Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues
Abstract
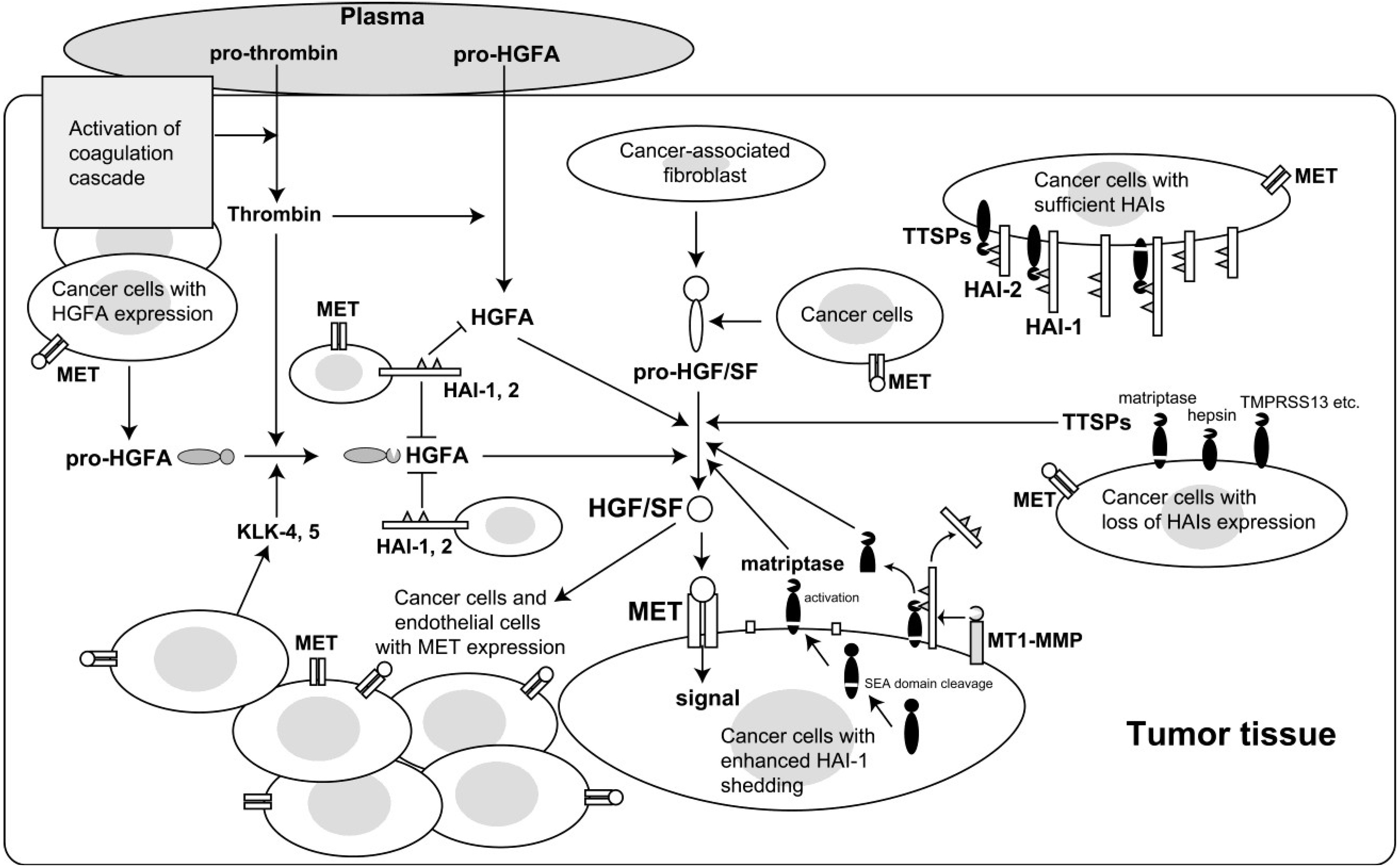
:1. Introduction
2. Enhanced Activation of HGF/SF in Cancer Tissues
3. Proteases Involved in Pro-HGF/SF Activation
| Proteases | Relative Activity * [Ref.] |
|---|---|
| Serum proteases | |
| HGFA | 1 |
| Factor XIIa | 0.018 [14]~0.02 [11] |
| Factor XIa | 0.011 [11]~0.015 [23] |
| tPA | <0.00001 [14] |
| Cellular proteases | |
| Matriptase | 2.07 [14] |
| Hepsin | 0.074 [14] |
| HAT | 0.024 [26] |
| TMPRSS13 | 0.012 [25] |
| uPA | <0.00001 [14] |
4. Roles for HGF/SF-Activating Proteases in Carcinogenesis and Malignant Progression
5. HAI Regulation of HGF/SF Activation
6. Reduced Cell Surface Expression of HAI-1 in Cancer Cells and Its Role in Cancer Progression
7. Epigenetic Silencing of HAI-2 in Cancer Cells and Its Role in Cancer Progression
8. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar]
- Matsumoto, K.; Nakamura, T. Hepatocyte growth factor and the Met system as a mediator of tumor-stromal interactions. Int. J. Cancer 2006, 119, 477–483. [Google Scholar]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET signalling: Principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar]
- Koeppen, H.; Rost, S.; Yauch, R.L. Developing biomarkers to predict benefit from HGF/MET pathway inhibitors. J. Pathol. 2014, 232, 210–218. [Google Scholar]
- Gak, E.; Taylor, W.G.; Chan, A.M.; Rubin, J.S. Processing of hepatocyte growth factor to the heterodimeric form is required for biological activity. FEBS Lett. 1992, 311, 17–21. [Google Scholar]
- Hartmann, G.; Naldini, L.; Weidner, K.M.; Sachs, M.; Vigna, E.; Comoglio, P.M.; Birchmeier, W. A functional domain in the heavy chain of scatter factor/hepatocyte growth factor binds the c-Met receptor and induces cell dissociation but not mitogenesis. Proc. Natl. Acad. Sci. USA 1992, 89, 11574–11578. [Google Scholar]
- Naka, D.; Ishii, T.; Yoshiyama, Y.; Miyazawa, K.; Hara, H.; Hishida, T.; Kidamura, N. Activation of hepatocyte growth factor by proteolytic conversion of a single chain form to a heterodimer. J. Biol. Chem. 1992, 267, 20114–20119. [Google Scholar]
- Naldini, L.; Tamagnone, L.; Vigna, E.; Sachs, M.; Hartmann, G.; Birchmeier, W.; Daikuhara, Y.; Tsubouchi, H.; Blasi, F.; Comoglio, P.M.; et al. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J. 1992, 11, 4825–4833. [Google Scholar]
- Kirchhofer, D.; Yao, X.; Peek, M.; Eigenbrot, C.; Lipari, M.T.; Billeci, K.L.; Maun, H.R.; Moran, P.; Santell, L.; Wiesmann, C.; et al. Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling. J. Biol. Chem. 2004, 279, 39915–39924. [Google Scholar]
- Lokker, N.A.; Mark, M.R.; Luis, E.A.; Bennett, G.L.; Robbins, K.A.; Baker, J.B.; Godowski, P.J. Structure-function analysis of hepatocyte growth factor: Identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992, 11, 2503–2510. [Google Scholar]
- Kataoka, H.; Miyata, S.; Uchinokura, S.; Itoh, H. Roles of hepatocyte growth factor (HGF) activator and HGF activator inhibitor in the pericellular activation of HGF/scatter factor. Cancer Metastasis Rev. 2003, 22, 223–236. [Google Scholar]
