Five Cellular Genes as Candidates for Cervical Adenocarcinoma Molecular Markers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Candidate Genes
2.2. Identification of Candidates from Datasets
2.3. Biological Samples
2.4. DNA/RNA Extraction and cDNA Synthesis
2.5. HPV Detection
2.6. Evaluation of Gene Expression
2.7. Statistical Analysis
3. Results
3.1. Overexpressed Gene Expression Identification from NCBI Analysis
3.2. Identification of Overexpressed Gene from Transcriptomic Libraries for CIN2+
3.3. Biological Samples and Characteristics
3.4. Selection of Differentially Expressed Genes in Cervical Cancer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Flores, H.; Apresa-García, T.; Garay-Villar, Ó.; Sánchez-Pérez, A.; Flores-Villegas, D.; Bandera-Calderón, A.; García-Palacios, R.; Rojas-Sánchez, T.; Romero-Morelos, P.; Sánchez-Albor, V.; et al. A non-invasive tool for detecting cervical cancer odor by trained scent dogs. BMC Cancer 2017, 17, 79. [Google Scholar] [CrossRef]
- Petersen, Z.; Jaca, A.; Ginindza, T.G.; Maseko, G.; Takatshana, S.; Ndlovu, P.; Zondi, N.; Zungu, N.; Varghese, C.; Hunting, G.; et al. Barriers to uptake of cervical cancer screening services in low-and-middle-income countries: A systematic review. BMC Womens Health 2022, 22, 486. [Google Scholar] [CrossRef]
- Mai, K.T. Differentiated cervical intraepithelial neoplasia-associated invasive cervical squamous cell carcinoma as a source of major cytopathological and surgical pathological discrepancy in Papanicolaou smear screening tests. Cytopathology 2018, 29, 143–149. [Google Scholar] [CrossRef]
- Pan American Health Organization. Integrating HPV Testing in Cervical Cancer Screening Programs; Pan American Health Organization: Washington, DC, USA, 2016; 68p. [Google Scholar]
- Lichtenberg, R. Human papillomavirus. Clevel. Clin. J. Med. 2019, 86, 300–301. [Google Scholar]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Rutgers, J.K.L.; Roma, A.A.; Park, K.J.; Zaino, R.J.; Johnson, A.; Alvarado, I.; Daya, D.; Rasty, G.; Longacre, T.A.; Ronnett, B.M.; et al. Pattern classification of endocervical adenocarcinoma: Reproducibility and review of criteria. Mod. Pathol. 2016, 29, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Adolph, L.; Mann, A.; Liu, X.Q.; Roberts, L.; Robinson, C.; Popowich, S.; Dean, E.; Kean, S.; Fischer, G.; Altman, A.D. Follow-up of women with cervical adenocarcinoma in situ treated by conization: A single centre clinical experience. Gynecol. Oncol. 2024, 187, 74–79. [Google Scholar] [CrossRef]
- Stolnicu, S.; Park, K.J.; Kiyokawa, T.; Oliva, E.; McCluggage, W.G.; Soslow, R.A. Tumor Typing of Endocervical Adenocarcinoma: Contemporary Review and Recommendations From the International Society of Gynecological Pathologists. Int. J. Gynecol. Pathol. 2021, 40, S75–S91. [Google Scholar] [CrossRef]
- He, Z.; Chen, R.; Hu, S.; Zhang, Y.; Liu, Y.; Li, C.; Lv, F.; Xiao, Z. The value of HPV genotypes combined with clinical indicators in the classification of cervical squamous cell carcinoma and adenocarcinoma. BMC Cancer 2022, 22, 776. [Google Scholar] [CrossRef]
- Nair, M.; Sandhu, S.S.; Sharma, A.K. Cancer molecular markers: A guide to cancer detection and management. Semin. Cancer Biol. 2018, 52, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Al-Samarai, F.; Al-Kazaz, A. Molecular Markers: An Introduction and Applications. Eur. J. Mol. Biotechnol. 2015, 9, 118–130. [Google Scholar] [CrossRef]
