNet, we further trained and validated the models on a newer version of the dataset named Plant Pathology 2021-FGVC-8, which includes 18,632 images divided into six classes (healthy, scab, rust, powdery mildew, frog eye spot, and multiple diseases). Our architecture demonstrated impressive performance, surpassing the state-of-the-art works and exhibiting excellent generalization capacity on a larger dataset with a greater variety of diseases. CTPlantNet achieved an average ACC of 95.96% and an average AUC of 99.17%, outperforming state-of-the-art works.
Author Contributions
Conceptualization, A.A.N. and M.A.A.; methodology, A.A.N. and M.A.A.; validation, A.A.N. and M.A.A.; formal analysis, A.A.N. and M.A.A.; writing—original draft preparation, A.A.N.; writing—review and editing, M.A.A.; funding acquisition, M.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was enabled in part by support provided by the Natural Sciences and Engineering Research Council of Canada (NSERC), funding reference number RGPIN-2018-06233.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data used in this work are available publicly; see Section 3.1 for details.
Acknowledgments
This research was enabled in part by support provided by ACENET (www.ace-net.ca (Accessed on 7 March 2024)) and the Digital Research Alliance of Canada (www.alliancecan.ca (Accessed on 7 March 2024)).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ML | machine learning |
| DL | deep learning |
| CNN | convolutional neural network |
| FAO | Food and Agriculture Organization |
| GPU | Graphical Processing Unit |
| SVM | Support Vector Machine |
| KNN | K-Nearest Neighbors |
| LR | Logistic Regression |
| RF | Random Forest |
| NB | Naïve Bayes |
| SMOTE | Synthetic Minority Over-Sampling |
| CIDA | Cornell Initiative for Digital Agriculture |
| SE | Squeeze-and-Excitation |
| GAP | global average pooling |
| FCL | fully connected layer |
| DP | Dropout |
| LRU | LeakyReLU |
| GD | Gaussian Dropout |
| SGD | Stochastic Gradient Descent |
| LR | learning rate |
| ACC | accuracy |
| PRE | precision |
| AUC | area under the curve |
| TP | true positive |
| TN | true negative |
| FP | false positive |
| FN | false negative |
| NSERC | Natural Sciences and Engineering Research Council of Canada |
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/##data/QCL (accessed on 7 March 2023).
- Thapa, R.; Zhang, K.; Snavely, N.; Belongie, S.; Khan, A. The Plant Pathology Challenge 2020 data set to classify foliar disease of apples. Appl. Plant Sci. 2020, 8, e11390. [Google Scholar] [CrossRef] [PubMed]
- Azgomi, H.; Haredasht, F.R.; Safari Motlagh, M.R. Diagnosis of some apple fruit diseases by using image processing and artificial neural network. Food Control 2023, 145, 109484. [Google Scholar] [CrossRef]
- Harvey, C.A.; Rakotobe, Z.L.; Rao, N.S.; Dave, R.; Razafimahatratra, H.; Rabarijohn, R.H.; Rajaofara, H.; MacKinnon, J.L. Extreme vulnerability of smallholder farmers to agricultural risks and climate change in Madagascar. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130089. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, K.; Perera, W.; Rupasinghe, T. Deep learning based classification of apple cedar rust using convolutional neural network. In Proceedings of the 4th International Conference on Computer Science and Information Technology, Samsun, Turkey, 11–15 September 2019; pp. 1–6. [Google Scholar]
- Agriculture Victoria (Victoria State Government). Pome Fruits. Available online: https://agriculture.vic.gov.au/biosecurity/plant-diseases/fruit-and-nut-diseases/pome-fruits (accessed on 7 March 2023).
