Studies of Tumor Suppressor Genes via Chromosome Engineering
Abstract
1. Introduction
2. Construction of Mouse A9 Clones Containing a Single Human Chromosome
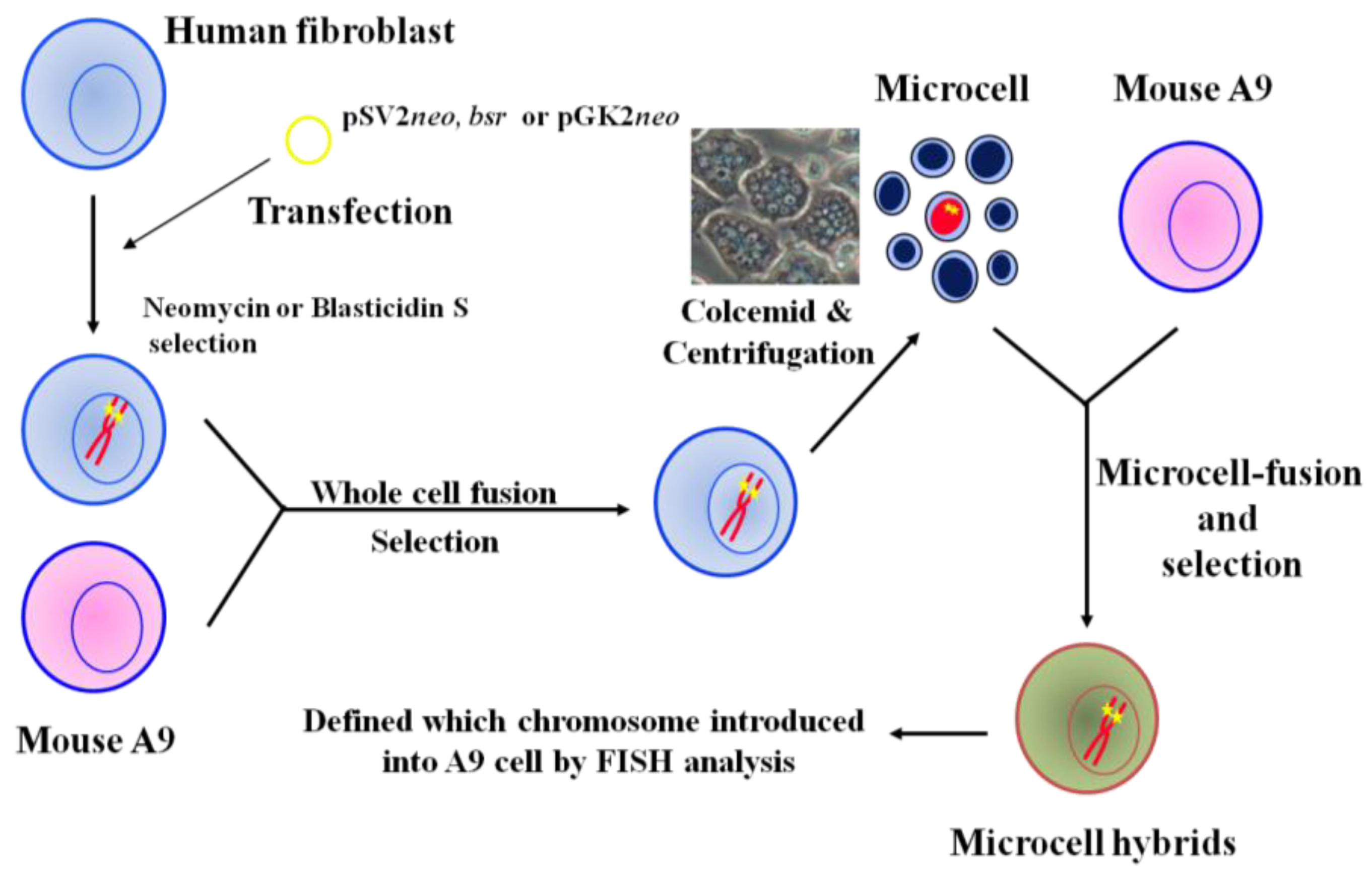
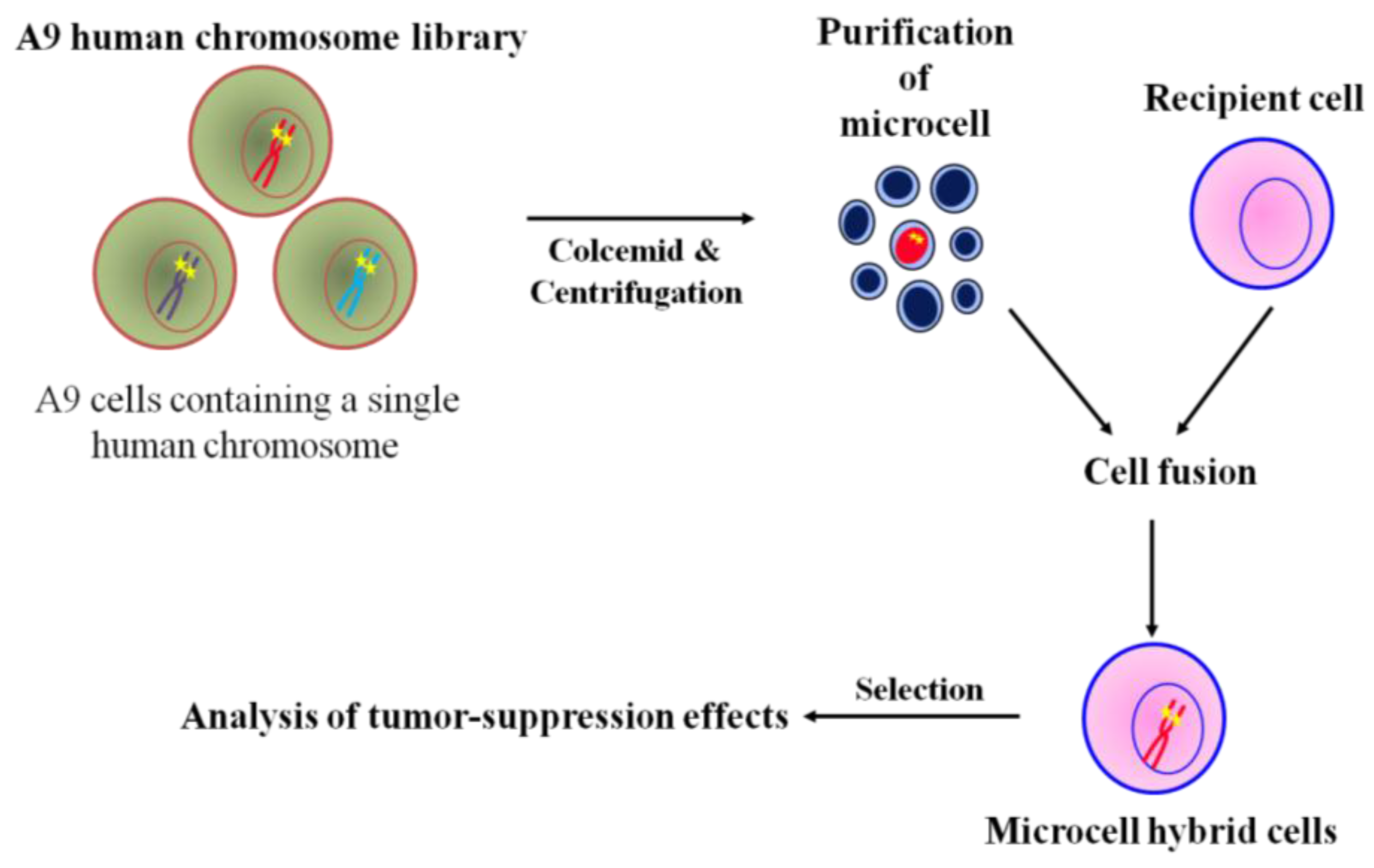
3. The Use of MMCT to Identify Human Chromosomes that Carry Tumor Suppressor Genes
| Transferred Chromosome | Cell Lines | Type | Mapping Region | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||||
| 1 | HT1080 (fibrosarcoma) | + | + | [30] | ||||
| TE85(osteosarcoma) | + | [44] | ||||||
| 143 B (TK-Ki-Ras-transformed TE85) | + | [44] | ||||||
| CMV-Mj-HEL-1 (CMV-transfermed lung fibroblasts) | + | [44] | ||||||
| 10W-2 (immortal Syrian hamster fibroblasts) | + | [51] | ||||||
| Isikawa (uterine endomerial carcinoma) | + | * | ||||||
| HHUA (uterine endometrial carcinoma) | + | + | [45] | |||||
| 2 | SiHa (cervical cancer) | + | 2q37 | [31,32] | ||||
| 3 | RCC23 (renal cell carcinoma) | + | + | 3p21.3 | [40,46,54] | |||
| KC12 (renal cell carcinoma) | + | + | 3p14.2-p21.1 | [33] | ||||
| HSC3 (oral squamous cell carcinoma) | + | + | + | 3p21.2-p21.3 or 3p25 | [41,55] | |||
| TS1 (lung adenocarcinoma) | + | * | ||||||
| 4 | HeLa (cevical cancer) | + | [47] | |||||
| J82 (baladder cancer) | + | [47] | ||||||
| T98G (glioblastoma) | + | [47] | ||||||
| 5 | A2058 (melanoma) | + | + | [26,39] | ||||
| 6 | HALneo (immortal fibroblasts) | + | [48] | |||||
| LCS-AF.1-3 (immortal fibroblasts) | + | 6p23-p24 | [56] | |||||
| 39neo (immortal fibroblasts) | + | [48] | ||||||
| SV/HF (SV40-tranformed fibroblasts) | + | [48] | ||||||
| HPV-16 (HPV-immortalized keratinocyte) | + | [53] | ||||||
| 7 | KMST-6 (immortal fibroblasts) | + | [49] | |||||
| SUSM-1 (immortal fibroblasts) | + | [49] | ||||||
| CC1 (choriocarcinoma) | + | * | ||||||
| R-3327 (rat prostatic cancer cells) | + | 7q21-22, 7q31.2-32 | [57] | |||||
| MeT5A (mesothelial cells) | + | + | [52] | |||||
| 8 | R-3327 (rat prostatic cancer cells) | + | 8p21-q12 | [57] | ||||
| 10 | Li7HM (hepatocellular carcinoma) | + | + | 10p15.1 | [35] | |||
| R-3327 (rat prostatic cancer cells) | + | 10q11-22 | [57] | |||||
| 11 | HeLa (cevical cancer) | + | [58] | |||||
| G401 (wilm’s tumor) | + | + | [59] | |||||
| SiHa (cervical cancer) | + | [29] | ||||||
| A204 (rhadboyomyosarcoma) | + | [29] | ||||||
| HHUA (uterine endometrial carcinoma) | + | [29] | ||||||
| HT1080 (fibrosarcoma) | + | [30] | ||||||
| RD (rhabdomyosarcoma) | + | [50] | ||||||
| H15 (bladder cancer) | + | * | ||||||
| R-3327 (rat prostatic cancer cells) | + | 11p13-11.2 | [57] | |||||
| 13 | R-3327 (rat prostatic cancer cells) | + | [60] | |||||
| 17 | R-3327 (rat prostatic cancer cells) | + | 12p11-q13, 12q24-ter | [57] | ||||