- Hirsch, B.; Endris, V.; Lassmann, S.; Weichert, W.; Pfarr, N.; Schirmacher, P.; Kovaleva, V.; Werner, M.; Bonzheim, I.; Fend, F.; et al. Multicenter validation of cancer gene panel-based next-generation sequencing for translational research and molecular diagnostics. Virchows Arch. 2018, 472, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Kisiel, J.B.; Ebbert, J.O.; Taylor, W.R.; Marinac, C.R.; Choudhry, O.A.; Rego, S.P.; Beer, T.M.; Beidelschies, M.A. Shifting the Cancer Screening Paradigm: Developing a Multi-Biomarker Class Approach to Multi-Cancer Early Detection Testing. Life 2024, 14, 925. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.; Cullinan, A.; Hogg, G.; Hosseini, D.; Ehrich, M. Mutation profiling in tumor samples using the Sequenom OncoCartaTM Panel. Nat. Methods 2009, 6, vii–viii. [Google Scholar] [CrossRef]
- Güzel, C.; van Sten-Van’t Hoff, J.; de Kok, I.M.C.M.; Govorukhina, N.I.; Boychenko, A.; Luider, T.M.; Bischoff, R. Molecular markers for cervical cancer screening. Expert Rev. Proteomics 2021, 18, 675–691. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Smyth, G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009, 25, 765–771. [Google Scholar] [CrossRef]
- Dalman, M.R.; Deeter, A.; Nimishakavi, G.; Duan, Z.-H. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics 2012, 13, S11. [Google Scholar] [CrossRef]
- De Roda Husman, A.M.; Walboomers, J.M.M.; Van den Brule, A.J.C.; Meijer, C.J.L.M.; Snijders, P.J.F. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995, 76, 1057–1062. [Google Scholar] [CrossRef]
- Qu, W.; Jiang, G.; Cruz, Y.; Chang, C.J.; Ho, G.Y.; Klein, R.S.; Burk, R.D. PCR detection of human papillomavirus: Comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 1997, 35, 1304–1310. [Google Scholar] [CrossRef]
- Alderete-Torres, L.C. Metilación y Expresión del gen Receptor del Ácido Reinoico (RAR) en el Desarrollo del Cancer Cervicouterino. Ph.D. Thesis, Universidad Autónoma de Ciudad Juárez, Chihuahua, Mexico, 2018. [Google Scholar]
- Picazo-Perez, L. Relación Citocinas pro Inflamatorias y Obesidad en el Riesgo y Progresión de Cancer Cervicouterino. Master’s Thesis, Universidad Autónoma de Ciudad Juárez, Chihuahua, Mexico, 2016. [Google Scholar]
- Liu, S.; Minaguchi, T.; Lachkar, B.; Zhang, S.; Xu, C.; Tenjimbayashi, Y.; Shikama, A.; Tasaka, N.; Akiyama, A.; Sakurai, M.; et al. Separate analysis of human papillomavirus E6 and E7 messenger RNAs to predict cervical neoplasia progression. PLoS ONE 2018, 13, e0193061. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Imoto, I.; Inoue, J.; Kozaki, K.; Tsuda, H.; Shimada, Y.; Aiko, S.; Yoshizumi, Y.; Iwai, T.; Kawano, T.; et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene 2007, 26, 6456–6468. [Google Scholar] [CrossRef]
- Arellano, A.L. Evaluación del Retino y su Participación en la Expresión y Metilación de los Genes CRABP1 y CRABP2 en Mujeres con Cancer Cervicouterino. Ph.D. Thesis, Universidad Autónoma de Ciudad Juárez, Chihuahua, Mexico, 2016. [Google Scholar]
- Park, S.; Lee, S.; Kim, J.; Kim, G.; Park, K.H.; Kim, T.U.; Chung, D.; Lee, H. ΔNp63 to TAp63 expression ratio as a potential molecular marker for cervical cancer prognosis. PLoS ONE 2019, 14, e0214867. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Prasad, S.B.; Yadav, S.S.; Govardhan, H.B.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. Over Expression of Minichromosome Maintenance Genes is Clinically Correlated to Cervical Carcinogenesis. PLoS ONE 2013, 8, e69607. [Google Scholar] [CrossRef]
