The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
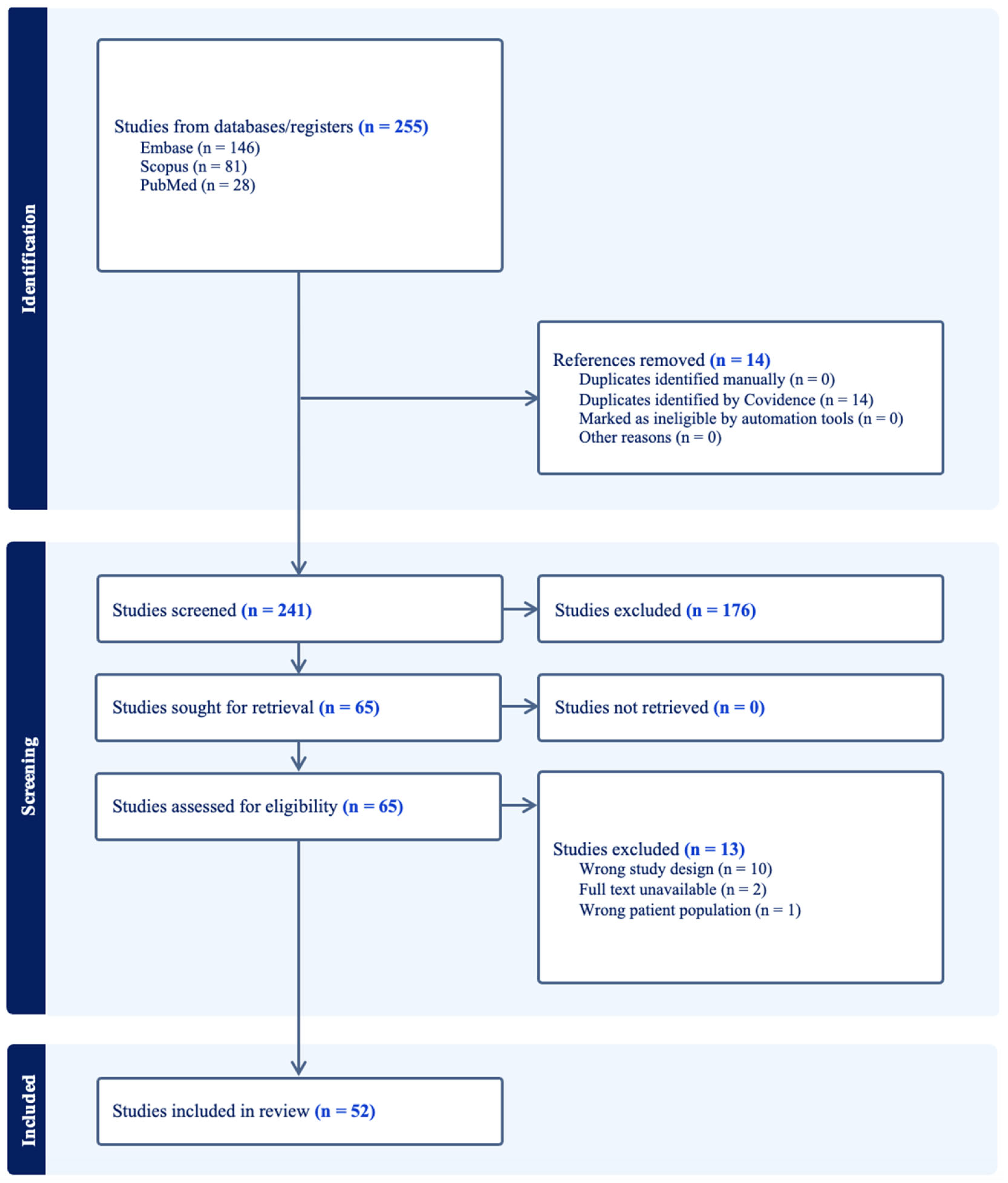
2.1. Literature Search Strategy and Data Extraction
2.2. Quality Assessment
2.3. Statistical Analysis
3. Results
3.1. Radiomics Studies over Time
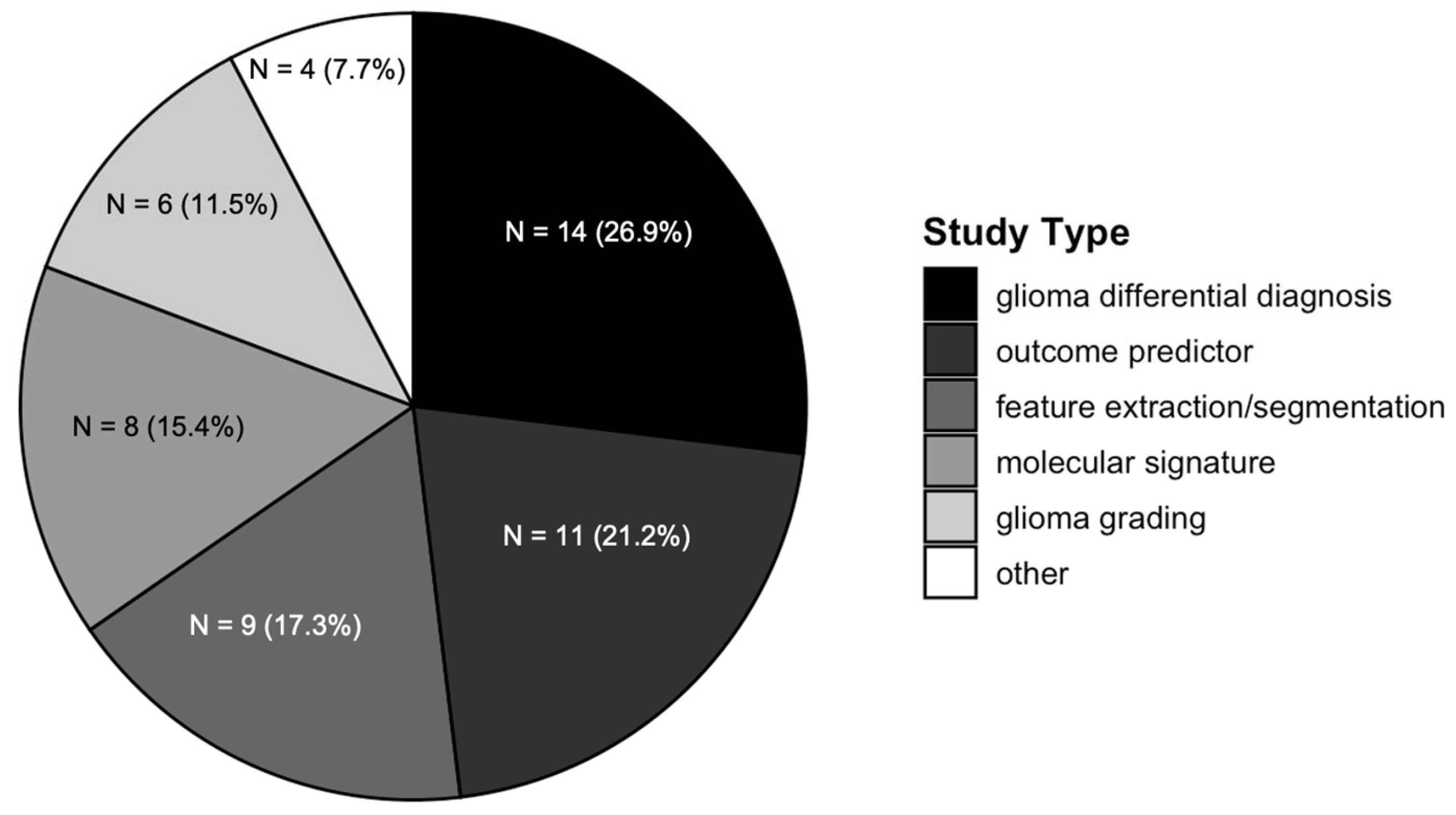
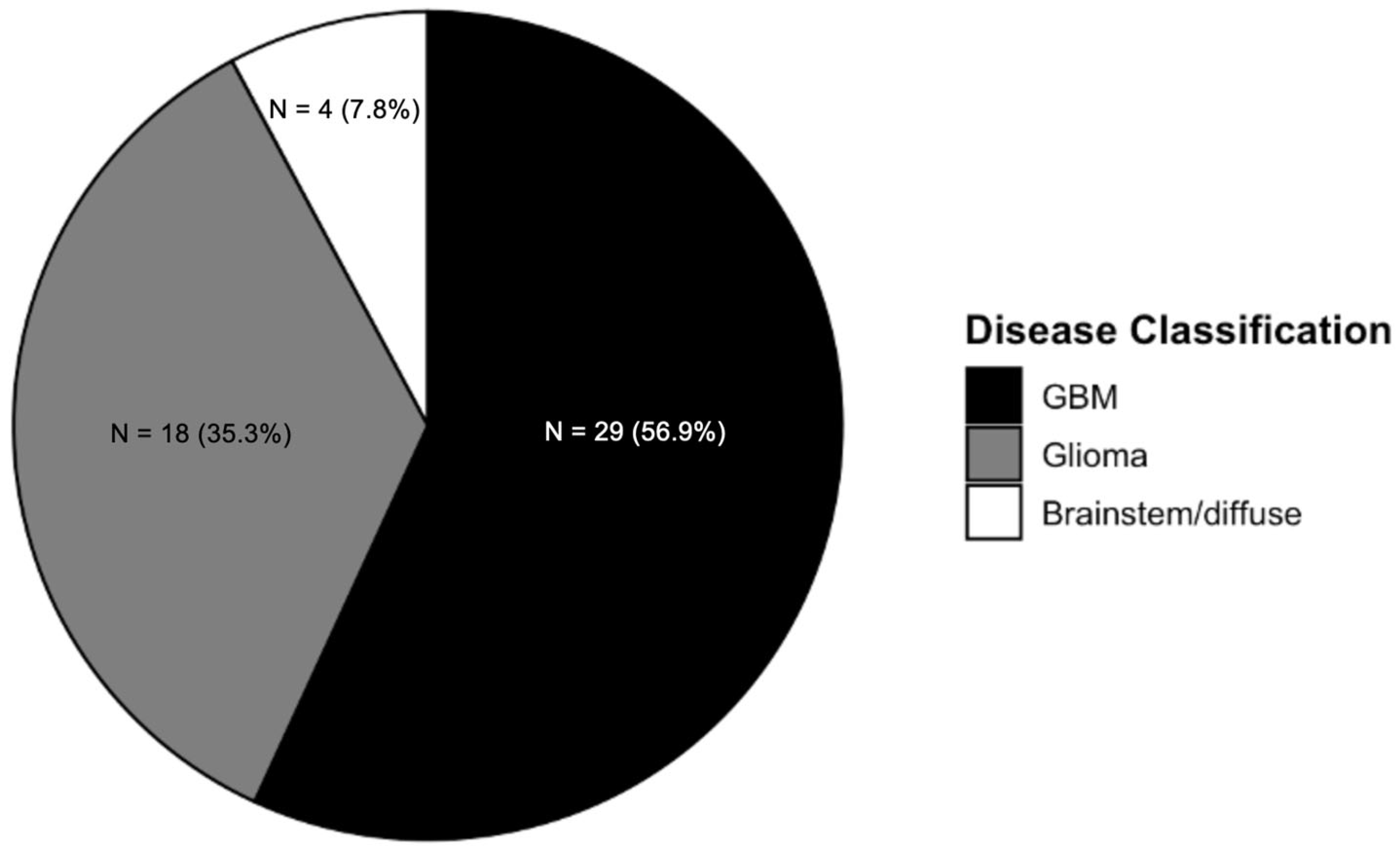
3.2. Radiomics Studies Characteristics
3.3. Image Sequences Utilized by Radiomics
3.4. Quality Assessment
4. Discussion
4.1. Radiomics Trends in Gliomas over a Decade
4.2. Radiomics for Gliomas Upfront and Longitudinal Differential Diagnosis
4.3. Can Radiomics Provide Patient-Specific Non-Invasive Tumor Molecular Signatures?
4.4. Frequently Selected Radiomics Features
4.5. Current Gaps and Future Opportunities in Radiomics
4.6. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barnholtz-Sloan, J.S.; Ostrom, Q.T.; Cote, D. Epidemiology of Brain Tumors. Neurol. Clin. 2018, 36, 395–419. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wen, P.Y.; Chang, S.M.; Dirven, L.; Lim, M.; Monje, M.; Reifenberger, G. Glioma. Nat. Rev. Dis. Primers 2024, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Verdugo, E.; Puerto, I.; Medina, M.Á. An Update on the Molecular Biology of Glioblastoma, with Clinical Implications and Progress in Its Treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; van den Bent, M.J.; Blumenthal, D.T.; Touat, M.; Peters, K.B.; Clarke, J.; Mendez, J.; Yust-Katz, S.; Welsh, L.; Mason, W.P.; et al. Vorasidenib in IDH1- or IDH2-Mutant Low-Grade Glioma. N. Engl. J. Med. 2023, 389, 589–601. [Google Scholar] [CrossRef]
- Kim, D.; Lee, S.-H.; Hwang, H.S.; Kim, S.J.; Yun, M. Recent Update on PET/CT Radiotracers for Imaging Cerebral Glioma. Nucl. Med. Mol. Imaging 2024, 58, 237–245. [Google Scholar] [CrossRef]
- Germano, I.M.; Johnson, D.R.; Patrick, H.H.; Goodman, A.L.; Ziu, M.; Ormond, D.R.; Olson, J.J. Congress of Neurological Surgeons Systematic Review and Evidence-Based Guidelines on the Management of Progressive Glioblastoma in Adults: Update of the 2014 Guidelines. Neurosurgery 2022, 90, 649. [Google Scholar] [CrossRef]
- Young, J.S.; Al-Adli, N.; Scotford, K.; Cha, S.; Berger, M.S. Pseudoprogression versus True Progression in Glioblastoma: What Neurosurgeons Need to Know. J. Neurosurg. 2023, 139, 748–759. [Google Scholar] [CrossRef]
- Nael, K.; Bauer, A.H.; Hormigo, A.; Lemole, M.; Germano, I.M.; Puig, J.; Stea, B. Multiparametric MRI for Differentiation of Radiation Necrosis From Recurrent Tumor in Patients With Treated Glioblastoma. AJR Am. J. Roentgenol. 2018, 210, 18–23. [Google Scholar] [CrossRef]
- Knitter, J.R.; Erly, W.K.; Stea, B.D.; Lemole, G.M.; Germano, I.M.; Doshi, A.H.; Nael, K. Interval Change in Diffusion and Perfusion MRI Parameters for the Assessment of Pseudoprogression in Cerebral Metastases Treated With Stereotactic Radiation. AJR Am. J. Roentgenol. 2018, 211, 168–175. [Google Scholar] [CrossRef]
- Oermann, E.K.; Germano, I.M. In Pursuit of Glioma Diagnosis: The Challenges and Opportunities of Deep Neural Network Augmented Analyses. Neuro Oncol. 2021, 23, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Speirs, C.K.; Simpson, J.R.; Robinson, C.G.; DeWees, T.A.; Tran, D.D.; Linette, G.; Chicoine, M.R.; Dacey, R.G.; Rich, K.M.; Dowling, J.L.; et al. Impact of 1p/19q Codeletion and Histology on Outcomes of Anaplastic Gliomas Treated With Radiation Therapy and Temozolomide. Int. J. Radiat. Oncol. Biol. Phys. 2015, 91, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.-H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; et al. A Novel Enhancer Regulates MGMT Expression and Promotes Temozolomide Resistance in Glioblastoma. Nat. Commun. 2018, 9, 2949. [Google Scholar] [CrossRef] [PubMed]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Grogan, P.T.; Lamont, J.D.; Decker, P.A.; Wu, W.; James, C.D.; Sarkaria, J.N. Induction of MGMT Expression Is Associated with Temozolomide Resistance in Glioblastoma Xenografts. Neuro-Oncol. 2009, 11, 281–291. [Google Scholar] [CrossRef]
- Grassl, N.; Poschke, I.; Lindner, K.; Bunse, L.; Mildenberger, I.; Boschert, T.; Jähne, K.; Green, E.W.; Hülsmeyer, I.; Jünger, S.; et al. A H3K27M-Targeted Vaccine in Adults with Diffuse Midline Glioma. Nat. Med. 2023, 29, 2586–2592. [Google Scholar] [CrossRef]