| 18 | HHUA (uterine endometrial carcinoma) | + | + | [45] | ||||
| X | HocB (ovarian carcinoma) | + | * | |||||
| ELCO (breast carcinoma) | + | * | ||||||
| Type 1 | Type 2 | Type 3 | Type 4 | Reference | |
|---|---|---|---|---|---|
| Transferred Chromosome | 1, 2 | 5, 7, 9, 10, 11, 15, 16, 19, 20 or 22 | 5, 7, 9, 10, 11, 13, 14, 15, 16, 19, 20, 22 or X | 5 | [36,37,38] |
4. Mapping of Tumor Suppressors on Human Chromosomes
4.1. Deletion Mapping Using Revertant Clones
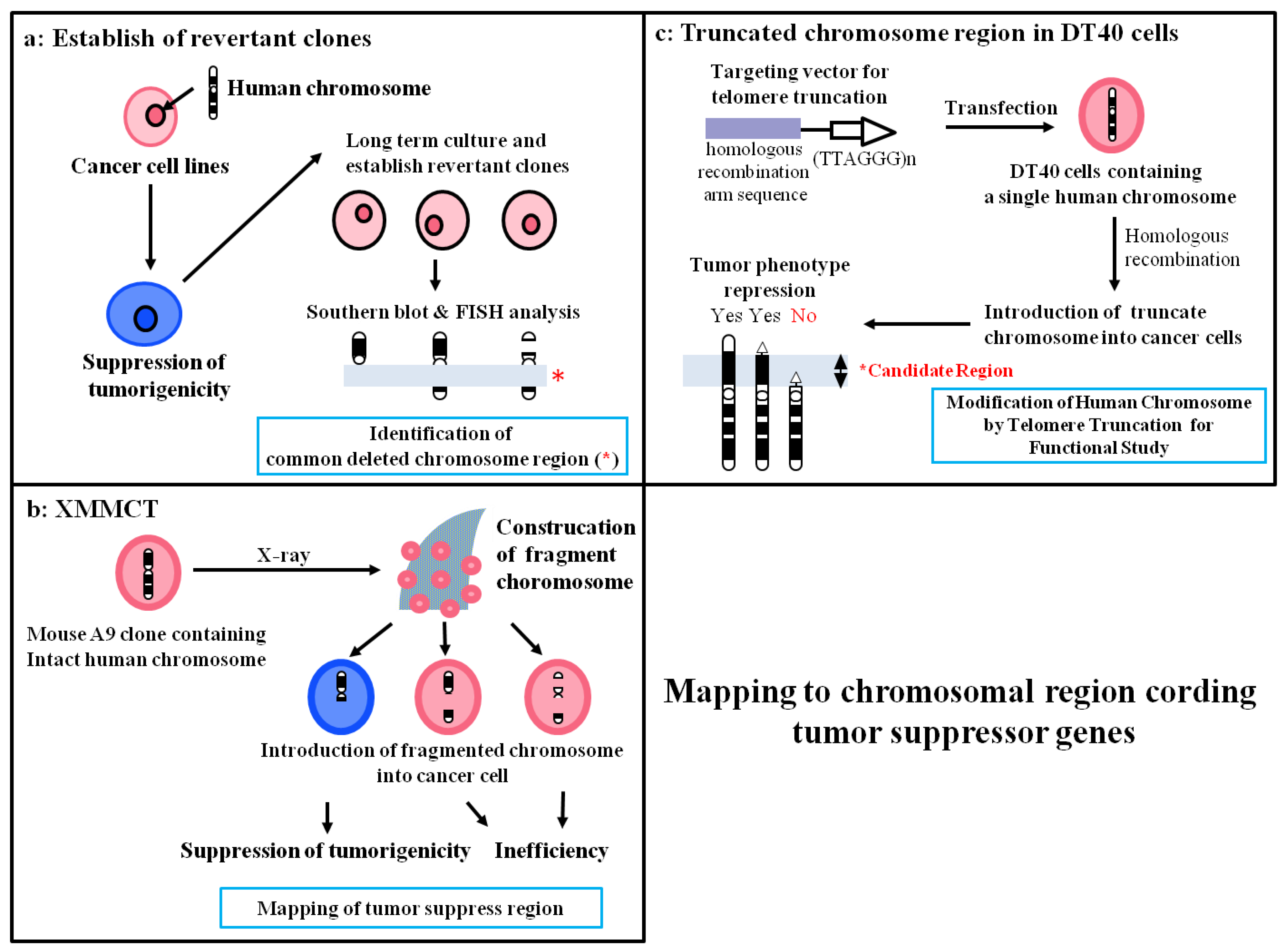
4.2. Construction of STFs by X-Irradiation Chromosome Transfer (XMMCT)
4.3. Construction of Truncated Chromosomes by Chromosome Engineering
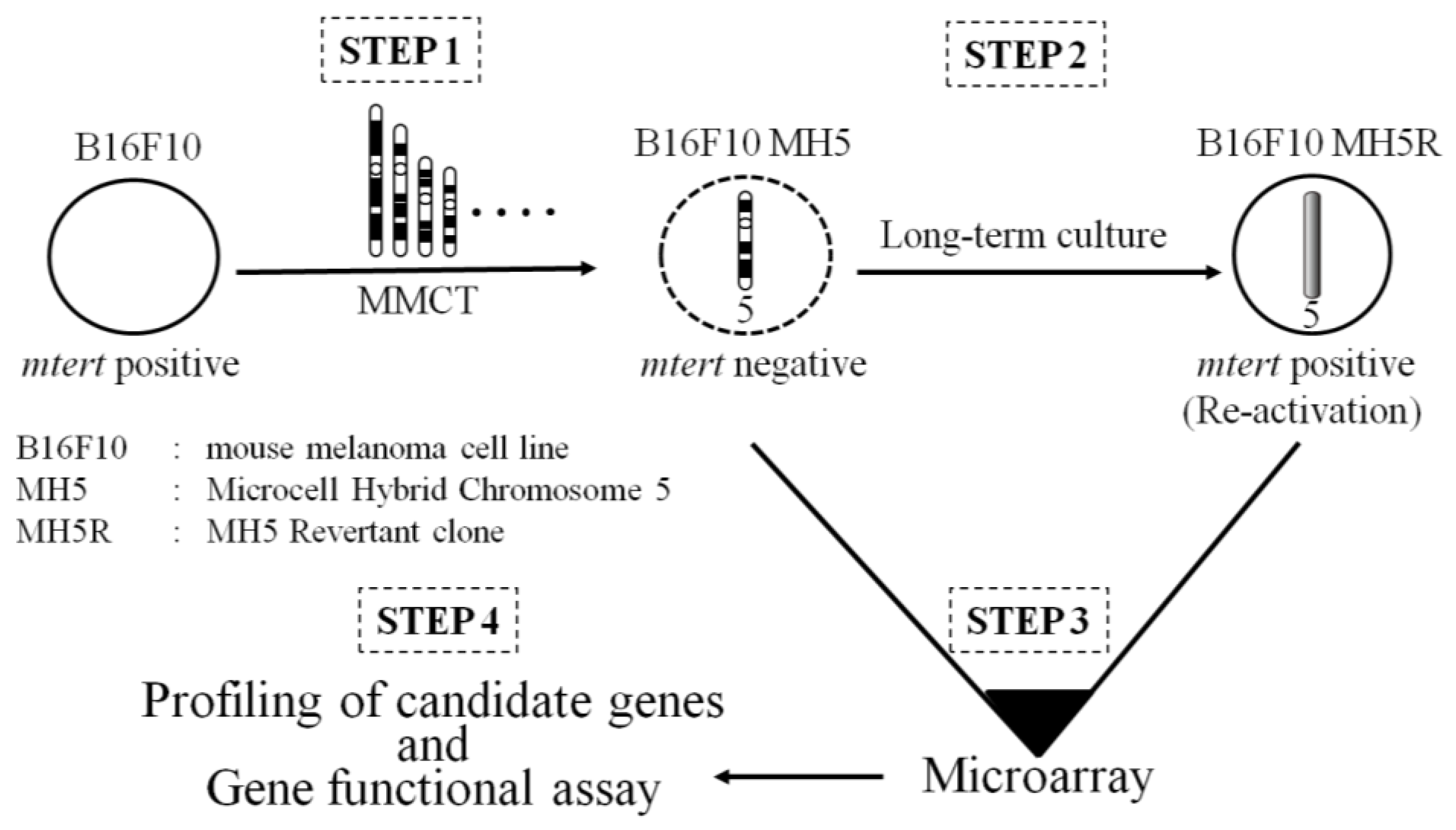
5. Identification of a Functional Suppressor Gene with a Combination of MMCT and Microarray
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Principles of tumor suppression. Cell 2004, 116, 235–246. [Google Scholar] [CrossRef]
- Krug, U.; Ganser, A.; Koeffler, H.P. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene 2002, 21, 3475–3495. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.C.; Weinberg, R.A. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer 2002, 2, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Sidebottom, E. The analysis of malignancy by cell fusion. In Vitro 1980, 16, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Bouck, N.; di Mayorca, G. Chemical carcinogens transform BHK cells by inducing a recessive mutation. Mol. Cell. Biol. 1982, 2, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Sidebottom, E.; Clark, S.R. Cell fusion segregates progressive growth from metastasis. Br. J. Cancer 1983, 47, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Smith, O.M.; Stein, G.H.; Robetorye, S.; Meyer-Demarest, S. Immortal phenotype of the HeLa variant D98 is recessive in hybrids formed with normal human fibroblasts. J. Cell Physiol. 1990, 143, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.L.; Moyer, M.P. Normal human colon cells suppress malignancy when fused with colon cancer cells. Vitr. Cell Dev. Biol. 1990, 26, 1095–1100. [Google Scholar] [CrossRef]
- Zajchowski, D.A.; Band, V.; Trask, D.K.; Kling, D.; Connolly, J.L.; Sager, R. Suppression of tumor-forming ability and related traits in MCF-7 human breast cancer cells by fusion with immortal mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1990, 87, 2314–2318. [Google Scholar] [CrossRef] [PubMed]
- Chuprin, A.; Gal, H.; Biron-Shental, T.; Biran, A.; Amiel, A.; Rozenblatt, S.; Krizhanovsky, V. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes. Dev. 2013, 27, 2356–2366. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, C.; Stenman, G.; Vojta, P.J.; Bongcam-Rudloff, E.; Barrett, J.C.; Westermark, B.; Paulsson, Y. Escape from senescence in hybrid cell clones involves deletions of two regions located on human chromosome 1q. Cancer Res. 1996, 56, 241–245. [Google Scholar] [PubMed]