- Li, H.; Harrison, M.D.; Avissar, P.L.; Malinowski, D.P. Quantitative molecular analysis of MCM6 and MCM7 and their association with HPV E6 and E7 mRNA expression, viral load and physical status in invasive cervical carcinoma. Cancer Res. 2006, 66 (Suppl. S8), 217. [Google Scholar]
- Steinau, M.; Rajeevan, M.S.; Lee, D.R.; Ruffin, M.T.; Horowitz, I.R.; Flowers, L.C.; Tadros, T.; Birdsong, G.; Husain, M.; Kmak, D.C.; et al. Evaluation of RNA Markers for Early Detection of Cervical Neoplasia in Exfoliated Cervical Cells. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 295–301. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, H.; Zhang, L.; Hou, T.; Wu, S.; Huang, Q.; Song, L.; Liu, J. URG4 overexpression is correlated with cervical cancer progression and poor prognosis in patients with early-stage cervical cancer. BMC Cancer 2014, 14, 885. [Google Scholar] [CrossRef]
- Geisen, C.; Denk, C.; Gremm, B.; Baust, C.; Karger, A.; Bollag, W.; Schwarz, E. High-Level Expression of the Retinoic Acid Receptor β Gene in Normal Cells of the Uterine Cervix Is Regulated by the Retinoic Acid Receptor α and Is Abnormally Down-Regulated in Cervical Carcinoma Cells1. Cancer Res. 1997, 57, 1460–1467. [Google Scholar]
- Hashimoto, I.; Kodama, J.; Seki, N.; Hongo, A.; Yoshinouchi, M.; Okuda, H.; Kudo, T. Vascular endothelial growth factor-C expression and its relationship to pelvic lymph node status in invasive cervical cancer. Br. J. Cancer 2001, 85, 93–97. [Google Scholar] [CrossRef]
- Min, Z.; Pu, X.; Gu, Z. Correlative analysis of the expression of IL-10 and Ki-67 in human cervical cancer and cervical intraepithelial neoplasias and human papillomavirus infection. Oncol. Lett. 2018, 16, 7189–7194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Barnie, P.A.; Yang, P.; Su, Z.; Chen, J.; Jiao, Z.; Lu, L.; Wang, S.; Xu, H. The Expression of Toll-like Receptor 8 and Its Relationship with VEGF and Bcl-2 in Cervical Cancer. Int. J. Med. Sci. 2014, 11, 608–613. [Google Scholar] [CrossRef]
- Liu, L.; Xia, M.; Wang, J.; Zhang, W.; Zhang, Y.; He, M. CISD2 expression is a novel marker correlating with pelvic lymph node metastasis and prognosis in patients with early-stage cervical cancer. Med. Oncol. 2014, 31, 183. [Google Scholar] [CrossRef]
- Dannenberg, A.J.; Howe, L.R. The role of COX-2 in breast and cervical cancer. Prog. Exp. Tumor Res. 2003, 37, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.K.; Lee, H.J.; Jeong, M.-H.; Rhee, M.; Mo, J.-W.; Song, E.H.; Lim, J.-Y.; Choi, K.-H.; Jo, I.; Park, S.I.; et al. Role of Activating Transcription Factor 3 on TAp73 Stability and Apoptosis in Paclitaxel-Treated Cervical Cancer Cells. Mol. Cancer Res. 2008, 6, 1232–1249. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Shin, H.-J.; Kim, T.-H.; Cho, K.-H.; Shin, K.-H.; Kim, B.-K.; Roh, J.-W.; Lee, S.; Park, S.-Y.; Hwang, Y.-J.; et al. Tumor-associated carbonic anhydrases are linked to metastases in primary cervical cancer. J. Cancer Res. Clin. Oncol. 2006, 132, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.-Q.; Lu, H.-S.; Zhang, L.; Chen, L.-L.; Gan, M.-F. MEKK3 and Survivin Expression in Cervical Cancer: Association with Clinicopathological Factors and Prognosis. Asian Pacific J. Cancer Prev. 2014, 15, 5271–5276. [Google Scholar] [CrossRef]