- Nakamura, T.; Sakai, K.; Nakamura, T.; Matsumoto, K. Hepatocyte growth factor twenty years on: Much more than a growth factor. J. Gastroenterol. Hepatol. 2011, 26, 188–202. [Google Scholar]
- Kataoka, H.; Kawaguchi, M. Hepatocyte growth factor activator (HGFA): Pathophysiological functions in vivo. FEBS J. 2010, 277, 2230–2237. [Google Scholar]
- Owen, K.A.; Qiu, D.; Alves, J.; Schumacher, A.M.; Kilpatrick, L.M.; Li, J.; Harris, J.L.; Ellis, V. Pericellular activation of hepatocyte growth factor by the transmembrane serine proteases matriptase and hepsin, but not by the membrane-associated protease uPA. Biochem. J. 2010, 426, 219–228. [Google Scholar]
- Nakamura, T.; Matsumoto, K.; Kiritoshi, A.; Tano, Y. Induction of hepatocyte growth factor in fibroblasts by tumor-derived factors affects invasive growth of tumor cells: In vitro analysis of tumor-stromal interactions. Cancer Res. 1997, 57, 3305–3313. [Google Scholar]
- Xie, Q.; Bradley, R.; Kang, L.; Koeman, J.; Ascierto, M.L.; Worschech, A.; de Giorgi, V.; Wang, E.; Kefene, L.; Su, Y.; et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc. Natl. Acad. Sci. USA 2012, 109, 570–575. [Google Scholar]
- Miyazawa, K.; Shimomura, T.; Naka, D.; Kitamura, N. Proteolytic activation of hepatocyte growth factor in response to tissue injury. J. Biol. Chem. 1994, 269, 8966–8970. [Google Scholar]
- Kataoka, H.; Hamasuna, R.; Itoh, H.; Kitamura, N.; Koono, M. Activation of hepatocyte growth factor/scatter factor in colorectal carcinoma. Cancer Res. 2000, 60, 6148–6159. [Google Scholar]
- Hoshiko, S.; Kawaguchi, M.; Fukushima, T.; Haruyama, Y.; Yorita, K.; Tanaka, H.; Seiki, M.; Inatsu, H.; Kitamura, K.; Kataoka, H.; et al. Hepatocyte growth factor activator inhibitor type 1 is a suppressor of intestinal tumorigenesis. Cancer Res. 2013, 73, 2659–2670. [Google Scholar]
- Olivero, M.; Rizzo, M.; Madeddu, R.; Casadio, C.; Pennacchietti, S.; Nicotra, M.; Prat, M.; Maggi, G.; Arena, N.; Natali, P.; et al. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br. J. Cancer 1996, 74, 1862–1868. [Google Scholar]
- Yamauchi, M.; Kataoka, H.; Itoh, H.; Seguchi, T.; Hasui, Y.; Osada, Y. Hepatocyte growth factor activator inhibitor types 1 and 2 are expressed by tubular epithelium in kidney and down-regulated in renal cell carcinoma. J. Urol. 2004, 171, 890–896. [Google Scholar]
- Kitajima, Y.; Ide, T.; Ohtsuka, T.; Miyazaki, K. Induction of hepatocyte growth factor activator gene expression under hypoxia activates the hepatocyte growth factor/c-Met system via hypoxia inducible factor-1 in pancreatic cancer. Cancer Sci. 2008, 99, 1341–1347. [Google Scholar]
- Peek, M.; Moran, P.; Mendoza, N.; Wickramasinghe, D.; Kirchhofer, D. Unusual proteolytic activation of pro-hepatocyte growth factor by plasma kallikrein and coagulation factor XIa. J. Biol. Chem. 2002, 277, 47804–47809. [Google Scholar]
- Kirchhofer, D.; Peek, M.; Lipari, M.T.; Billeci, K.; Fan, B.; Moran, P. Hepsin activates pro-hepatocyte growth factor and is inhibited by hepatocyte growth factor activator inhibitor-1B (HAI-1B) and HAI-2. FEBS Lett. 2005, 579, 1945–1950. [Google Scholar]