- Oshimura, M.; Koi, M.; Ozawa, N.; Sugawara, O.; Lamb, P.W.; Barrett, J.C. Role of chromosome loss in ras/myc-induced Syrian hamster tumors. Cancer Res. 1988, 48, 1623–1632. [Google Scholar] [PubMed]
- Knudson, A.G. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.; Zhao, X.; Meyerson, M. Somatic alterations in the human cancer genome. Cancer Cell 2004, 6, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Balmain, A.; Gray, J.; Ponder, B. The genetics and genomics of cancer. Nat. Genet. 2003, 33, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Cavenee, W.K.; Dryja, T.P.; Phillips, R.A.; Benedict, W.F.; Godbout, R.; Gallie, B.L.; Murphree, A.L.; Strong, L.C.; White, R.L. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 1983, 305, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Takayama, H.; Hamaguchi, M.; Takano, H.; Yamaguchi, N.; Tsuda, H.; Hirohashi, S.; Vissing, H.; Shimizu, M.; Oshimura, M. Deletion mapping of chromosome 3p in human uterine cervical cancer. Oncogene 1993, 8, 1825–1832. [Google Scholar] [PubMed]
- Tomlinson, I.P.; Lambros, M.B.; Roylance, R.R. Loss of heterozygosity analysis: Practically and conceptually flawed? Genes Chromosomes Cancer 2002, 34, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Muzumdar, M.D.; Luo, L.; Zong, H. Modeling sporadic loss of heterozygosity in mice by using mosaic analysis with double markers (MADM). Proc. Natl. Acad. Sci. USA 2007, 104, 4495–4500. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.H.; Perez-Losada, J.; Wu, D.; Delrosario, R.; Tsunematsu, R.; Nakayama, K.I.; Brown, K.; Bryson, S.; Balmain, A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 2004, 432, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Sibley, K.; Cuthbert-Heavens, D.; Knowles, M.A. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene 2001, 20, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Mazelin, L.; Bernet, A.; Bonod-Bidaud, C.; Pays, L.; Arnaud, S.; Gespach, C.; Bredesen, D.E.; Scoazec, J.Y.; Mehlen, P. Netrin-1 controls colorectal tumorigenesis by regulating apoptosis. Nature 2004, 431, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.C.; Bailey, D.; Burkhardt, T.; Gardina, P.; Turpaz, Y.; Cowell, J.K. Comprehensive analysis of loss of heterozygosity events in glioblastoma using the 100K SNP mapping arrays and comparison with copy number abnormalities defined by BAC array comparative genomic hybridization. Genes Chromosomes Cancer 2008, 47, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.L.; Ohhira, T.; Fujisaki, C.; Inoue, T.; Ohta, T.; Osaki, M.; Ohshiro, E.; Seko, T.; Aoki, S.; Oshimura, M.; et al. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol. Cell. Biol. 2011, 31, 1624–1636. [Google Scholar] [CrossRef] [PubMed]