- Yadav, M.; Sharma, P.; Singh, V.; Tewari, R.; Mishra, P.S.; Roy, K. An Audit of Diagnostic Disparity between Intraoperative Frozen Section Diagnosis and Final Histopathological Diagnosis of Central Nervous System Lesions at a Tertiary Care Center. J. Lab. Physicians 2022, 14, 384–393. [Google Scholar] [CrossRef]
- Sarkiss, C.A.; Germano, I.M. Machine Learning in Neuro-Oncology: Can Data Analysis From 5346 Patients Change Decision-Making Paradigms? World Neurosurg 2019, 124, 287–294. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and Challenges for the Implementation of Computational Medical Imaging (Radiomics) in Oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Koh, D.-M.; Papanikolaou, N.; Bick, U.; Illing, R.; Kahn, C.E.; Kalpathi-Cramer, J.; Matos, C.; Martí-Bonmatí, L.; Miles, A.; Mun, S.K.; et al. Artificial Intelligence and Machine Learning in Cancer Imaging. Commun. Med. 2022, 2, 133. [Google Scholar] [CrossRef]
- Khalighi, S.; Reddy, K.; Midya, A.; Pandav, K.B.; Madabhushi, A.; Abedalthagafi, M. Artificial Intelligence in Neuro-Oncology: Advances and Challenges in Brain Tumor Diagnosis, Prognosis, and Precision Treatment. npj Precis. Oncol. 2024, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, M.; Suman, A.A.; Suero Molina, E.; Pan, E.; Di Ieva, A.; Liu, S. Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review. Cancers 2023, 15, 3845. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Yu, Z.; Sun, L.; Zhu, L.; Geng, D. A Brain Tumor Computer-Aided Diagnosis Method with Automatic Lesion Segmentation and Ensemble Decision Strategy. Front. Med. 2023, 10, 1232496. [Google Scholar] [CrossRef]
- Wang, Z.; He, C.; Hu, Y.; Luo, H.; Li, C.; Wu, X.; Zhang, Y.; Li, J.; Cai, J. A Hybrid Deep Learning Scheme for MRI-Based Preliminary Multiclassification Diagnosis of Primary Brain Tumors. Front. Oncol. 2024, 14, 1363756. [Google Scholar] [CrossRef]
- Bathla, G.; Dhruba, D.D.; Soni, N.; Liu, Y.; Larson, N.B.; Kassmeyer, B.A.; Mohan, S.; Roberts-Wolfe, D.; Rathore, S.; Le, N.H.; et al. AI-Based Classification of Three Common Malignant Tumors in Neuro-Oncology: A Multi-Institutional Comparison of Machine Learning and Deep Learning Methods. J. Neuroradiol. 2024, 51, 258–264. [Google Scholar] [CrossRef]
- Felefly, T.; Roukoz, C.; Fares, G.; Achkar, S.; Yazbeck, S.; Meyer, P.; Kordahi, M.; Azoury, F.; Nasr, D.N.; Nasr, E.; et al. An Explainable MRI-Radiomic Quantum Neural Network to Differentiate Between Large Brain Metastases and High-Grade Glioma Using Quantum Annealing for Feature Selection. J. Digit. Imaging 2023, 36, 2335–2346. [Google Scholar] [CrossRef]
- Li, T.; Wang, L.; Wang, X.; Sun, H. Differential Diagnosis of Radiation Encephalopathy and Post-Radiation Brain Tumor Recurrence by Machine Learning Models Based on Contrast-Enhanced MRI. In Proceedings of the 2024 4th International Conference on Bioinformatics and Intelligent Computing, Beijing, China, 26–28 January 2024; Association for Computing Machinery: New York, NY, USA, 2024; pp. 317–322. [Google Scholar]
- Zhang, J.; Wu, Y.; Wang, Y.; Zhang, X.; Lei, Y.; Zhu, G.; Mao, C.; Zhang, L.; Ma, L. Diffusion-Weighted Imaging and Arterial Spin Labeling Radiomics Features May Improve Differentiation between Radiation-Induced Brain Injury and Glioma Recurrence. Eur. Radiol. 2023, 33, 3332–3342. [Google Scholar] [CrossRef]
- Yan, Q.; Li, F.; Cui, Y.; Wang, Y.; Wang, X.; Jia, W.; Liu, X.; Li, Y.; Chang, H.; Shi, F.; et al. Discrimination Between Glioblastoma and Solitary Brain Metastasis Using Conventional MRI and Diffusion-Weighted Imaging Based on a Deep Learning Algorithm. J. Digit. Imaging 2023, 36, 1480–1488. [Google Scholar] [CrossRef]
- Biggs, M.; Wang, Y.; Soni, N.; Priya, S.; Bathla, G.; Canahuate, G. Evaluating Autoencoders for Dimensionality Reduction of MRI-Derived Radiomics and Classification of Malignant Brain Tumors. In Proceedings of the 35th International Conference on Scientific and Statistical Database Management, Los Angeles, CA, USA, 10–12 July 2023; Association for Computing Machinery: New York, NY, USA, 2023; pp. 1–11. [Google Scholar]
- Zhang, Y.; Zhang, H.; Zhang, H.; Ouyang, Y.; Su, R.; Yang, W.; Huang, B. Glioblastoma and Solitary Brain Metastasis: Differentiation by Integrating Demographic-MRI and Deep-Learning Radiomics Signatures. J. Magn. Reson. Imaging 2024, 60, 909–920. [Google Scholar] [CrossRef]