- Hashimoto, T.; Kato, M.; Shimomura, T.; Kitamura, N. TMPRSS13, a type II transmembrane serine protease, is inhibited by hepatocyte growth factor activator inhibitor type 1 and activates pro-hepatocyte growth factor. FEBS J. 2010, 277, 4888–4900. [Google Scholar]
- Kato, M.; Hashimoto, T.; Shimomura, T.; Kataoka, H.; Ohi, H.; Kitamura, N. Hepatocyte growth factor activator inhibitor type 1 inhibits protease activity and proteolytic activation of human airway trypsin-like protease. J. Biochem. 2012, 151, 179–187. [Google Scholar]
- Mars, W.M.; Zarnegar, R.; Michalopoulos, G.K. Activation of hepatocyte growth factor by the plasminogen activators uPA and tPA. Am. J. Pathol. 1993, 143, 949–958. [Google Scholar]
- Naldini, L.; Vigna, E.; Bardelli, A.; Follenzi, A.; Galimi, F.; Comoglio, P.M. Biological activation of pro-HGF (hepatocyte growth factor) by urokinase is controlled by a stoichiometric reaction. J. Biol. Chem. 1995, 270, 603–611. [Google Scholar]
- Kawaguchi, M.; Orikawa, H.; Baba, T.; Fukushima, T.; Kataoka, H. Hepatocyte growth factor activator is a serum activator of single-chain precursor macrophage-stimulating protein. FEBS J. 2009, 276, 3481–3490. [Google Scholar]
- Dvorak, H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar]
- Nierodzik, M.L.; Karpatkin, S. Thrombin induces tumor growth, metastasis, and angiogenesis: Evidence for a thrombin-regulated dormant tumor phenotype. Cancer Cell 2006, 10, 355–362. [Google Scholar]
- Mukai, S.; Fukushima, T.; Naka, D.; Tanaka, H.; Osada, Y.; Kataoka, H. Activation of hepatocyte growth factor activator zymogen (pro-HGFA) by human kallikrein 1-related peptidases. FEBS J. 2008, 275, 1003–1017. [Google Scholar]
- Moriyama, T.; Kataoka, H.; Tsubouchi, H.; Koono, M. Concomitant expression of hepatocyte growth factor (HGF), HGF activator and c-met genes in human glioma cells in vitro. FEBS Lett. 1995, 372, 78–82. [Google Scholar]
- Parr, C.; Jiang, W.G. Expression of hepatocyte growth factor/scatter factor, its activator, inhibitors and the c-Met receptor in human cancer cells. Int. J. Oncol. 2001, 19, 857–863. [Google Scholar]
- Tjin, E.P.M.; Groen, R.W.J.; Vogelzang, I.; Derksen, P.W.B.; Klok, M.D.; Meijer, H.P.; van Eeden, S.; Pals, S.T.; Spaargaren, M. Functional analysis of HGF/MET signaling and aberrant HGF-activator expression in diffuse large B-cell lymphoma. Blood 2006, 107, 760–768. [Google Scholar]
- Tjin, E.P.M.; Derksen, P.W.B.; Kataoka, H.; Spaargaren, M.; Pals, S.T. Multiple myeloma cells catalyze hepatocyte growth factor (HGF) activation by secreting the serine protease HGF-activator. Blood 2004, 104, 2172–2175. [Google Scholar]
- Wader, K.F.; Fagerli, U.M.; Holt, R.U.; Stordal, B.; Børset, M.; Sundan, A.; Waage, A. Elevated serum concentrations of activated hepatocyte growth factor activator in patients with multiple myeloma. Eur. J. Haematol. 2008, 81, 380–383. [Google Scholar]
- Nagakawa, O.; Yamagishi, T.; Fujiuchi, Y.; Junicho, A.; Akashi, T.; Nagaike, K.; Fuse, H. Serum hepatocyte growth factor activator (HGFA) in benign prostatic hyperplasia and prostate cancer. Eur. Urol. 2005, 48, 686–690. [Google Scholar]
- Parr, C.; Watkins, G.; Mansel, R.E.; Jiang, W.G. The hepatocyte growth factor regulatory factors in human breast cancer. Clin. Cancer Res. 2004, 10, 202–211. [Google Scholar]