- Branca, M.; Giorgi, C.; Santini, D.; Di Bonito, L.; Ciotti, M.; Costa, S.; Benedetto, A.; Casolati, E.A.; Favalli, C.; Paba, P.; et al. Survivin as a Marker of Cervical Intraepithelial Neoplasia and High-Risk Human Papillomavirus and a Predictor of Virus Clearance and Prognosis in Cervical Cancer. Am. J. Clin. Pathol. 2005, 124, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Branca, M.; Giorgi, C.; Ciotti, M.; Santini, D.; Di Bonito, L.; Costa, S.; Benedetto, A.; Bonifacio, D.; Di Bonito, P.; Paba, P.; et al. Relationship of Up-Regulation of 67-kd Laminin Receptor to Grade of Cervical Intraepithelial Neoplasia and to High-Risk HPV Types and Prognosis in Cervical Cancer. Acta Cytol. 2006, 50, 6–15. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X. Evaluating the expression of MACC1 and c-myc in cervical cancer and their correlation. Int. J. Clin. Exp. Med. 2018, 11, 5960–5966. [Google Scholar]
- Abba, M.C.; Laguens, R.M.; Dulout, F.N.; Golijow, C.D. The c-myc activation in cervical carcinomas and HPV 16 infections. Mutat. Res. Toxicol. Environ. Mutagen. 2004, 557, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, Y.; Nishi, H.; Isaka, K.; Fujito, A.; Takayama, M. The correlation of TERT expression with c-myc expression in cervical cancer. Cancer Lett. 2001, 168, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Chen, L.; Liu, C.; Jiang, Y.; Rong, J. Elevated CTHRC1 expression is an indicator for poor prognosis and lymph node metastasis in cervical squamous cell carcinoma. Hum. Pathol. 2019, 85, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, Q.; Tian, P.; Jia, Y. [Highly expressed CHAF1A and PCNA are positively associated with malignancy of cervical squamous cell carcinoma]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi = Chin. J. Cell. Mol. Immunol. 2017, 33, 1696–1701. [Google Scholar]
- Guo, L.; Lu, W.; Zhang, X.; Luo, D.; Zhang, H. Metastasis-associated colon cancer-1 is a novel prognostic marker for cervical cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 4150–4155. [Google Scholar]
- Zhang, F.; Ren, C.-C.; Liu, L.; Chen, Y.-N.; Yang, L.; Zhang, X.-A. HOXC6 gene silencing inhibits epithelial-mesenchymal transition and cell viability through the TGF-β/smad signaling pathway in cervical carcinoma cells. Cancer Cell Int. 2018, 18, 204. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Liu, N.; Hou, J.; Xiao, W.; Wang, H. HOXC6 promotes cervical cancer progression via regulation of Bcl-2. FASEB J. 2019, 33, 3901–3911. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, L.; Guo, A. Upregulated expression of HOXC8 is associated with poor prognosis of cervical cancer. Oncol. Lett. 2018, 15, 7291–7296. [Google Scholar] [CrossRef]
- Arellano-Ortiz, A.L.; Salcedo-Vargas, M.; Vargas-Requena, C.L.; López-Díaz, J.A.; De la Mora-Covarrubias, A.; Silva-Espinoza, J.C.; Jimέnez-Vega, F. DNA Methylation of Cellular Retinoic Acid-Binding Proteins in Cervical Cancer. Genet. Epigenet. 2016, 8, GEG.S40847. [Google Scholar] [CrossRef]
- den Boon, J.A.; Pyeon, D.; Wang, S.S.; Horswill, M.; Schiffman, M.; Sherman, M.; Zuna, R.E.; Wang, Z.; Hewitt, S.M.; Pearson, R.; et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proc. Natl. Acad. Sci. USA 2015, 112, E3255–E3264. [Google Scholar] [CrossRef]
- Bachtiary, B.; Boutros, P.C.; Pintilie, M.; Shi, W.; Bastianutto, C.; Li, J.-H.; Schwock, J.; Zhang, W.; Penn, L.Z.; Jurisica, I.; et al. Gene Expression Profiling in Cervical Cancer: An Exploration of Intratumor Heterogeneity. Clin. Cancer Res. 2006, 12, 5632–5640. [Google Scholar] [CrossRef] [PubMed]