- Oshimura, M.; Uno, N.; Kazuki, Y.; Katoh, M.; Inoue, T. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Res. 2015, 23, 111–133. [Google Scholar] [CrossRef] [PubMed]
- Oshimura, M.; Kugoh, H.M.; Shimizu, M.; Yamada, H.; Hashiba, H.; Horikawa, I.; Sasaki, M. Multiple chromosomes carrying tumor suppressor activity, via microcell-mediated chromosome transfer, for various tumor cell lines. Princess Takamatsu Symp. 1989, 20, 249–257. [Google Scholar] [PubMed]
- Oshimura, M.; Kugoh, H.; Koi, M.; Shimizu, M.; Yamada, H.; Satoh, H.; Barrett, J.C. Transfer of a normal human chromosome 11 suppresses tumorigenicity of some but not all tumor cell lines. J. Cell. Biochem. 1990, 42, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Kugoh, H.M.; Hashiba, H.; Shimizu, M.; Oshimura, M. Suggestive evidence for functionally distinct, tumor-suppressor genes on chromosomes 1 and 11 for a human fibrosarcoma cell line, HT1080. Oncogene 1990, 5, 1637–1644. [Google Scholar] [PubMed]
- Uejima, H.; Mitsuya, K.; Kugoh, H.; Horikawa, I.; Oshimura, M. Normal human chromosome 2 induces cellular senescence in the human cervical carcinoma cell line SiHa. Genes Chromosomes Cancer 1995, 14, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Uejima, H.; Shinohara, T.; Nakayama, Y.; Kugoh, H.; Oshimura, M. Mapping a novel cellular-senescence gene to human chromosome 2q37 by irradiation microcell-mediated chromosome transfer. Mol. Carcinog. 1998, 22, 34–45. [Google Scholar] [CrossRef]
- Tanaka, H.; Shimizu, M.; Horikawa, I.; Kugoh, H.; Yokota, J.; Barrett, J.C.; Oshimura, M. Evidence for a putative telomerase repressor gene in the 3p14.2-p21.1 region. Genes Chromosomes Cancer 1998, 23, 123–133. [Google Scholar] [CrossRef]
- Kugoh, H.; Fujiwara, M.; Kihara, K.; Fukui, I.; Horikawa, I.; Schulz, T.C.; Oshimura, M. Cellular senescence of a human bladder carcinoma cell line (JTC-32) induced by a normal chromosome 11. Cancer Genet. Cytogenet. 2000, 116, 158–163. [Google Scholar] [CrossRef]
- Nishimoto, A.; Miura, N.; Horikawa, I.; Kugoh, H.; Murakami, Y.; Hirohashi, S.; Kawasaki, H.; Gazdar, A.F.; Shay, J.W.; Barrett, J.C.; et al. Functional evidence for a telomerase repressor gene on human chromosome 10p15.1. Oncogene 2001, 20, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Kugoh, H.; Nakamoto, H.; Inoue, J.; Funaki, K.; Barrett, J.C.; Oshimura, M. Multiple human chromosomes carrying tumor-suppressor functions for the mouse melanoma cell line B16-F10, identified by microcell-mediated chromosome transfer. Mol. Carcinog. 