- Lin, C.Y.; Wang, J.K.; Torri, J.; Dou, L.; Sang, Q.A.; Dickson, R.B. Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells. Monoclonal antibody production, isolation, and localization. J. Biol. Chem. 1997, 272, 9147–9152. [Google Scholar]
- Takeuchi, T.; Shuman, M.A.; Craik, C.S. Reverse biochemistry: Use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc. Natl. Acad. Sci. USA 1999, 96, 11054–11061. [Google Scholar]
- Oberst, M.D.; Singh, B.; Ozdemirli, M.; Dickson, R.B.; Johnson, M.D.; Lin, C.-Y. Characterizationof matriptase expression in normal human tissues. J. Histochem. Cytochem. 2003, 51, 1017–1025. [Google Scholar]
- List, K.; Szabo, R.; Molinolo, A.; Nielsen, B.S.; Bugge, T.H. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am. J. Pathol. 2006, 168, 1513–1525. [Google Scholar]
- List, K.; Bugge, T.H.; Szabo, R. Matriptase: Potent proteolysis on the cell surface. Mol. Med. 2006, 12, 1–7. [Google Scholar]
- Inouye, K.; Yasumoto, M.; Tsuzuki, S.; Mochida, S.; Fushiki, T. The optimal activity of a pseudozymogen form of recombinant matriptase under the mildly acidic pH and low ionic strength conditions. J. Biochem. 2010, 147, 485–492. [Google Scholar]
- Wang, J.-K.; Teng, I.-J.; Lo, T.-J.; Moore, S.; Yeo, Y.H.; Teng, Y.-C.; Kaul, M.; Chen, C.-C.; Zuo, A.H.; Chou, F.-P.; et al. Matriptase autoactivation is tightly regulated by the cellular chemical environments. PLoS One 2014, 9, e93899. [Google Scholar]
- Tannock, I.F.; Rotin, D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989, 49, 4373–4384. [Google Scholar]
- Antalis, T.M.; Buzza, M.S.; Hodge, K.M.; Hooper, J.D.; Netzel-Arnett, S. The cutting edge: Membrane-anchored serine protease activities in the pericellular microenvironment. Biochem. J. 2010, 428, 325–346. [Google Scholar]
- Oberst, M.D.; Johnson, M.D.; Dickson, R.B.; Lin, C.-Y.; Singh, B.; Stewart, M.; Williams, A.; al-Nafussi, A.; Smyth, J.F.; Gabra, H.; et al. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: Correlation with clinical outcome and tumor clinicopathological parameters. Clin. Cancer Res. 2002, 8, 1101–1107. [Google Scholar]
- Kang, J.Y.; Dolled-Filhart, M.; Ocal, I.T.; Singh, B.; Lin, C.-Y.; Dickson, R.B.; Rimm, D.L.; Camp, R.L. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003, 63, 1101–1105. [Google Scholar]
- Jarzab, B.; Wiench, M.; Fujarewicz, K.; Simek, K.; Jarzab, M.; Oczko-Wojciechowska, M.; Wloch, J.; Czarniecka, A.; Chmielik, E.; Lange, D.; et al. Gene expression profile of papillary thyroid cancer: Sources of variability and diagnostic implications. Cancer Res. 2005, 65, 1587–1597. [Google Scholar]
- Lee, J.-W.; Yong Song, S.; Choi, J.-J.; Lee, S.-J.; Kim, B.-G.; Park, C.-S.; Lee, J.-H.; Lin, C.-Y.; Dickson, R.B.; Bae, D.-S.; et al. Increased expression of matriptase is associated with histopathologic grades of cervical neoplasia. Hum. Pathol. 2005, 36, 626–633. [Google Scholar]
- Saleem, M.; Adhami, V.M.; Zhong, W.; Longley, B.J.; Lin, C.-Y.; Dickson, R.B.; Reagan-Shaw, S.; Jarrard, D.F.; Mukhtar, H. A novel biomarker for staging human prostate adenocarcinoma: Overexpression of matriptase with concomitant loss of its inhibitor, hepatocyte growth factor activator inhibitor-1. Cancer Epidemiol. Biomark. Prev. 2006, 15, 217–227. [Google Scholar]