- Battalapalli, D.; Safai, A.; Ismail, M.; Hill, V.; Statsevych, V.; Huang, R.; Ahluwalia, M.S.; Tiwari, P. Graph-Radiomics Learning (GrRAiL): Application to Distinguishing Glioblastoma Recurrence from Pseudo-Progression on Structural MRI. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–5. [Google Scholar]
- Bai, J.; He, M.; Gao, E.; Yang, G.; Zhang, C.; Yang, H.; Dong, J.; Ma, X.; Gao, Y.; Zhang, H.; et al. High-Performance Presurgical Differentiation of Glioblastoma and Metastasis by Means of Multiparametric Neurite Orientation Dispersion and Density Imaging (NODDI) Radiomics. Eur. Radiol. 2024, 34, 6616–6628. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Gong, J.; Su, X.; Chen, N.; Li, S.; Yang, X.; Zhang, S.; Huang, Z.; Hu, W.; Gong, Q.; et al. MRI Characteristics of H3 G34-Mutant Diffuse Hemispheric Gliomas and Possible Differentiation from IDH-Wild-Type Glioblastomas in Adolescents and Young Adults. J. Neurosurg. Pediatr. 2024, 33, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Tippareddy, C.; Onyewadume, L.; Sloan, A.E.; Wang, G.-M.; Patil, N.T.; Hu, S.; Barnholtz-Sloan, J.S.; Boyacıoğlu, R.; Gulani, V.; Sunshine, J.; et al. Novel 3D Magnetic Resonance Fingerprinting Radiomics in Adult Brain Tumors: A Feasibility Study. Eur. Radiol. 2023, 33, 836–844. [Google Scholar] [CrossRef]
- Lin, J.; Su, C.-Q.; Tang, W.-T.; Xia, Z.-W.; Lu, S.-S.; Hong, X.-N. Radiomic Features on Multiparametric MRI for Differentiating Pseudoprogression from Recurrence in High-Grade Gliomas. Acta Radiol. 2024, 65, 1390–1400. [Google Scholar] [CrossRef]
- Kaur, G.; Rana, P.S.; Arora, V. Automated Neural Network-Based Survival Prediction of Glioblastoma Patients Using Pre-Operative MRI and Clinical Data. IETE J. Res. 2024, 70, 3614–3630. [Google Scholar] [CrossRef]
- Glory Precious, J.; Keren Evangeline, I.; Kirubha, S.P.A. Brain Tumour Segmentation and Survival Prognostication Using 3D Radiomics Features and Machine Learning Algorithms. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2023, 11, 1803–1817. [Google Scholar] [CrossRef]
- Bathla, G.; Soni, N.; Ward, C.; Pillenahalli Maheshwarappa, R.; Agarwal, A.; Priya, S. Clinical and Magnetic Resonance Imaging Radiomics-Based Survival Prediction in Glioblastoma Using Multiparametric Magnetic Resonance Imaging. J. Comput. Assist. Tomogr. 2023, 47, 919–923. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Tan, Q.; Sun, H.; Chen, D.; Chen, Y.; Zhang, H.; Yang, Y.; Gong, Q.; Yue, Q. Cortical Myelin and Thickness Mapping Provide Insights into Whole-Brain Tumor Burden in Diffuse Midline Glioma. Cereb. Cortex 2024, 34, bhad491. [Google Scholar] [CrossRef]
- Yun, J.; Yun, S.; Park, J.E.; Cheong, E.-N.; Park, S.Y.; Kim, N.; Kim, H.S. Deep Learning of Time-Signal Intensity Curves from Dynamic Susceptibility Contrast Imaging Enables Tissue Labeling and Prediction of Survival in Glioblastoma. AJNR Am. J. Neuroradiol. 2023, 44, 543–552. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, Z.; Roth, H.R.; Anwar, S.M.; Bonner, E.R.; Mahtabfar, A.; Packer, R.J.; Kazerooni, A.F.; Bornhorst, M.; Linguraru, M.G. Early Prognostication of Overall Survival for Pediatric Diffuse Midline Gliomas Using MRI Radiomics and Machine Learning: A Two-Center Study. Neuro-Oncol. Adv. 2024, 6, vdae108. [Google Scholar] [CrossRef]
- Joo, B.; Ahn, S.S.; An, C.; Han, K.; Choi, D.; Kim, H.; Park, J.E.; Kim, H.S.; Lee, S.-K. Fully Automated Radiomics-Based Machine Learning Models for Multiclass Classification of Single Brain Tumors: Glioblastoma, Lymphoma, and Metastasis. J. Neuroradiol. 2023, 50, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Cheng, J.; Qiu, A.; Zhao, D.; Wang, J.; Liu, J. Magnetic Resonance Imaging (MRI)-Based Intratumoral and Peritumoral Radiomics for Prognosis Prediction in Glioma Patients. Clin. Radiol. 2024, 79, e1383–e1393. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, S.; Ibrar, N.; Majid, M.; Anwar, S. Overall Survial Prediction from Brain MRI in Glioblastoma. In Proceedings of the 2023 International Conference on Robotics and Automation in Industry (ICRAI), Peshawar, Pakistan, 3–5 March 2023; pp. 1–6. [Google Scholar]
- Karabacak, M.; Patil, S.; Gersey, Z.C.; Komotar, R.J.; Margetis, K. Radiomics-Based Machine Learning with Natural Gradient Boosting for Continuous Survival Prediction in Glioblastoma. Cancers 2024, 16, 3614. [Google Scholar] [CrossRef] [PubMed]