2002, 35, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Kugoh, H.; Shigenami, K.; Funaki, K.; Barrett, J.C.; Oshimura, M. Human chromosome 5 carries a putative telomerase repressor gene. Genes Chromosomes Cancer 2003, 36, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Yawata, T.; Kamino, H.; Kugoh, H.; Katoh, M.; Nomura, N.; Oishi, M.; Horikawa, I.; Barrett, J.C.; Oshimura, M. Identification of a ≤ 600-kb region on human chromosome 1q42.3 inducing cellular senescence. Oncogene 2003, 22, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.L.; Ohhira, T.; Oshimura, M.; Kugoh, H. Human chromosome 5 carries a transcriptional regulator of human telomerase reverse transcriptase (hTERT). Biochem. Biophys. Res. Commun. 2010, 398, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Tanaka, H.; Notsu, T.; Horike, S.; Fujisaki, C.; Qi, D.L.; Ohhira, T.; Gilley, D.; Oshimura, M.; Kugoh, H. Localization of an hTERT repressor region on human chromosome 3p21.3 using chromosome engineering. Genome. Integr. 2010, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Nishio, S.; Ohira, T.; Sunamura, N.; Oshimura, M.; Ryoke, K.; Kugoh, H. Repression of hTERT transcription by the introduction of chromosome 3 into human oral squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2015, 466, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Koi, M.; Morita, H.; Shimizu, M.; Oshimura, M. Construction of mouse A9 clones containing a single human chromosome (X/autosome translocation) via micro-cell fusion. Jpn. J. Cancer Res. 1989, 80, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Koi, M.; Shimizu, M.; Morita, H.; Yamada, H.; Oshimura, M. Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn. J. Cancer Res. 1989, 80, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Hensler, P.J.; Annab, L.A.; Barrett, J.C.; Pereira-Smith, O.M. A gene involved in control of human cellular senescence on human chromosome 1q. Mol. Cell. Biol. 1994, 14, 2291–2297. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Honda, T.; Yamada, H.; Wake, N.; Barrett, J.C.; Oshimura, M. Evidence for multiple pathways to cellular senescence. Cancer Res. 1994, 54, 6090–6093. [Google Scholar] [PubMed]
- Ohmura, H.; Tahara, H.; Suzuki, M.; Ide, T.; Shimizu, M.; Yoshida, M.A.; Tahara, E.; Shay, J.W.; Barrett, J.C.; Oshimura, M. Restoration of the cellular senescence program and repression of telomerase by human chromosome 3. Jpn. J. Cancer Res. 1995, 86, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Weber, J.L.; Killary, A.M.; Ledbetter, D.H.; Smith, J.R.; Pereira-Smith, O.M. Genetic analysis of indefinite division in human cells: Evidence for a cell senescence-related gene(s) on human chromosome 4. Proc. Natl. Acad. Sci. USA 1991, 88, 5635–5639. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, A.K.; Hubbard, K.; Kaur, G.P.; Jha, K.K.; Ozer, H.L.; Athwal, R.S. Senescence of immortal human fibroblasts by the introduction of normal human chromosome 6. Proc. Natl. Acad. Sci. USA 1994, 91, 5498–5502. [Google Scholar] [CrossRef] [PubMed]