- Nakamura, K.; Hongo, A.; Kodama, J.; Abarzua, F.; Nasu, Y.; Kumon, H.; Hiramatsu, Y. Expression of matriptase and clinical outcome of human endometrial cancer. Anticancer Res. 2009, 29, 1685–1690. [Google Scholar]
- Ha, S.Y.; Kim, K.Y.; Lee, N.K.; Kim, M.G.; Kim, S.-H. Overexpression of matriptase correlates with poor prognosis in esophageal squamous cell carcinoma. Virchows Arch. 2014, 464, 19–27. [Google Scholar]
- Cheng, M.-F.; Huang, M.-S.; Lin, C.-S.; Lin, L.-H.; Lee, H.-S.; Jiang, J.-C.; Hsia, K.-T. Expression of matriptase correlates with tumour progression and clinical prognosis in oral squamous cell carcinoma. Histopathology 2014, 65, 24–34. [Google Scholar]
- Chou, F.-P.; Chen, Y.-W.; Zhao, X.F.; Xu-Monette, Z.Y.; Young, K.H.; Gartenhaus, R.B.; Wang, J.-K.; Kataoka, H.; Zuo, A.H.; Barndt, R.J.; et al. Imbalanced matriptase pericellular proteolysis contributes to the pathogenesis of malignant B-cell lymphomas. Am. J. Pathol. 2013, 183, 1306–1317. [Google Scholar]
- List, K.; Szabo, R.; Molinolo, A.; Sriuranpong, V.; Redeye, V.; Murdock, T.; Burke, B.; Nielsen, B.S.; Gutkind, J.S.; Bugge, T.H.; et al. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005, 19, 1934–1950. [Google Scholar]
- Szabo, R.; Rasmussen, A.L.; Moyer, A.B.; Kosa, P.; Schafer, J.M.; Molinolo, A.A.; Gutkind, J.S.; Bugge, T.H. c-Met-induced epithelial carcinogenesis is initiated by the serine protease matriptase. Oncogene 2011, 30, 2003–2016. [Google Scholar]
- Kosa, P.; Szabo, R.; Molinolo, A.A.; Bugge, T.H. Suppression of Tumorigenicity-14, encoding matriptase, is a critical suppressor of colitis and colitis-associated colon carcinogenesis. Oncogene 2012, 31, 3679–3695. [Google Scholar]
- Zhang, Y.; Cai, X.; Schlegelberger, B.; Zheng, S. Assignment1 of human putative tumor suppressor genes ST13 (alias SNC6) and ST14 (alias SNC19) to human chromosome bands 22q13 and 11q24→q25 by in situ hybridization. Cytogenet. Cell Genet. 1998, 83, 56–57. [Google Scholar]
- Shimomura, T.; Denda, K.; Kitamura, A.; Kawaguchi, T.; Kito, M.; Kondo, J.; Kagaya, S.; Qin, L.; Takata, H.; Miyazawa, K.; et al. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J. Biol. Chem. 1997, 272, 6370–6376. [Google Scholar]
- Kawaguchi, T.; Qin, L.; Shimomura, T.; Kondo, J.; Matsumoto, K.; Denda, K.; Kitamura, N. Purification and cloning of hepatocyte growth factor activator inhibitor type 2, a Kunitz-type serine protease inhibitor. J. Biol. Chem. 1997, 272, 27558–27564. [Google Scholar]
- Marlor, C.W.; Delaria, K.A.; Davis, G.; Muller, D.K.; Greve, J.M.; Tamburini, P.P. Identification and cloning of human placental bikunin, a novel serine protease inhibitor containing two Kunitz domains. J. Biol. Chem. 1997, 272, 12202–12208. [Google Scholar]
- Denda, K.; Shimomura, T.; Kawaguchi, T.; Miyazawa, K.; Kitamura, N. Functional characterization of Kunitz domains in hepatocyte growth factor activator inhibitor type 1. J. Biol. Chem. 2002, 277, 14053–14059. [Google Scholar]