- Hajianfar, G.; Haddadi Avval, A.; Hosseini, S.A.; Nazari, M.; Oveisi, M.; Shiri, I.; Zaidi, H. Time-to-Event Overall Survival Prediction in Glioblastoma Multiforme Patients Using Magnetic Resonance Imaging Radiomics. Radiol. Med. 2023, 128, 1521–1534. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, J.; Wu, Y.; Bai, Y.; Meng, N.; Wu, Q.; Jin, S.; Liu, H.; Li, P.; Wang, M. Assessment of MGMT Promoter Methylation Status in Glioblastoma Using Deep Learning Features from Multi-Sequence MRI of Intratumoral and Peritumoral Regions. Cancer Imaging 2024, 24, 172. [Google Scholar] [CrossRef]
- Medeiros, T.A.; Saraiva Junior, R.G.; de Souza e Cassia, G.; Nascimento, F.A.d.O.; de Carvalho, J.L.A. Classification of 1p/19q Status in Low-Grade Gliomas: Experiments with Radiomic Features and Ensemble-Based Machine Learning Methods. Braz. Arch. Biol. Technol. 2023, 66, e23230002. [Google Scholar] [CrossRef]
- Yang, N.; Xiao, X.; Gu, G.; Wang, X.; Zhang, L.; Liao, H. Combined Evaluation of T1 and Diffusion MRI Improves the Noninvasive Prediction of H3K27M Mutation in Brainstem Gliomas. In Proceedings of the 12th Asian-Pacific Conference on Medical and Biological Engineering, Suzhou, China, 18–21 May 2023; Wang, G., Yao, D., Gu, Z., Peng, Y., Tong, S., Liu, C., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 197–204. [Google Scholar]
- Yang, N.; Xiao, X.; Gu, G.; Wang, X.; Zhang, X.; Wang, Y.; Pan, C.; Zhang, P.; Ma, L.; Zhang, L.; et al. Diffusion MRI-Based Connectomics Features Improve the Noninvasive Prediction of H3K27M Mutation in Brainstem Gliomas. Radiother. Oncol. 2023, 186, 109789. [Google Scholar] [CrossRef]
- Saxena, S.; Jena, B.; Mohapatra, B.; Gupta, N.; Kalra, M.; Scartozzi, M.; Saba, L.; Suri, J.S. Fused Deep Learning Paradigm for the Prediction of O6-Methylguanine-DNA Methyltransferase Genotype in Glioblastoma Patients: A Neuro-Oncological Investigation. Comput. Biol. Med. 2023, 153, 106492. [Google Scholar] [CrossRef]
- Wang, N.C.; Gagnon-Bartsch, J.; Srinivasan, A.; Kim, M.M.; Noll, D.C.; Rao, A. Radiomic Features of Contralateral and Ipsilateral Hemispheres for Prediction of Glioma Genetic Markers. Neurosci. Inform. 2023, 3, 100116. [Google Scholar] [CrossRef]
- Liang, H.-X.; Wang, Z.-Y.; Li, Y.; Ren, A.-N.; Chen, Z.-F.; Wang, X.-Z.; Wang, X.-M.; Yuan, Z.-G. The Application Value of Support Vector Machine Model Based on Multimodal MRI in Predicting IDH-1mutation and Ki-67 Expression in Glioma. BMC Med. Imaging 2024, 24, 244. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, D.; Sun, H.; Kemp, G.J.; Chen, Y.; Tan, Q.; Yang, Y.; Gong, Q.; Yue, Q. Whole Brain Morphologic Features Improve the Predictive Accuracy of IDH Status and VEGF Expression Levels in Gliomas. Cereb. Cortex 2024, 34, bhae151. [Google Scholar] [CrossRef] [PubMed]
- Papi, Z.; Fathi, S.; Dalvand, F.; Vali, M.; Yousefi, A.; Tabatabaei, M.H.; Amouheidari, A.; Abedi, I. Auto-Segmentation and Classification of Glioma Tumors with the Goals of Treatment Response Assessment Using Deep Learning Based on Magnetic Resonance Imaging. Neuroinformatics 2023, 21, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Beser-Robles, M.; Castellá-Malonda, J.; Martínez-Gironés, P.M.; Galiana-Bordera, A.; Ferrer-Lozano, J.; Ribas-Despuig, G.; Teruel-Coll, R.; Cerdá-Alberich, L.; Martí-Bonmatí, L. Deep Learning Automatic Semantic Segmentation of Glioblastoma Multiforme Regions on Multimodal Magnetic Resonance Images. Int. J. CARS 2024, 19, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, H.; Li, T.; Yang, J.; Zhou, Y.; Wang, J.; Saidaer, T.; Liu, X.; Wang, L.; Wang, Y. Distinguishing Tumor Cell Infiltration and Vasogenic Edema in the Peritumoral Region of Glioblastoma at the Voxel Level via Conventional MRI Sequences. Acad. Radiol. 2024, 31, 1082–1090. [Google Scholar] [CrossRef]
- Muthusivarajan, R.; Celaya, A.; Yung, J.P.; Long, J.P.; Viswanath, S.E.; Marcus, D.S.; Chung, C.; Fuentes, D. Evaluating the Relationship between Magnetic Resonance Image Quality Metrics and Deep Learning-Based Segmentation Accuracy of Brain Tumors. Med. Phys. 2024, 51, 4898–4906. [Google Scholar] [CrossRef]
- Falcó-Roget, J.; Cacciola, A.; Sambataro, F.; Crimi, A. Functional and Structural Reorganization in Brain Tumors: A Machine Learning Approach Using Desynchronized Functional Oscillations. Commun. Biol. 2024, 7, 419. [Google Scholar] [CrossRef]