- Ogata, T.; Ayusawa, D.; Namba, M.; Takahashi, E.; Oshimura, M.; Oishi, M. Chromosome 7 suppresses indefinite division of nontumorigenic immortalized human fibroblast cell lines KMST-6 and SUSM-1. Mol. Cell. Biol. 1993, 13, 6036–6043. [Google Scholar] [CrossRef] [PubMed]
- Koi, M.; Johnson, L.A.; Kalikin, L.M.; Little, P.F.; Nakamura, Y.; Feinberg, A.P. Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science 1993, 260, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, O.; Oshimura, M.; Koi, M.; Annab, L.A.; Barrett, J.C. Induction of cellular senescence in immortalized cells by human chromosome 1. Science 1990, 247, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, K.; Ogino, H.; Michishita, E.; Satoh, N.; Ayusawa, D. Introduction of chromosome 7 suppresses telomerase with shortening of telomeres in a human mesothelial cell line. Exp. Cell Res. 1999, 252, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, R.D.; Kramer, D.; Meijer, C.J.; Walboomers, J.M.; Trott, D.A.; Cuthbert, A.P.; Newbold, R.F.; Overkamp, W.J.; Zdzienicka, M.Z.; Snijders, P.J. Telomerase suppression by chromosome 6 in a human papillomavirus type 16-immortalized keratinocyte cell line and in a cervical cancer cell line. J. Natl. Cancer Inst. 2001, 93, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, I.; Oshimura, M.; Barrett, J.C. Repression of the telomerase catalytic subunit by a gene on human chromosome 3 that induces cellular senescence. Mol. Carcinog. 1998, 22, 65–72. [Google Scholar] [CrossRef]
- Uzawa, N.; Yoshida, M.A.; Hosoe, S.; Oshimura, M.; Amagasa, T.; Ikeuchi, T. Functional evidence for involvement of multiple putative tumor suppressor genes on the short arm of chromosome 3 in human oral squamous cell carcinogenesis. Cancer Genet. Cytogenet. 1998, 107, 125–131. [Google Scholar] [CrossRef]
- Kumata, M.; Shimizu, M.; Oshimura, M.; Uchida, M.; Tsutsui, T. Induction of cellular senescence in a telomerase negative human immortal fibroblast cell line, LCS-AF.1–3, by human chromosome 6. Int. J. Oncol. 2002, 21, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Hosoki, S.; Suzuki, H.; Akakura, K.; Igarashi, T.; Furuya, Y.; Oshimura, M.; Rinker-Schaeffer, C.W.; Nihei, N.; Barrett, J.C.; et al. Mapping of metastasis suppressor genes for prostate cancer by microcell-mediated chromosome transfer. Asian J. Androl. 2000, 2, 167–171. [Google Scholar] [PubMed]
- Saxon, P.J.; Srivatsan, E.S.; Stanbridge, E.J. Introduction of human chromosome 11 via microcell transfer controls tumorigenic expression of HeLa cells. EMBO. J. 1986, 5, 3461–3466. [Google Scholar] [PubMed]