- Itoh, H.; Kataoka, H.; Hamasuna, R.; Kitamura, N.; Koono, M. Hepatocyte growth factor activator inhibitor type 2 lacking the first Kunitz-type serine proteinase inhibitor domain is a predominant product in mouse but not in human. Biochem. Biophys. Res. Commun. 1999, 255, 740–748. [Google Scholar]
- Qin, L.; Denda, K.; Shimomura, T.; Kawaguchi, T.; Kitamura, N. Functional characterization of Kunitz domains in hepatocyte growth factor activator inhibitor type 2. FEBS Lett. 1998, 436, 111–114. [Google Scholar]
- Kataoka, H.; Itoh, H.; Nuki, Y.; Hamasuna, R.; Naganuma, S.; Kitamura, N.; Shimomura, T. Mouse hepatocyte growth factor (HGF) activator inhibitor type 2 lacking the first Kunitz domain potently inhibits the HGF activator. Biochem. Biophys. Res. Commun. 2002, 290, 1096–1100. [Google Scholar]
- Kataoka, H.; Suganuma, T.; Shimomura, T.; Itoh, H.; Kitamura, N.; Nabeshima, K.; Koono, M. Distribution of hepatocyte growth factor activator inhibitor type 1 (HAI-1) in human tissues. Cellular surface localization of HAI-1 in simple columnar epithelium and its modulated expression in injured and regenerative tissues. J. Histochem. Cytochem. 1999, 47, 673–682. [Google Scholar]
- Szabo, R.; Hobson, J.P.; List, K.; Molinolo, A.; Lin, C.-Y.; Bugge, T.H. Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity. J. Biol. Chem. 2008, 283, 29495–29504. [Google Scholar]
- Zeng, L.; Cao, J.; Zhang, X. Expression of serine protease SNC19/matriptase and its inhibitor hepatocyte growth factor activator inhibitor type 1 in normal and malignant tissues of gastrointestinal tract. World J. Gastroenterol. 2005, 11, 6202–6207. [Google Scholar]
- Nakamura, K.; Abarzua, F.; Kodama, J.; Hongo, A.; Nasu, Y.; Kumon, H.; Hiramatsu, Y. Expression of hepatocyte growth factor activator inhibitors (HAI-1 and HAI-2) in ovarian cancer. Int. J. Oncol. 2009, 34, 345–353. [Google Scholar]
- Nakamura, K.; Hongo, A.; Kodama, J.; Hiramatsu, Y. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int. J. Cancer 2011, 128, 2613–2624. [Google Scholar]
- Vogel, L.K.; Saebø, M.; Skjelbred, C.F.; Abell, K.; Pedersen, E.D.K.; Vogel, U.; Kure, E.H. The ratio of Matriptase/HAI-1 mRNA is higher in colorectal cancer adenomas and carcinomas than corresponding tissue from control individuals. BMC Cancer 2006. [Google Scholar] [CrossRef]
- Ye, J.; Cheng, H.; Wang, Y.; Cao, J. Down-regulation of HAI-1 is associated with poor-differentiation status of colorectal cancer. Hum. Cell 2013, 26, 162–169. [Google Scholar]
- Baba, T.; Kawaguchi, M.; Fukushima, T.; Sato, Y.; Orikawa, H.; Yorita, K.; Tanaka, H.; Lin, C.-Y.; Sakoda, S.; Kataoka, H.; et al. Loss of membrane-bound serine protease inhibitor HAI-1 induces oral squamous cell carcinoma cells’ invasiveness. J. Pathol. 2012, 228, 181–192. [Google Scholar]
- Cheng, H.; Fukushima, T.; Takahashi, N.; Tanaka, H.; Kataoka, H. Hepatocyte growth factor activator inhibitor type 1 regulates epithelial to mesenchymal transition through membrane-bound serine proteinases. Cancer Res. 2009, 69, 1828–1835. [Google Scholar]
- Fukushima, T.; Kawaguchi, M.; Yamasaki, M.; Tanaka, H.; Yorita, K.; Kataoka, H. Hepatocyte growth factor activator inhibitor type 1 suppresses metastatic pulmonary colonization of pancreatic carcinoma cells. Cancer Sci. 2011, 102, 407–413. [Google Scholar]