- Chilaca-Rosas, M.-F.; Contreras-Aguilar, M.-T.; Garcia-Lezama, M.; Salazar-Calderon, D.-R.; Vargas-Del-Angel, R.-G.; Moreno-Jimenez, S.; Piña-Sanchez, P.; Trejo-Rosales, R.-R.; Delgado-Martinez, F.-A.; Roldan-Valadez, E. Identification of Radiomic Signatures in Brain MRI Sequences T1 and T2 That Differentiate Tumor Regions of Midline Gliomas with H3.3K27M Mutation. Diagnostics 2023, 13, 2669. [Google Scholar] [CrossRef]
- Parvaze, P.S.; Bhattacharjee, R.; Verma, Y.K.; Singh, R.K.; Yadav, V.; Singh, A.; Khanna, G.; Ahlawat, S.; Trivedi, R.; Patir, R.; et al. Quantification of Radiomics Features of Peritumoral Vasogenic Edema Extracted from Fluid-Attenuated Inversion Recovery Images in Glioblastoma and Isolated Brain Metastasis, Using T1-Dynamic Contrast-Enhanced Perfusion Analysis. NMR Biomed. 2023, 36, e4884. [Google Scholar] [CrossRef]
- Parvaze, S.; Bhattacharjee, R.; Singh, A.; Ahlawat, S.; Patir, R.; Vaishya, S.; Shah, T.J.; Gupta, R.K. Radiomics-Based Evaluation and Possible Characterization of Dynamic Contrast Enhanced (DCE) Perfusion Derived Different Sub-Regions of Glioblastoma. Eur. J. Radiol. 2023, 159, 110655. [Google Scholar] [CrossRef]
- Vossough, A.; Khalili, N.; Familiar, A.M.; Gandhi, D.; Viswanathan, K.; Tu, W.; Haldar, D.; Bagheri, S.; Anderson, H.; Haldar, S.; et al. Training and Comparison of nnU-Net and DeepMedic Methods for Autosegmentation of Pediatric Brain Tumors. AJNR Am. J. Neuroradiol. 2024, 45, 1081–1089. [Google Scholar] [CrossRef]
- Ubaldi, L.; Saponaro, S.; Giuliano, A.; Talamonti, C.; Retico, A. Deriving Quantitative Information from Multiparametric MRI via Radiomics: Evaluation of the Robustness and Predictive Value of Radiomic Features in the Discrimination of Low-Grade versus High-Grade Gliomas with Machine Learning. Phys. Med. 2023, 107, 102538. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, S.; Ma, C.; Fang, W.; Jing, X.; Yang, C.; Li, H.; Zhang, X.; Ge, C.; Liu, B.; et al. Glioma Subtype Prediction Based on Radiomics of Tumor and Peritumoral Edema under Automatic Segmentation. Sci. Rep. 2024, 14, 27471. [Google Scholar] [CrossRef] [PubMed]
- Maskani, M.; Abbasi, S.; Etemad-Rezaee, H.; Abdolahi, H.; Zamanpour, A.; Montazerabadi, A. Grading of Gliomas by Contrast-Enhanced CT Radiomics Features. J. Biomed. Phys. Eng. 2024, 14, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Renugadevi, M.; Narasimhan, K.; Ravikumar, C.V.; Anbazhagan, R.; Pau, G.; Ramkumar, K.; Abbas, M.; Raju, N.; Sathish, K.; Sevugan, P. Machine Learning Empowered Brain Tumor Segmentation and Grading Model for Lifetime Prediction. IEEE Access 2023, 11, 120868–120880. [Google Scholar] [CrossRef]
- Lin, D.; Liu, J.; Ke, C.; Chen, H.; Li, J.; Xie, Y.; Ma, J.; Lv, X.; Feng, Y. Radiomics Analysis of Quantitative Maps from Synthetic MRI for Predicting Grades and Molecular Subtypes of Diffuse Gliomas. Clin. Neuroradiol. 2024, 34, 817–826. [Google Scholar] [CrossRef]
- Zhu, F.-Y.; Sun, Y.-F.; Yin, X.-P.; Wang, T.-D.; Zhang, Y.; Xing, L.-H.; Xue, L.-Y.; Wang, J.-N. Use of Radiomics Models in Preoperative Grading of Cerebral Gliomas and Comparison with Three-Dimensional Arterial Spin Labelling. Clin. Oncol. 2023, 35, 726–735. [Google Scholar] [CrossRef]
- Nalepa, J.; Kotowski, K.; Machura, B.; Adamski, S.; Bozek, O.; Eksner, B.; Kokoszka, B.; Pekala, T.; Radom, M.; Strzelczak, M.; et al. Deep Learning Automates Bidimensional and Volumetric Tumor Burden Measurement from MRI in Pre- and Post-Operative Glioblastoma Patients. Comput. Biol. Med. 2023, 154, 106603. [Google Scholar] [CrossRef]
- Kwak, S.; Akbari, H.; Garcia, J.A.; Mohan, S.; Dicker, Y.; Sako, C.; Matsumoto, Y.; Nasrallah, M.P.; Shalaby, M.; O’Rourke, D.M.; et al. Predicting Peritumoral Glioblastoma Infiltration and Subsequent Recurrence Using Deep-Learning-Based Analysis of Multi-Parametric Magnetic Resonance Imaging. J. Med. Imaging 2024, 11, 054001. [Google Scholar] [CrossRef]
- Danilov, G.V.; Kalayeva, D.B.; Vikhrova, N.B.; Konakova, T.A.; Zagorodnova, A.I.; Popova, A.A.; Postnov, A.A.; Shugay, S.V.; Pronin, I.N. Radiomics in Determining Tumor-to-Normal Brain SUV Ratio Based on 11C-Methionine PET/CT in Glioblastoma. Sovrem. Tekhnologii Med. 2023, 15, 5–11. [Google Scholar] [CrossRef]
- Danilov, G.; Kalaeva, D.; Vikhrova, N.; Konakova, T.; Zagorodnova, A.; Popova, A.; Postnov, A.; Shugay, S.; Shifrin, M.; Pronin, I. The Assessment of Glioblastoma Metabolic Activity via 11C-Methionine PET and Radiomics. Stud. Health Technol. Inform. 2023, 302, 972–976. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Florkowski, C.M. Sensitivity, Specificity, Receiver-Operating Characteristic (ROC) Curves and Likelihood Ratios: Communicating the Performance of Diagnostic Tests. Clin. Biochem. Rev. 2008, 29, S83–S87. [Google Scholar] [PubMed]