- Legendre, R.; Basinger, N.T.; van Iersel, M.W. Low-Cost Chlorophyll Fluorescence Imaging for Stress Detection. Sensors 2021, 21, 2055. [Google Scholar] [CrossRef] [PubMed]
- Gudkov, S.V.; Matveeva, T.A.; Sarimov, R.M.; Simakin, A.V.; Stepanova, E.V.; Moskovskiy, M.N.; Dorokhov, A.S.; Izmailov, A.Y. Optical Methods for the Detection of Plant Pathogens and Diseases (Review). AgriEngineering 2023, 5, 1789–1812. [Google Scholar] [CrossRef]
- Pham, T.C.; Nguyen, V.D.; Le, C.H.; Packianather, M.; Hoang, V.D. Artificial intelligence-based solutions for coffee leaf disease classification. IOP Conf. Ser. Earth Environ. Sci. 2023, 1278, 012004. [Google Scholar] [CrossRef]
- Kirola, M.; Singh, N.; Joshi, K.; Chaudhary, S.; Gupta, A. Plants Diseases Prediction Framework: A Image-Based System Using Deep Learning. In Proceedings of the 2022 IEEE World Conference on Applied Intelligence and Computing (AIC), Sonbhadra, India, 17–19 June 2022; pp. 307–313. [Google Scholar]
- Ahmed, I.; Yadav, D.P.K. A systematic analysis of machine learning and deep learning based approaches for identifying and diagnosing plant diseases. Sustain. Oper. Comput. 2023, 4, 96–104. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic Study of Automated Diagnosis of Viral Plant Diseases Using Convolutional Neural Networks. In Proceedings of the Advances in Visual Computing, Las Vegas, NV, USA, 14–16 December 2015; pp. 638–645. [Google Scholar]
- Ma, J.; Du, K.; Zheng, F.; Zhang, L.; Gong, Z.; Sun, Z. A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network. Comput. Electron. Agric. 2018, 154, 18–24. [Google Scholar] [CrossRef]
- Chen, H.C.; Widodo, A.M.; Wisnujati, A.; Rahaman, M.; Lin, J.C.W.; Chen, L.; Weng, C.E. AlexNet Convolutional Neural Network for Disease Detection and Classification of Tomato Leaf. Electronics 2022, 11, 951. [Google Scholar] [CrossRef]
- Sulistyowati, T.; Purwanto, P.; Alzami, F.; Pramunendar, R.A. VGG16 Deep Learning Architecture Using Imbalance Data Methods for The Detection of Apple Leaf Diseases. Monet. J. Keuang. Dan Perbank. 2023, 11, 41–53. [Google Scholar] [CrossRef]
- Yadav, A.; Thakur, U.; Saxena, R.; Pal, V.; Bhateja, V.; Lin, J.C.W. AFD-Net: Apple Foliar Disease multi classification using deep learning on plant pathology dataset. Plant Soil 2022, 477, 595–611. [Google Scholar] [CrossRef]
- Alsayed, A.; Alsabei, A.; Arif, M. Classification of apple tree leaves diseases using deep learning methods. Int. J. Comput. Sci. Netw. Secur. 2021, 21, 324–330. [Google Scholar]
- Bansal, P.; Kumar, R.; Kumar, S. Disease detection in apple leaves using deep convolutional neural network. Agriculture 2021, 11, 617. [Google Scholar] [CrossRef]
- Subetha, T.; Khilar, R.; Christo, M.S. A comparative analysis on plant pathology classification using deep learning architecture—Resnet and VGG19. Mater. Today Proc. 2021; in press. [Google Scholar]
- Kejriwal, S.; Patadia, D.; Sawant, V. Apple Leaves Diseases Detection Using Deep Convolutional Neural Networks and Transfer Learning. In Computer Vision and Machine Learning in Agriculture; Uddin, M.S., Bansal, J.C., Eds.; Springer: Singapore, 2022; Volume 2, pp. 207–227. [Google Scholar]
- Thapa, R.; Zhang, K.; Snavely, N.; Belongie, S.; Khan, A. Plant Pathology 2020-FGVC7. Available online: https://www.kaggle.com/competitions/plant-pathology-2020-fgvc7/ (accessed on 7 March 2023).
- Fruit Pathology, S.D. Plant Pathology 2021-FGVC8. 2021. Available online: https://www.kaggle.com/competitions/plant-pathology-2021-fgvc8 (accessed on 27 March 2024).
- Nasser, A.A.; Akhloufi, M.A. CTPlantNet: A Hybrid CNN-Transformer Architecture for Plant Disease Classification. In Proceedings of the 2022 International Conference on Microelectronics (ICM), Casablanca, Morocco, 4–7 December 2022; pp. 156–159. [Google Scholar] [CrossRef]
- Ghali, R.; Akhloufi, M.A.; Mseddi, W.S. Deep learning and transformer approaches for UAV-based wildfire detection and segmentation. Sensors 2022, 22, 1977. [Google Scholar] [CrossRef] [PubMed]
- Nasser, A.A.; Akhloufi, M. Chest Diseases Classification Using CXR and Deep Ensemble Learning. In Proceedings of the 19th International Conference on Content-based Multimedia Indexing, Graz, Austria, 14–16 September 2022; pp. 116–120. [Google Scholar]
- Nasser, A.A.; Akhloufi, M. Classification of CXR Chest Diseases by Ensembling Deep Learning Models. In Proceedings of the 23rd International Conference on Information Reuse and Integration for Data Science (IRI), San Diego, CA, USA, 9–11 August 2022; pp. 250–255. [Google Scholar]
- Lee, D.I.; Lee, J.H.; Jang, S.H.; Oh, S.J.; Doo, I.C. Crop Disease Diagnosis with Deep Learning-Based Image Captioning and Object Detection. Appl. Sci. 2023, 13, 3148. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Tan, M.; Le, Q. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Volume 139, pp. 10096–10106. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lu Lu, Y.S.; Yanhui Su, G.E.K. Dying ReLU and Initialization: Theory and Numerical Examples. arXiv 2019, arXiv:1903.06733. [Google Scholar] [CrossRef]
- Wang, S.; Manning, C. Fast dropout training. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; Volume 28, pp. 118–126. [Google Scholar]
- Seetala, K.; Birdsong, W.; Reddy, Y.B. Image classification using tensorflow. In Proceedings of the 16th International Conference on Information Technology-New Generations (ITNG 2019), Las Vegas, NV, USA, 1–3 April 2019; pp. 485–488. [Google Scholar]
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
- Hoo, Z.H.; Candlish, J.; Teare, D. What is an ROC Curve? Emerg. Med. J. 2017, 34, 357–359. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).