- Ye, J.; Kawaguchi, M.; Haruyama, Y.; Kanemaru, A.; Fukushima, T.; Yamamoto, K.; Lin, C.-Y.; Kataoka, H. Loss of hepatocyte growth factor activator inhibitor type 1 participates in metastatic spreading of human pancreatic cancer cells in a mouse orthotopic transplantation model. Cancer Sci. 2014, 105, 44–51. [Google Scholar]
- Tomioka, D.; Maehara, N.; Kuba, K.; Mizumoto, K.; Tanaka, M.; Matsumoto, K.; Nakamura, T. Inhibition of growth, invasion, and metastasis of human pancreatic carcinoma cells by NK4 in an orthotopic mouse model. Cancer Res. 2001, 61, 7518–7524. [Google Scholar]
- Kohama, K.; Kawaguchi, M.; Fukushima, T.; Lin, C.-Y.; Kataoka, H. Regulation of pericellular proteolysis by hepatocyte growth factor activator inhibitor type 1 (HAI-1) in trophoblast cells. Hum. Cell 2012, 25, 100–110. [Google Scholar]
- Parr, C.; Jiang, W.G. Hepatocyte growth factor activation inhibitors (HAI-1 and HAI-2) regulate HGF-induced invasion of human breast cancer cells. Int. J. Cancer 2006, 119, 1176–1183. [Google Scholar]
- Nagaike, K.; Kohama, K.; Uchiyama, S.; Tanaka, H.; Chijiiwa, K.; Itoh, H.; Kataoka, H. Paradoxically enhanced immunoreactivity of hepatocyte growth factor activator inhibitor type 1 (HAI-1) in cancer cells at the invasion front. Cancer Sci. 2004, 95, 728–735. [Google Scholar]
- Komaki, W.; Fukushima, T.; Tanaka, H.; Itoh, H.; Chosa, E.; Kataoka, H. Expression of hepatocyte growth factor activator inhibitor type 1 on the epithelial cell surface is regulated by hypoxic and oxidative stresses. Virchows Arch. 2008, 453, 347–357. [Google Scholar]
- Kataoka, H.; Uchino, H.; Denda, K.; Kitamura, N.; Itoh, H.; Tsubouchi, H.; Nabeshima, K.; Koono, M. Evaluation of hepatocyte growth factor activator inhibitor expression in normal and malignant colonic mucosa. Cancer Lett. 1998, 128, 219–227. [Google Scholar]
- Domoto, T.; Takino, T.; Guo, L.; Sato, H. Cleavage of hepatocyte growth factor activator inhibitor-1 by membrane-type MMP-1 activates matriptase. Cancer Sci. 2012, 103, 448–454. [Google Scholar]
- Sato, H.; Takino, T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010, 101, 843–847. [Google Scholar]
- Hamasuna, R.; Kataoka, H.; Meng, J.Y.; Itoh, H.; Moriyama, T.; Wakisaka, S.; Koono, M. Reduced expression of hepatocyte growth factor activator inhibitor type-2/placental bikunin (HAI-2/PB) in human glioblastomas: Implication for anti-invasive role of HAI-2/PB in glioblastoma cells. Int. J. Cancer 2001, 93, 339–345. [Google Scholar]
- Kongkham, P.N.; Northcott, P.A.; Ra, Y.S.; Nakahara, Y.; Mainprize, T.G.; Croul, S.E.; Smith, C.A.; Taylor, M.D.; Rutka, J.T. An epigenetic genome-wide screen identifies SPINT2 as a novel tumor suppressor gene in pediatric medulloblastoma. Cancer Res. 2008, 68, 9945–9953. [Google Scholar]
- Betsunoh, H.; Mukai, S.; Akiyama, Y.; Fukushima, T.; Minamiguchi, N.; Hasui, Y.; Osada, Y.; Kataoka, H. Clinical relevance of hepsin and hepatocyte growth factor activator inhibitor type 2 expression in renal cell carcinoma. Cancer Sci. 2007, 98, 491–498. [Google Scholar]
- Morris, M.R.; Gentle, D.; Abdulrahman, M.; Maina, E.N.; Gupta, K.; Banks, R.E.; Wiesener, M.S.; Kishida, T.; Yao, M.; Teh, B.; et al. Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Res. 2005, 65, 4598–4606. [Google Scholar]