- Weissman, B.E.; Saxon, P.J.; Pasquale, S.R.; Jones, G.R.; Geiser, A.G.; Stanbridge, E.J. Introduction of a normal human chromosome 11 into a Wilms’ tumor cell line controls its tumorigenic expression. Science 1987, 236, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, S.; Ota, S.; Ichikawa, Y.; Suzuki, H.; Ueda, T.; Naya, Y.; Akakura, K.; Igarashi, T.; Oshimura, M.; Nihei, N.; et al. Suppression of metastasis of rat prostate cancer by introduction of human chromosome 13. Asian J. Androl. 2002, 4, 131–136. [Google Scholar] [PubMed]
- De Bernardes Jesus, B.; Blasco, M.A. Telomerase at the intersection of cancer and aging. Trends Genet. 2013, 29, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 2005, 6, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.P.; Harley, C.B. Replicative senescence and cell immortality: The role of telomeres and telomerase. Proc. Soc. Exp. Biol. Med. 1997, 214, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, Y.; Shinohara, T.; Notsu, T.; Tomizuka, K.; Yoshida, H.; Takeda, S.; Oshimura, M.; Ishida, I. Efficient modification of a human chromosome by telomere-directed truncation in high homologous recombination-proficient chicken DT40 cells. Nucleic Acids Res. 1998, 26, 3447–3448. [Google Scholar] [CrossRef] [PubMed]
- Lamonerie, T.; Tremblay, J.J.; Lanctôt, C.; Therrien, M.; Gauthier, Y.; Drouin, J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996, 10, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Lanctôt, C.; Moreau, A.; Chamberland, M.; Tremblay, M.L.; Drouin, J. Hindlimb patterning and mandible development require the Ptx1 gene. Development 1999, 126, 1805–1810. [Google Scholar] [PubMed]
- Shang, J.; Luo, Y.; Clayton, D.A. Backfoot is a novel homeobox gene expressed in the mesenchyme of developing hind limb. Dev. Dyn. 1997, 209, 242–253. [Google Scholar] [CrossRef]
- Kolfschoten, I.G.; van Leeuwen, B.; Berns, K.; Mullenders, J.; Beijersbergen, R.L.; Bernards, R.; Voorhoeve, P.M.; Agami, R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 2005, 121, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Lobie, P.E. Transcriptional activation of p53 by Pitx1. Cell Death Differ. 2007, 14, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Knösel, T.; Ye, F.; Pacyna-Gengelbach, M.; Deutschmann, N.; Petersen, I. Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer 2007, 55, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Chen, H.; Xu, Y.; Zhang, X.; Luo, Y. Expression of pituitary homeobox 1 gene in human gastric carcinogenesis and its clinicopathological significance. World J. Gastroenterol. 