- Who Team Management-Screening, Diagnosis and Treatment (MND), Noncommunicable Diseases R and D (NCD). WHO Cervical Cancer Elimination Initiative: From Call to Action to Global Movement; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- Bhattacharjee, R.; Das, S.S.; Biswal, S.S.; Nath, A.; Das, D.; Basu, A.; Malik, S.; Kumar, L.; Kar, S.; Singh, S.K.; et al. Mechanistic role of HPV-associated early proteins in cervical cancer: Molecular pathways and targeted therapeutic strategies. Crit. Rev. Oncol. Hematol. 2022, 174, 103675. [Google Scholar] [CrossRef]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Van Ostade, X.; Abebe, T. HPV E6/E7 mRNA test for the detection of high grade cervical intraepithelial neoplasia (CIN2+): A systematic review. Infect. Agent. Cancer 2020, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Ran, T.; Wei, Q.; Pan, R.; Chen, S.; Luo, J. Diagnostic value of HPV E6/E7 mRNA in screening for cervical intraepithelial neoplasia grade 2 or worse: A systematic review and meta-analysis. Oncol. Lett. 2024, 27, 231. [Google Scholar] [CrossRef]
- Burk, R.D.; Chen, Z.; Saller, C.; Tarvin, K.; Carvalho, A.L.; Scapulatempo-Neto, C.; Silveira, H.C.; Fregnani, J.H.; Creighton, C.J.; Anderson, M.L.; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Okada, P.A.; Mitrat, S.; Rojanawiwat, A. External quality assessment program for human papillomaviruses DNA testing in Thailand. Pract. Lab. Med. 2024, 38, e00352. [Google Scholar] [CrossRef]
- Samuel, R.; Francois, C.; Dan, F.; James, B.; Nicholas, E.; Prafull, G.; Veeresh, G.; Glen, H.; Elias, B.; Nick, K.; et al. Clinical Performance of the PreTect HPV-Proofer E6/E7 mRNA Assay in Comparison with That of the Hybrid Capture 2 Test for Identification of Women at Risk of Cervical Cancer. J. Clin. Microbiol. 2010, 48, 2779–2785. [Google Scholar] [CrossRef]
- Gutiérrez, J.; García-Villa, E.; Ocadiz-Delgado, R.; Cortés-Malagón, E.M.; Vázquez, J.; Roman-Rosales, A.; Alvarez-Rios, E.; Celik, H.; Romano, M.C.; Üren, A.; et al. Human papillomavirus type 16 E7 oncoprotein upregulates the retinoic acid receptor-beta expression in cervical cancer cell lines and K14E7 transgenic mice. Mol. Cell. Biochem. 2015, 408, 261–272. [Google Scholar] [CrossRef]
- Alami, Y.; Castronovo, V.; Belotti, D.; Flagiello, D.; Clausse, N. HOXC5 and HOXC8 expression are selectively turned on in human cervical cancer cells compared to normal keratinocytes. Biochem. Biophys. Res. Commun. 1999, 257, 738–745. [Google Scholar] [CrossRef]
- López-Romero, R.; Marrero-Rodríguez, D.; Romero-Morelos, P.; Villegas, V.; Valdivia, A.; Arreola, H.; Huerta-Padilla, V.; Salcedo, M. El papel de los genes del desarrollo tipo HOX en el cáncer cervicouterino. Rev. Med. Inst. Mex. Seguro Soc. 2015, 53, S188–S192. [Google Scholar]
- Giannella, L.; Di Giuseppe, J.; Delli Carpini, G.; Grelloni, C.; Fichera, M.; Sartini, G.; Caimmi, S.; Natalini, L.; Ciavattini, A. HPV-Negative Adenocarcinomas of the Uterine Cervix: From Molecular Characterization to Clinical Implications. Int. J. Mol. Sci. 2022, 23, 5022. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.S.; Chowdhury, R.R.; Mondal, N.R.; Roy, S.; Sengupta, S. Expression signatures of HOX cluster genes in cervical cancer pathogenesis: Impact of human papillomavirus type 16 oncoprotein E7. Oncotarget 2017, 8, 36591–36602. [Google Scholar] [CrossRef]