- Morris, M.R.; Gentle, D.; Abdulrahman, M.; Clarke, N.; Brown, M.; Kishida, T.; Yao, M.; Teh, B.T.; Latif, F.; Maher, E.R.; et al. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br. J. Cancer 2008, 98, 496–501. [Google Scholar]
- Fukai, K.; Yokosuka, O.; Chiba, T.; Hirasawa, Y.; Tada, M.; Imazeki, F.; Kataoka, H.; Saisho, H. Hepatocyte growth factor activator inhibitor 2/placental bikunin (HAI-2/PB) gene is frequently hypermethylated in human hepatocellular carcinoma. Cancer Res. 2003, 63, 8674–8679. [Google Scholar]
- Dong, W.; Chen, X.; Xie, J.; Sun, P.; Wu, Y. Epigenetic inactivation and tumor suppressor activity of HAI-2/SPINT2 in gastric cancer. Int. J. Cancer 2010, 127, 1526–1534. [Google Scholar]
- Yue, D.; Fan, Q.; Chen, X.; Li, F.; Wang, L.; Huang, L.; Dong, W.; Chen, X.; Zhang, Z.; Liu, J.; et al. Epigenetic inactivation of SPINT2 is associated with tumor suppressive function in esophageal squamous cell carcinoma. Exp. Cell Res. 2014, 322, 149–158. [Google Scholar]
- Bergum, C.; List, K. Loss of the matriptase inhibitor HAI-2 during prostate cancer progression. Prostate 2010, 70, 1422–1428. [Google Scholar]
- Hurst, N.J.; Najy, A.J.; Ustach, C.V.; Movilla, L.; Kim, H.-R.C. Platelet-derived growth factor-C (PDGF-C) activation by serine proteases: Implications for breast cancer progression. Biochem. J. 2012, 441, 909–918. [Google Scholar]
- Ustach, C.V.; Huang, W.; Conley-LaComb, M.K.; Lin, C.-Y.; Che, M.; Abrams, J.; Kim, H.-R.C. A novel signaling axis of matriptase/PDGF-D/β-PDGFR in human prostate cancer. Cancer Res. 2010, 70, 9631–9640. [Google Scholar]
- Li, W.; Wang, B.-E.; Moran, P.; Lipari, T.; Ganesan, R.; Corpuz, R.; Ludlam, M.J.C.; Gogineni, A.; Koeppen, H.; Bunting, S.; et al. Pegylated kunitz domain inhibitor suppresses hepsin-mediated invasive tumor growth and metastasis. Cancer Res. 2009, 69, 8395–8402. [Google Scholar]
- Galkin, A.V.; Mullen, L.; Fox, W.D.; Brown, J.; Duncan, D.; Moreno, O.; Madison, E.L.; Agus, D.B. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate 2004, 61, 228–235. [Google Scholar]
- Gray, K.; Elghadban, S.; Thongyoo, P.; Owen, K.A.; Szabo, R.; Bugge, T.H.; Tate, E.W.; Leatherbarrow, R.J.; Ellis, V. Potent and specific inhibition of the biological activity of the type-II transmembrane serine protease matriptase by the cyclic microprotein MCoTI-II. Thromb. Haemost. 2014, 112, 1–10. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, M.; Kataoka, H. Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues. Cancers 2014, 6, 1890-1904. https://doi.org/10.3390/cancers6041890
Kawaguchi M, Kataoka H. Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues. Cancers. 2014; 6(4):1890-1904. https://doi.org/10.3390/cancers6041890
Chicago/Turabian StyleKawaguchi, Makiko, and Hiroaki Kataoka. 2014. "Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues" Cancers 6, no. 4: 1890-1904. https://doi.org/10.3390/cancers6041890
APA StyleKawaguchi, M., & Kataoka, H. (2014). Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues. Cancers, 6(4), 1890-1904. https://doi.org/10.3390/cancers6041890