- Huang, J.; Ling, C.X. Using AUC and Accuracy in Evaluating Learning Algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar] [CrossRef]
- Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. [Google Scholar] [CrossRef]
- Arrieta, V.A.; Dmello, C.; McGrail, D.J.; Brat, D.J.; Lee-Chang, C.; Heimberger, A.B.; Chand, D.; Stupp, R.; Sonabend, A.M. Immune Checkpoint Blockade in Glioblastoma: From Tumor Heterogeneity to Personalized Treatment. J. Clin. Invest. 2023, 133. [Google Scholar] [CrossRef]
- Yip, S.; Butterfield, Y.S.; Morozova, O.; Chittaranjan, S.; Blough, M.D.; An, J.; Birol, I.; Chesnelong, C.; Chiu, R.; Chuah, E.; et al. Concurrent CIC Mutations, IDH Mutations, and 1p/19q Loss Distinguish Oligodendrogliomas from Other Cancers. J. Pathol. 2012, 226, 7–16. [Google Scholar] [CrossRef]
- Petrelli, F.; De Stefani, A.; Ghidini, A.; Bruschieri, L.; Riboldi, V.; Dottorini, L.; Iaculli, A.; Zaniboni, A.; Trevisan, F. Steroids Use and Survival in Patients with Glioblastoma Multiforme: A Pooled Analysis. J. Neurol. 2021, 268, 440–447. [Google Scholar] [CrossRef]
- Ohmura, K.; Tomita, H.; Hara, A. Peritumoral Edema in Gliomas: A Review of Mechanisms and Management. Biomedicines 2023, 11, 2731. [Google Scholar] [CrossRef]
- Albert, N.L.; Le Rhun, E.; Minniti, G.; Mair, M.J.; Galldiks, N.; Tolboom, N.; Jakola, A.S.; Niyazi, M.; Smits, M.; Verger, A.; et al. Translating the Theranostic Concept to Neuro-Oncology: Disrupting Barriers. Lancet Oncol. 2024, 25, e441–e451. [Google Scholar] [CrossRef]

| Study | N | Features Extracted | Validation Method | Performance |
|---|---|---|---|---|
| ||||
| A brain tumor computer-aided diagnosis method with automatic lesion segmentation and ensemble decision strategy [24] | 1022 | ET, NCR, PTE | 10 fold | AUC = 0.9 |
| A hybrid deep learning scheme for MRI-based preliminary multiclassification diagnosis of primary brain tumors [25] | 66 | Tumor | 5 fold | AUC = 0.9 |
| AI-based classification of three common malignant tumors in neuro-oncology: A multi-institutional comparison of machine learning and deep learning methods [26] | 208 | ET, NCR, PTE | 5 fold | AUC = 0.919 |
| An explainable MRI-radiomic quantum neural network to differentiate between large brain metastases and high-grade glioma using quantum annealing for feature selection [27] | 72 | PTE, tumor | 5 fold | AUC = 0.95 |
| Differential diagnosis of radiation encephalopathy and post-radiation brain tumor recurrence by machine learning models based on contrast-enhanced MRI [28] | 67 | PTE, Tumor | 3 fold | AUC = 0.9805 |
| Diffusion-weighted imaging and arterial spin-labeling radiomics features may improve differentiation between radiation-induced brain injury and glioma recurrence [29] | 66 | Solid tumor | 10 fold | AUC = 0.96 |
| Discrimination between glioblastoma and solitary brain metastasis using conventional MRI and diffusion-weighted imaging based on a deep learning algorithm [30] | 123 | PTE, tumor | No CV | AUC = 0.956 |
| Evaluating autoencoders for dimensionality reduction of MRI-derived radiomics and classification of malignant brain tumors [31] | 93 | Whole tumor, ET, NCR, PTE | 5 × 5 fold | AUC = 0.91 |
| Glioblastoma and solitary brain metastasis: differentiation by integrating demographic MRI and deep learning radiomics signatures [32] | 115 | ET, PTE | No CV | AUC = 0.999 |
| Graph-radiomics learning (GrRAiL): Application to distinguishing glioblastoma recurrence from pseudo-progression on structural MRI [33] | 106 | ET, NCR, PTE | CV | AUC = 0.85 |
| High-performance presurgical differentiation of glioblastoma and metastasis by means of multiparametric neurite orientation dispersion and density imaging (NODDI) radiomics [34] | 57 | NEC, solid tumor, PTE | 5 fold | AUC = 0.701 (nec), 0.820 (PTE), 0.904 (whole tumor) |
| MRI characteristics of H3 G34 mutant diffuse hemispheric gliomas and possible differentiation from IDH wild-type glioblastomas in adolescents and young adults [35] | 53 | ET, NET, PTE | 5 fold | AUC = 0.925 |
| Novel 3D magnetic resonance fingerprinting radiomics in adult brain tumors: a feasibility study [36] | 33 | ET, NET, PTE | No CV | AUC = 0.85 |