2008, 14, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Lord, R.V.; Brabender, J.; Wickramasinghe, K.; DeMeester, S.R.; Holscher, A.; Schneider, P.M.; Danenberg, P.V.; DeMeester, T.R. Increased CDX2 and decreased PITX1 homeobox gene expression in Barrett’s esophagus and Barrett’s-associated adenocarcinoma. Surgery 2005, 138, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, M.; Osaki, M.; Kodani, I.; Okada, F.; Ryoke, K.; Oshimura, M.; Ito, H.; Kugoh, H. PITX1 is a reliable biomarker for predicting prognosis in patients with oral epithelial dysplasia. Oncol. Lett. 2014, 7, 750–754. [Google Scholar] [PubMed]
- Osaki, M.; Chinen, H.; Yoshida, Y.; Ohhira, T.; Sunamura, N.; Yamamoto, O.; Ito, H.; Oshimura, M.; Kugoh, H. Decreased PITX1 gene expression in human cutaneous malignant melanoma and its clinicopathological significance. Eur. J. Dermatol. 2013, 23, 344–349. [Google Scholar] [PubMed]
- Ohira, T.; Naohiro, S.; Nakayama, Y.; Osaki, M.; Okada, F.; Oshimura, M.; Kugoh, H. miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci. Rep. 2015, 5, 8201. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Kazuki, Y.; Kazuki, K.; Kajitani, N.; Takiguchi, M.; Nakayama, Y.; Nakamura, T.; Oshimura, M. Exploitation of the interaction of measles virus fusogenic envelope proteins with the surface receptor CD46 on human cells for microcell-mediated chromosome transfer. BMC. Biotechnol. 2015, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, M.; Ueda, K.; Uno, N.; Uno, K.; Fukuhara, S.; Kurosaki, H.; Takehara, S.; Osaki, M.; Kazuki, Y.; Kurosawa, Y.; et al. Retargeting of microcell fusion towards recipient cell-oriented transfer of human artificial chromosome. BMC. Biotechnol. 2015, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Fatima, R.; Akhade, V.S.; Pal, D.; Rao, S.M. Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 2015, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Valeri, N.; Braconi, C.; Gasparini, P.; Murgia, C.; Lampis, A.; Paulus-Hock, V.; Hart, J.R.; Ueno, L.; Grivennikov, S.I.; Lovat, F.; et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014, 25, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kugoh, H.; Ohira, T.; Oshimura, M. Studies of Tumor Suppressor Genes via Chromosome Engineering. Cancers 2016, 8, 4. https://doi.org/10.3390/cancers8010004
Kugoh H, Ohira T, Oshimura M. Studies of Tumor Suppressor Genes via Chromosome Engineering. Cancers. 2016; 8(1):4. https://doi.org/10.3390/cancers8010004
Chicago/Turabian StyleKugoh, Hiroyuki, Takahito Ohira, and Mitsuo Oshimura. 2016. "Studies of Tumor Suppressor Genes via Chromosome Engineering" Cancers 8, no. 1: 4. https://doi.org/10.3390/cancers8010004
APA StyleKugoh, H., Ohira, T., & Oshimura, M. (2016). Studies of Tumor Suppressor Genes via Chromosome Engineering. Cancers, 8(1), 4. https://doi.org/10.3390/cancers8010004