- Hartl, M.; Bister, K. MYC Analysis in Cancer and Evolution BT. In The Myc Gene: Methods and Protocols; Soucek, L., Whitfield, J., Eds.; Springer: New York, NY, USA, 2021; pp. 87–117. [Google Scholar] [CrossRef]
- Salcedo, M.; Taja, L.; Utrera, D.; Chávez, P.; Hidalgo, A.; Pérez, C.; Benítez, L.; Castañeda, C.; Delgado, R.; Gariglio, P. Changes in retinoblastoma gene expression during cervical cancer progression. Int. J. Exp. Pathol. 2002, 83, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Lashen, A.G.; Toss, M.S.; Rutland, C.S.; Green, A.R.; Mongan, N.P.; Rakha, E. Prognostic and Clinical Significance of the Proliferation Marker MCM7 in Breast Cancer. Pathobiology 2025, 92, 18–27. [Google Scholar] [CrossRef]
- Liu, X.; Clements, A.; Zhao, K.; Marmorstein, R. Structure of the Human Papillomavirus E7 Oncoprotein and Its Mechanism for Inactivation of the Retinoblastoma Tumor Suppressor. J. Biol. Chem. 2006, 281, 578–586. [Google Scholar] [CrossRef]
- Campbell, K.J.; Tait, S.W.G. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Crawford, R.A.; Caldwell, C.; Iles, R.K.; Lowe, D.; Shepherd, J.H.; Chard, T. Prognostic significance of the bcl-2 apoptotic family of proteins in primary and recurrent cervical cancer. Br. J. Cancer 1998, 78, 210–214. [Google Scholar] [CrossRef]
- Turker, P.; Segersten, U.; Malmström, P.-U.; Hemdan, T. Is Bcl-2 a predictive marker of neoadjuvant chemotherapy response in patients with urothelial bladder cancer undergoing radical cystectomy? Scand. J. Urol. 2019, 53, 45–50. [Google Scholar] [CrossRef]
- Zhu, T.; Xu, F.; Zhang, L.; Zhang, Y.; Yang, C.; Cheng, M.; Chen, F.; Wang, K. Measurement of molecular biomarkers that predict the tumor response in estrogen receptor-positive breast cancers after dose-dense (biweekly) paclitaxel/carboplatin neoadjuvant chemotherapy. Oncotarget 2017, 8, 101087–101094. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Z.; Liu, M.; Ye, H.; He, J.; Chen, J. CD34 and Bcl-2 as predictors for the efficacy of neoadjuvant chemotherapy in cervical cancer. Arch. Gynecol. Obstet. 2021, 304, 495–501. [Google Scholar] [CrossRef]
- Qu, J.-H.; Chang, X.-J.; Lu, Y.-Y.; Bai, W.-L.; Chen, Y.; Zhou, L.; Zeng, Z.; Wang, C.-P.; An, L.-J.; Hao, L.-Y.; et al. Overexpression of metastasis-associated in colon cancer 1 predicts a poor outcome of hepatitis B virus-related hepatocellular carcinoma. World J. Gastroenterol. 2012, 18, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- Kobelt, D.; Zhang, C.; Clayton-Lucey, I.A.; Glauben, R.; Voss, C.; Siegmund, B.; Stein, U. Pro-inflammatory TNF-α and IFN-γ Promote Tumor Growth and Metastasis via Induction of MACC1. Front. Immunol. 2020, 11, 980. [Google Scholar] [CrossRef] [PubMed]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 61, 344–349. [Google Scholar] [CrossRef] [PubMed]
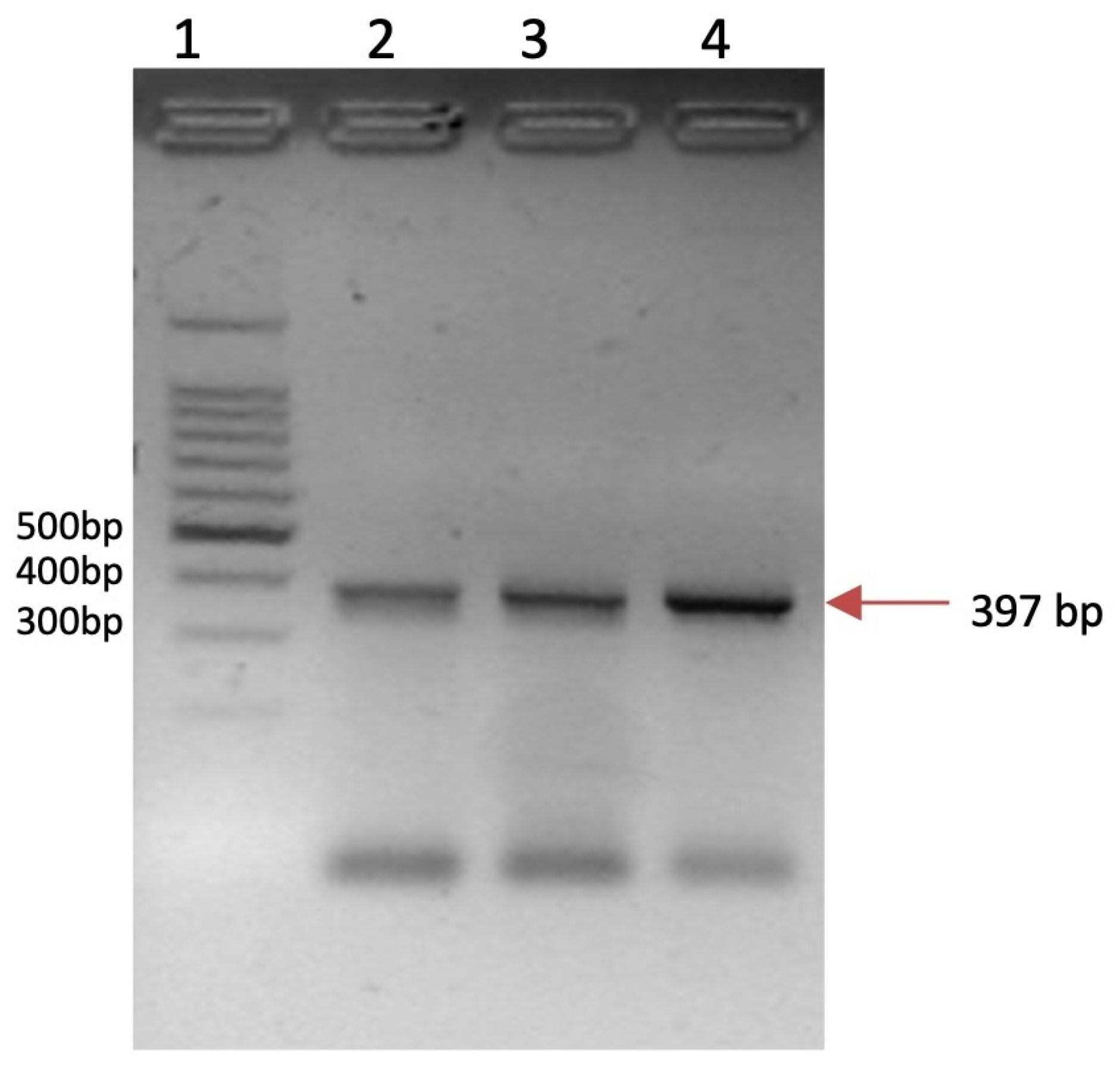
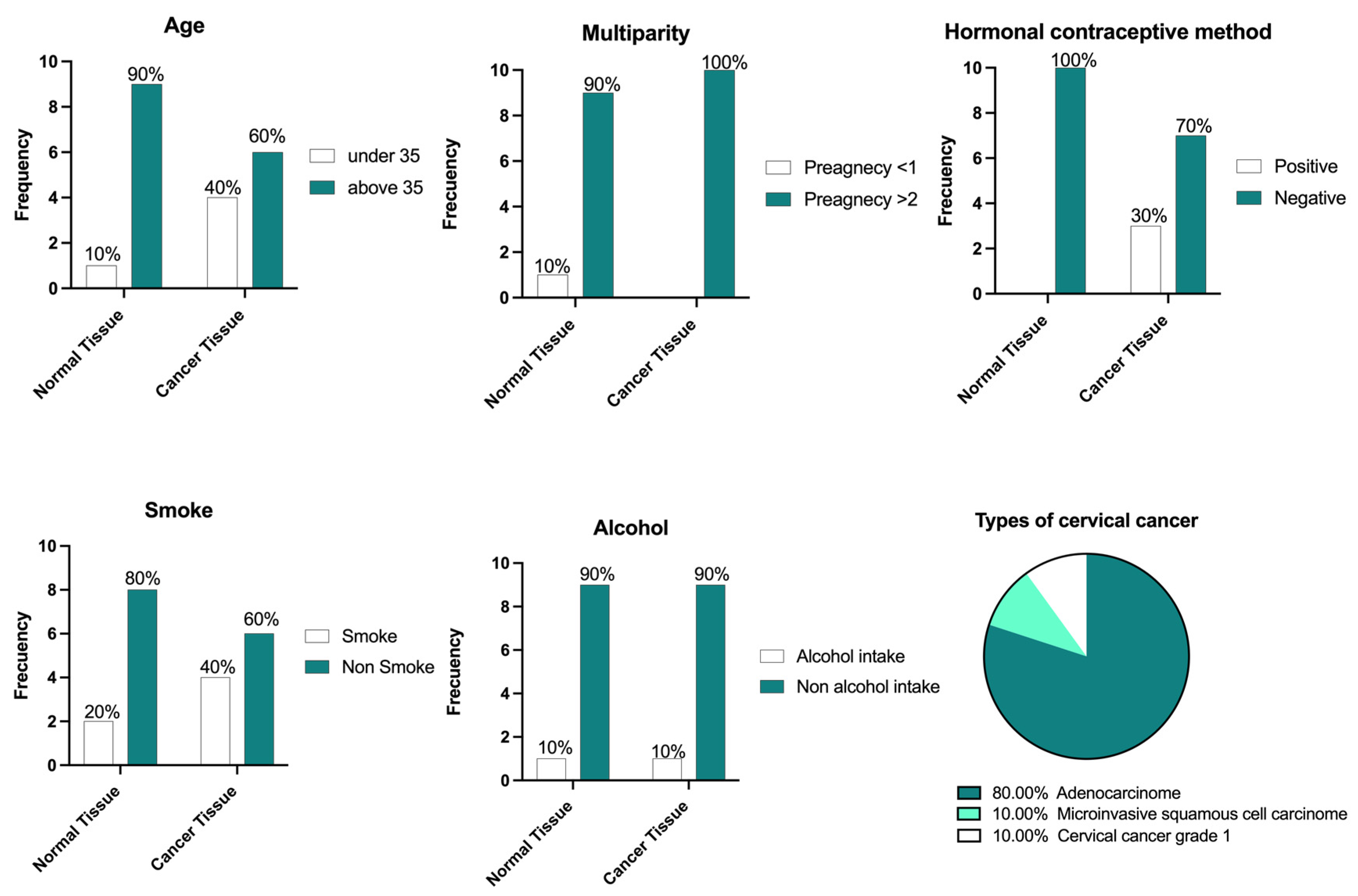

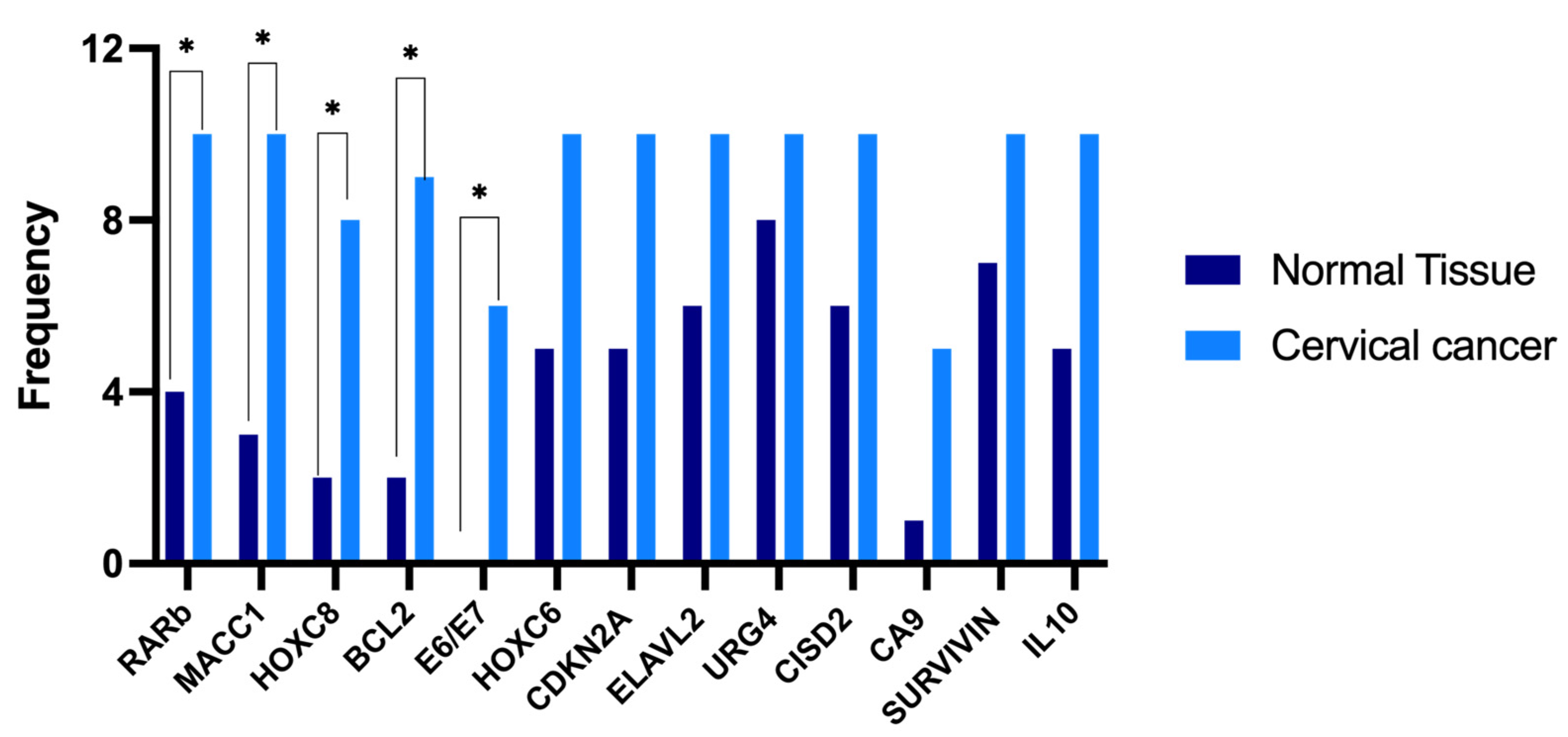
| Gene Symbol | Primer Sequence (5′-3′) | Tm °C | Amplicon Length (pb) | Identification Number/Reference |
|---|---|---|---|---|
| URG4 | Fw GCATCAGAGAGACGAACAGC | 62 | 180 | NM_017920.4 |
| Rv GCACGTCCAGCACCATAG | ||||
| P63 | Fw GAGCTGAGCCGTGAATTC | 55 | 319 | AB082923.1 |
| Rv CCTTCCTGTCTCTTCCTGG | ||||
| HOXC6 | Fw GAGGAAAAGCGGGAAGAG | 60 | 250 | NM_004503.4 |
| Rv CGTGGTGAAAGAGAGTTGTG | ||||
| RARβ | Fw GTGTCCTTCCTGATTCATGC | 62 | 163 | [23] |
| Rv CCACTCTACCACAGCTTTCAC | ||||