| Radiomic features on multiparametric MRI for differentiating pseudo-progression from recurrence in high-grade gliomas [37] | 109 | Whole tumor, PTE | 10 fold | AUC = 0.841 |
| ||||
| Automated neural network-based survival prediction of glioblastoma patients using preoperative MRI and clinical data [38] | 369 | NCR, NET, ED, ET | 5 fold | 51.7% accuracy |
| Brain tumor segmentation and survival prognostication using 3D radiomics features and machine learning algorithms [39] | 325 | Tumor, PTR | No CV | 73.8% accuracy |
| Clinical and magnetic resonance imaging radiomics-based survival prediction in glioblastoma using multiparametric magnetic resonance imaging [40] | 93 | Tumor, ED | 5 fold | C index = 0.7 |
| Cortical myelin and thickness mapping provide insights into whole-brain tumor burden in diffuse midline glioma [41] | 154 | Whole tumor | No CV | AUC = 0.84 |
| Deep learning of time–signal intensity curves from dynamic susceptibility contrast imaging enables tissue labeling and prediction of survival in glioblastoma [42] | 272 | NET, ET | 10 fold | C index = 0.72 |
| Early prognostication of overall survival for pediatric diffuse midline gliomas using MRI radiomics and machine learning: A two-center study [43] | 69 | Whole tumor, tumor core | 5 fold | 77% accuracy |
| Fully automated radiomics-based machine learning models for multiclass classification of single brain tumors: glioblastoma, lymphoma, and metastasis [44] | 401 | ET, NET | 10 fold | AUC = 0.878 |
| Magnetic resonance imaging (MRI)-based intratumoral and peritumoral radiomics for prognosis prediction in glioma patients [45] | 163 | ET, NET, PTE | 5 fold | AUC = 0.91 |
| Overall survival prediction from brain MRI in glioblastoma [46] | 285 | ET, NET, PTE | 2 fold | 82% accuracy |
| Radiomics-based machine learning with natural gradient boosting for continuous survival prediction in glioblastoma [47] | 865 | ET, NCR | No CV | AUC = 0.791 |
| Time-to-event overall survival prediction in glioblastoma multiforme patients using magnetic resonance imaging radiomics [48] | 119 | Core tumor, ET, NCR | 3 fold | C index = 0.77 |
| ||||
| Assessment of MGMT promoter methylation status in glioblastoma using deep learning features from multi-sequence MRI of intratumoral and peritumoral regions [49] | 356 | NCR, ET, PTE | 5 fold | AUC = 0.923 |
| Classification of 1p/19q status in low-grade gliomas: experiments with radiomic features and ensemble-based machine learning methods [50] | 159 | Whole tumor | No CV | AUC = 0.846 |
| Combined evaluation of T1 and diffusion MRI improves the non-invasive prediction of H3K27M mutation in brainstem gliomas [51] | 126 | Whole tumor | 10 fold | AUC = 0.9246 |
| Diffusion MRI-based connectomics features improve the non-invasive prediction of H3K27M mutation in brainstem gliomas [52] | 133 | Whole tumor | 10 fold | AUC = 0.9136 |
| Fused deep learning paradigm for the prediction of o6-methylguanine-DNA methyltransferase genotype in glioblastoma patients: A neuro-oncological investigation [53] | 585 | PTE, Tumor core, ET | 5 fold | AUC = 0.753 |
| Radiomic features of contralateral and ipsilateral hemispheres for prediction of glioma genetic markers [54] | 143 | ET, NET, PTE | 5 fold | IDH AUC = 0.72 |
| The application value of the support vector machine model based on multimodal MRI in predicting IDH-1mutation and Ki-67 expression in glioma [55] | 309 | Solid tumor | 10 fold | IDHAUC = 0.997 KI67AUC = 0.965 |
| Whole-brain morphologic features improve the predictive accuracy of IDH status and VEGF expression levels in gliomas [56] | 182 | ED, ET, NCR | 10 fold | AUC = 0.88 |
| ||||
| Auto-segmentation and classification of glioma tumors with the goals of treatment response assessment using deep learning based on magnetic resonance imaging [57] | 285 | Whole tumor, ET, NEC, PTE | No CV | 99.1% accuracy |
| Deep learning automatic semantic segmentation of glioblastoma multiforme regions on multimodal magnetic resonance images [58] | 1251 | ET, PTE, NET, whole tumor | No CV | Precision = 9.3 |
| Distinguishing tumor cell infiltration and vasogenic edema in the peritumoral region of glioblastoma at the voxel level via conventional MRI sequences [59] | 28 | NET, PTE | 5 fold | AUC = 0.93 |
| Evaluating the relationship between magnetic resonance image quality metrics and deep learning-based segmentation accuracy of brain tumors [60] | 306 | ET, ED, NCR | 5 fold | Dicescore = 0.7280, |