| MCM7 | Fw GCTGCATTGATGAGTTCG | 45 | 271 | NM_005916.4 |
| Rv CGTAGGTCATTGTCTCGG | ||||
| PCNA | Fw CTCCCAAGATCGAGGATG | 55 | 249 | NM_002592.2 |
| Rv GACCAGATCTGACTTTGGAC | ||||
| CISD2 | Fw GGCTGCTGCAATTTGAAG | 57 | 264 | NM_001008388.5 |
| Rv GTGTACGGAGGGTCAACTG | ||||
| IL-10 | Fw CCATTCCAAGCCTGACCAC | 60 | 181 | [24] |
| Rv GAATCCCTCCGAGACACTG | ||||
| E6/E7 | Fw ACCGAAAACGGTTGAACCGAAAACGGT | 60 | 500 | [25] |
| Rv GAG CTG TCG CTT AAT TGC TC | ||||
| TAP73 | Fw GAGCAGTACCGCATGACC | 65 | 290 | NM_005427.4 |
| Rv CGTGAACTCCTCCTTGATG | ||||
| COX2 | Fw GCTGTATCCTGCCCTTCTG | 55 | 291 | AY462100.1 |
| Rv CGGGAAGAACTTGCATTG | ||||
| CA9 | Fw CGGCTACAGCTGAACTTCC | 60 | 238 | NM_001216.3 |
| Rv GTAGCTCACACCCCCTTTG | ||||
| MACC1 | Fw CAATGGAAGCCCTTTTGC | 60 | 247 | NM_182762.4 |
| Rv GGTGACGGAAGAGCTTTAGC | ||||
| HOXC8 | Fw GAGCTCCTACTTCGTCAACC | 55 | 250 | NM_022658.4 |
| Rv GTCTCCGTGGCAGCTAAG | ||||
| CTHRC1 | Fw GGACACCCAACTACAAGCAG | 55 | 380 | NM_138455.4 |
| Rv CCAGCACCAATTCCTTCAC | ||||
| BCL2 | Fw CGACTCCTGATTCATTGGG | 55 | 550 | NM_000633.2 |
| Rv GCTTTGCATTCTTGGACG | ||||
| VEGF | Fw CTTCAAGCCATCCTGTGTGC | 55 | 147 | [24] |
| Rv GCTCATCTCTCCTATGTGC | ||||
| CRABP1 | Fw GCACGCAAACTCTTCTTGAAG | 60 | 133 | [26] |
| Rv CGGACATAAATTCTGGTGCAG | ||||
| cMYC | Fw CCTCAACGTTAGCTTCACC | 65 | 242 | NM_002467.6 |
| Rv GAAGGGAGAAGGGTGTGAC | ||||
| SURVIVIN | Fw GTCCCTGGCTCCTCTACTG | 65 | 222 | NM_001168.3 |
| Rv CACTGGGCCTGTCTAATCAC | ||||
| 67LR | Fw GGCTGTGCTGAAGTTTGC | 57 | 216 | NM_002295.6 |
| Rv CCACATAGCGCAGAGGAG | ||||
| CDKN2A | Fw GAAGGTCCTACAGGGCCACA | 68 | 211 | NM_000077.4 |
| Rv CAACACAGTGAAAAGGCAGAAGC | ||||
| ELAVL2 | Fw GACAAACTATGATGAGGCTGC | 68.1 | 330 | NM_004432.5 |
| Rv CCCTGTCCTCTTGTCCATATTC | ||||
| HS6ST2 | Fw CGTACCGCTCGGAGGATG | 63.5 | 313 | NM_001077188.2 |
| Rv GTGAGCTCGGTCCAGTCG | ||||
| ZIC2 | Fw GGAGCAGAGCAACCACGTC | 64.5 | 268 | NM_007129.5 |
| Rv GTGCATGTGCTTCTTCCTGTC | ||||
| 18S | Fw TTTGCGAGTACTCAACACCA | 60 | 280 | [27] |
| Rv GTTGTCCSGSCCSTTGGCTA |
| Protein | Gene Name | Molecular Function/Biological Process | Type of Cancer | p-Value | Reference |
|---|---|---|---|---|---|
| Tumor protein p63 | p63 | DNA binding/transcription, transcription regulation | Cervical cancer | 0.001 | [28] |
| Minichromosome maintenance complex component 7 | MCM7 | DNA binding/cell cycle | Cervical cancer, CIN 3, invasive cancer | 0.002, 0.035 | [29,30,31] |
| Upregulator of cell proliferation | URG4 | Proliferation | Cervical cancer | 0.0001 | [32] |
| Retinoic acid receptor beta | RARβ | DNA binding/transcription, transcription regulation | Cervical cancer | NR | [33] |
| Vascular endothelial growth factor C | VEGFC | Growth factor/angiogenesis | Cervical cancer | 0.002 | [34] |
| Interleukine 10 | IL-10 | Cytokine | Invasive squamous cell carcinoma of the cervix | <0.05 | [35] |
| BCL2 apoptosis regulator | BCL-2 | Apoptosis | Cervical cancer | <0.001 | [36] |
| CDGSH Iron-Sulfur Domain-Containing Protein 2 | CISD2 | RNA binding/Autophagy | Cervical cancer | <0.001 | [37] |
| Cyclooxygenase 2 | COX-2 | Angiogenesis | Cervical cancer | 0.0152 | [38] |
| Tumor suppression protein P73 | TAP73 | P53binding/positive regulation apoptosis process | Cervical cancer | 0.001 | [39] |
| Carbonic anhydrase 9 | CA9 | Proliferation | Uterine cervical cancer | 0.008 | [40] |
| Survivin | SURVIVIN | Apoptosis | Cervical cancer, squamous cell carcinomas | 0.0001 <0.05 | [41,42] |
| Laminin Receptor 67 kD, Ribosomal Protein SA | 67LR | Laminin binding/cell adhesion | Squamous cell carcinomas, carcinoma in situ | 0.0001 | [43] |
| Myc proto-oncogene protein | cMYC | DNA binding transcription factor/proliferation | Cervical cancer, squamous cell carcinoma | <0.0001 <0.05 | [44,45,46] |
| Collagen triple helix repeat-containing | CTHRC1 | Cell migration | Squamous cell carcinoma | <0.001 | [47] |
| Proliferating cell nuclear antigen | PCNA | DNA binding/DNA replication | Squamous cell carcinoma | NR | [48] |
| MET Transcriptional Regulator MACC1 | MACC1 | Growth factor activity/transcription regulator | Cervical cancer | 0.039 | [49] |
| Homeobox protein Hox-C6 | HOXC6 | DNA binding/transcription regulator | Cervical cancer | 0.016 | [50,51] |
| Homeobox protein Hox-C8 | HOXC8 | DNA binding/transcription regulator | Cervical cancer | <0.0001 | [52] |
| Cellular retinoic acid-binding protein 1 | CRABP 1 | Cell cycle | Cervical cancer | <0.001 | [53] |
| Proteins E6/E7 | E6/E7 | DNA binding/transcription regulation, modulation of host cell apoptosis | Cervical cancer | 0.034 | [25] |
| Classification | ||||
|---|---|---|---|---|
| Gene | CIN I | CIN II | CIN III | Cancer |
| CDKN2A | 2.91 | 7.99 | 11.11 | 12.49 |
| ZIC2 | 1.38 | 2.12 | 4.05 | 13.15 |
| ELAVL2 | 2.11 | 2.92 | 4.37 | 7.29 |
| HS6ST2 | 2.67 | 2.90 | 6.28 | 6.51 |
| Hallmark of Cancer | Gene | p-Value |
|---|---|---|
| Cell cycle, cell division, and proliferation | URG4 | 0.0395 * |
| P63 | 0.3191 | |
| MCM7 | 0.1041 | |
| PCNA | 0.0974 | |
| Tap73 | 0.0889 | |
| CRABP1 | 0.4246 | |
| 67LR | 0.0680 | |
| HS6ST2 | 0.0511 | |
| ZIC2 | 0.1618 | |
| HOXC6 | 0.0060 * | |
| HOXC8 | 0.0373 * | |
| RARβ | 0.0031 * | |
| E6/E7 | 0.0078 * | |
| CDKN2A | 0.0001 * | |
| ELAVL2 | 0.0013 * | |
| Immune system | IL-10 | 0.0190 * |
| Apoptosis | Survivin | 0.0047 * |
| BCL2 | 0.0001 * | |
| CISD2 | 0.0086 * | |
| Angiogenesis | COX2 | 0.0524 |
| CTHRC1 | 0.3900 | |
| VEGF | 0.4728 | |
| cMYC | 0.0859 | |
| Invasion and metastasis | MACC1 | 0.0024 * |
| CA9 | 0.0326 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Montoya, I.A.; López-Córdova, K.B.; Marrero-Rodríguez, D.; Salcedo-Vargas, M.; Vargas-Requena, C.L.; Escárcega-Avila, A.M.; Martel-Estrada, S.A.; Jiménez-Vega, F. Five Cellular Genes as Candidates for Cervical Adenocarcinoma Molecular Markers. Cancers 2025, 17, 1558. https://doi.org/10.3390/cancers17091558
García-Montoya IA, López-Córdova KB, Marrero-Rodríguez D, Salcedo-Vargas M, Vargas-Requena CL, Escárcega-Avila AM, Martel-Estrada SA, Jiménez-Vega F. Five Cellular Genes as Candidates for Cervical Adenocarcinoma Molecular Markers. Cancers. 2025; 17(9):1558. https://doi.org/10.3390/cancers17091558
Chicago/Turabian StyleGarcía-Montoya, Isui Abril, Karla Berenice López-Córdova, Daniel Marrero-Rodríguez, Mauricio Salcedo-Vargas, Claudia Lucía Vargas-Requena, Angélica Maria Escárcega-Avila, Santos Adriana Martel-Estrada, and Florinda Jiménez-Vega. 2025. "Five Cellular Genes as Candidates for Cervical Adenocarcinoma Molecular Markers" Cancers 17, no. 9: 1558. https://doi.org/10.3390/cancers17091558
APA StyleGarcía-Montoya, I. A., López-Córdova, K. B., Marrero-Rodríguez, D., Salcedo-Vargas, M., Vargas-Requena, C. L., Escárcega-Avila, A. M., Martel-Estrada, S. A., & Jiménez-Vega, F. (2025). Five Cellular Genes as Candidates for Cervical Adenocarcinoma Molecular Markers. Cancers, 17(9), 1558. https://doi.org/10.3390/cancers17091558