| Functional and structural reorganization in brain tumors: a machine learning approach using desynchronized functional oscillations [61] | 11 | ET, NET, PTE, NCR | 19 fold | Correlation = 0.795 |
| Identification of radiomic signatures in brain MRI sequences T1 and T2 that differentiate tumor regions of midline gliomas with H3.3K27M mutation [62] | 12 | Tumor, PTE | No CV | 5% ofcharacteristic |
| Quantification of radiomics features of peritumoral vasogenic edema extracted from fluid-attenuated inversion recovery images in glioblastoma and isolated brain metastasis, using T1-dynamic contrast-enhanced perfusion analysis [63] | 48 | NET, PTE | No CV | AUC = 0.84 |
| Radiomics-based evaluation and possible characterization of dynamic contrast-enhanced (DCE) perfusion derived different sub-regions of glioblastoma [64] | 89 | ET, NET, ED, NCR | No CV | AUC = 0.89 |
| Training and comparison of nnU-Net and deepmedic methods for auto-segmentation of pediatric brain tumors [65] | 339 | ET, NET, PTE, NCR | 5 fold | Dicescore = 0.9 |
| ||||
| Deriving quantitative information from multiparametric MRI via radiomics: evaluation of the robustness and predictive value of radiomic features in the discrimination of low-grade versus high-grade gliomas with machine learning [66] | 158 | ET, NET, ED | 5 fold | AUC = 0.92 |
| Glioma subtype prediction based on radiomics of tumor and peritumoral edema under automatic segmentation [67] | 424 | Tumor, PTE | 5 fold | AUC = 0.945 |
| Grading of gliomas by contrast-enhanced CT radiomics features [68] | 62 | Whole tumor | 5 fold | AUC = 0.98 |
| Machine learning-empowered brain tumor segmentation and grading model for lifetime prediction [69] | 369 | NCR, NET, ED, ET | No CV | 98% accuracy |
| Radiomics analysis of quantitative maps from synthetic MRI for predicting grades and molecular subtypes of diffuse gliomas [70] | 124 | ET, NET, PTE | 10 fold | AUC = 0.92 |
| Use of radiomics models in preoperative grading of cerebral gliomas and comparison with three-dimensional arterial spin labeling [71] | 105 | Whole tumor | No CV | AUC = 0.929 |
| Other | ||||
| Deep learning automates bidimensional and volumetric tumor burden measurement from MRI in pre- and post-operative glioblastoma patients [72] | 1264 | ET, PTE | 5 fold | Coefficient = 0.959 |
| Predicting peritumoral glioblastoma infiltration and subsequent recurrence using deep-learning-based analysis of multiparametric magnetic resonance imaging [73] | 229 | NET, ET, PTE | 10 fold | OR = 6.90 to 12.63 |
| Radiomics in determining tumor-to-normal brain SUV ratio based on11C-Methionine PET/CT in glioblastoma [74] | 40 | Whole tumor | No CV | Spearman = 0.58 |
| The assessment of glioblastoma metabolic activity via 11C-Methionine PET and radiomics [75] | 40 | active region | No CV | Spearman = 0.7 |
| Total N= 12,482 | 12,482 | |||
| Author | Year | T1w | T1wCE | T2w | FLAIR | DTI | Molecular Signature |
|---|---|---|---|---|---|---|---|
| Molecular Signature (n = 10) | |||||||
| Liang et al. [55] | 2024 | 1 | 1 | 1 | 1 | 1 | IDH, Ki-67 |
| Yang et al. [51] | 2024 | 1 | 0 | 0 | 0 | 1 | H3K27M |
| Yu et al. [49] | 2024 | 1 | 1 | 0 | 0 | 0 | MGMT |
| Zhang et al. [56] | 2024 | 1 | 1 | 1 | 1 | 0 | IDH, VEGF |
| Medeiros et al. [50] | 2023 | 0 | 0 | 1 | 0 | 0 | 1p/19q |
| Saxena et al. [53] | 2023 | 1 | 1 | 1 | 1 | 0 | MGMT |
| Wang et al. [54] | 2023 | 1 | 1 | 1 | 1 | 0 | IDH, MGMT, TERT, and ATRX |
| Yang et al. [52] | 2023 | 1 | 1 | 1 | 0 | 1 | H3K27M |
| Total | 8 | 8 | 8 | 5 | 4 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dedhia, M.; Germano, I.M. The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends. Cancers 2025, 17, 1582. https://doi.org/10.3390/cancers17091582
Dedhia M, Germano IM. The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends. Cancers. 2025; 17(9):1582. https://doi.org/10.3390/cancers17091582
Chicago/Turabian StyleDedhia, Mehek, and Isabelle M. Germano. 2025. "The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends" Cancers 17, no. 9: 1582. https://doi.org/10.3390/cancers17091582
APA StyleDedhia, M., & Germano, I. M. (2025). The Evolving Landscape of Radiomics in Gliomas: Insights into Diagnosis, Prognosis, and Research Trends. Cancers, 17(9), 1582. https://doi.org/10.3390/cancers17091582






