A Multiparametric MRI and Baseline-Clinical-Feature-Based Dense Multimodal Fusion Artificial Intelligence (MFAI) Model to Predict Castration-Resistant Prostate Cancer Progression
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Image Acquisition
2.3. Data Analysis
2.3.1. α-Mapping Acquisition
2.3.2. T2 Mapping Acquisition
2.3.3. Apparent Diffusion Coefficient Mapping Acquisition
2.3.4. Long-Run High Gray-Level Emphasis Mapping Acquisition
2.4. Data Preparation
2.5. Deep Learning Model Construction
2.6. Multimodal Fusion Artificial Intelligence Model Construction
2.7. Quantitative Assessment Metrics
3. Results
3.1. Patient Characteristics
3.2. Parameter Settings for Each Model
3.3. Predictive Performance of MFAI Models
3.4. Effectiveness of Radiomics Feature Mapping at Predicting CRPC Progression
3.5. Importance of Baseline Clinical Features in Predicting CRPC Progression and Validation of Visualization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCa | Prostate cancer |
| CRPC | Castration-resistant prostate cancer |
| PSA | Prostate-specific antigen |
| mpMRI | Multiparametric magnetic resonance imaging |
| DCE-MRI | Dynamic contrast-enhanced MRI |
| MFAI | Multimodal fusion artificial intelligence |
| DWI | Diffusion-weighted imaging |
| ADC | Apparent diffusion coefficient |
| FOV | Field of view |
| TE | Echo time |
| TR | Repetition time |
| T2WI | T2-weighted imaging |
| PSE | Percentage signal enhancement |
| LRHGLE | Long-run high gray-level emphasis |
| ROI | Region of interest |
| SE block | Squeeze-and-excitation block |
| SPP layer | Spatial pyramid pooling layer |
| BN | Batch normalization |
| ReLU | Rectified linear units |
| Conv | Convolutional layer |
| GAP | Global average pooling |
| FC | Fully connected |
| DenseNet | Dense convolutional network |
| ResNet | Deep residual network |
| PPV | Positive predictive value |
| NPV | Negative predictive value |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| AI | Artificial intelligence |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sharma, R. The burden of prostate cancer is associated with human development index: Evidence from 87 countries, 1990–2016. EPMA J. 2019, 10, 137–152. [Google Scholar] [CrossRef]
- Khauli, R.; Ferrigno, R.; Guimarães, G.; Bulbulan, M.; Uson Junior, P.L.S.; Salvajoli, B.; Palhares, D.M.F.; Racy, D.; Gil, E.; de Arruda, F.F.; et al. Treatment of localized and locally advanced, high-risk prostate cancer: A report from the First Prostate Cancer Consensus Conference for Developing Countries. JCO Glob. Oncol. 2021, 7, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Pernigoni, N.; Zagato, E.; Calcinotto, A.; Troiani, M.; Mestre, R.P.; Calì, B.; Attanasio, G.; Troisi, J.; Minini, M.; Mosole, S.; et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 2021, 374, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Nagappa, A.N.; Bhatt, S.; Kanoujia, J. Studies on Structures and Functions of Kinases leading to Prostate Cancer and Their Inhibitors. Curr. Enzym. Inhib. 2020, 16, 90–105. [Google Scholar] [CrossRef]
- Park, J.; Rho, M.J.; Moon, H.W.; Lee, J.Y. Castration-Resistant Prostate Cancer Outcome Prediction Using Phased Long Short-Term Memory with Irregularly Sampled Serial Data. Appl. Sci. 2020, 10, 2000. [Google Scholar] [CrossRef]
- Ryan, C.J.; Smith, M.R.; Fizazi, K.; Saad, F.; Mulders, P.F.; Sternberg, C.N.; Miller, K.; Logothetis, C.J.; Shore, N.D.; Small, E.J.; et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): Final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015, 16, 152–160. [Google Scholar] [CrossRef]
- de Jong, A.C.; Danyi, A.; van Riet, J.; de Wit, R.; Sjöström, M.; Feng, F.; de Ridder, J.; Lolkema, M.P. Predicting response to enzalutamide and abiraterone in metastatic prostate cancer using whole-omics machine learning. Nat. Commun. 2023, 14, 1968. [Google Scholar] [CrossRef]
- Cornford, P.; Bellmunt, J.; Bolla, M.; Briers, E.; De Santis, M.; Gross, T.; Henry, A.M.; Joniau, S.; Lam, T.B.; Mason, M.D.; et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part II: Treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur. Urol. 2017, 71, 630–642. [Google Scholar] [CrossRef]
- Cornford, P.; van den Bergh, R.C.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer. Part II—2020 update: Treatment of relapsing and metastatic prostate cancer. Eur. Urol. 2021, 79, 263–282. [Google Scholar] [CrossRef]
- Roest, C.; Kwee, T.C.; de Jong, I.J.; Schoots, I.G.; van Leeuwen, P.; Heijmink, S.W.; van der Poel, H.G.; Fransen, S.J.; Saha, A.; Huisman, H.; et al. Development and Validation of a Deep Learning Model Based on MRI and Clinical Characteristics to Predict Risk of Prostate Cancer Progression. Radiol. Imaging Cancer 2025, 7, e240078. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.; Armstrong, A.; Schonhoft, J.; Gill, A.; Zhao, J.; Barnett, E.; Carbone, E.; Lu, J.; Antonarakis, E.; Luo, J.; et al. Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur. J. Cancer 2021, 150, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-H.; Kyung, D.; Yuk, H.D.; Jeong, C.W.; Lee, W.; Yoon, J.-K.; Kim, H.-P.; Bang, D.; Kim, T.-Y.; Lim, Y.; et al. Practical Utility of Liquid Biopsies for Evaluating Genomic Alterations in Castration-Resistant Prostate Cancer. Cancers 2023, 15, 2847. [Google Scholar] [CrossRef] [PubMed]
- McGrath, S.; Christidis, D.; Perera, M.; Hong, S.K.; Manning, T.; Vela, I.; Lawrentschuk, N. Prostate cancer biomarkers: Are we hitting the mark? Prostate Int. 2016, 4, 130–135. [Google Scholar] [CrossRef]
- A, J.; Zhang, B.; Zhang, Z.; Hu, H.; Dong, J.-T. Novel Gene Signatures Predictive of Patient Recurrence-Free Survival and Castration Resistance in Prostate Cancer. Cancers 2021, 13, 917. [Google Scholar] [CrossRef]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Hara, N.; Okuizumi, M.; Koike, H.; Kawaguchi, M.; Bilim, V. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is a useful modality for the precise detection and staging of early prostate cancer. Prostate 2005, 62, 140–147. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Liberini, V.; Laudicella, R.; Balma, M.; Nicolotti, D.G.; Buschiazzo, A.; Grimaldi, S.; Lorenzon, L.; Bianchi, A.; Peano, S.; Bartolotta, T.V.; et al. Radiomics and artificial intelligence in prostate cancer: New tools for molecular hybrid imaging and theragnostics. Eur. Radiol. Exp. 2022, 6, 27. [Google Scholar] [CrossRef]
- Esteva, A.; Feng, J.; van der Wal, D.; Huang, S.-C.; Simko, J.P.; DeVries, S.; Chen, E.; Schaeffer, E.M.; Morgan, T.M.; Sun, Y.; et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. npj Digit. Med. 2022, 5, 71. [Google Scholar] [CrossRef]
- Chatterjee, A.; He, D.; Fan, X.; Antic, T.; Jiang, Y.; Eggener, S.; Karczmar, G.S.; Oto, A. Diagnosis of prostate cancer by use of MRI-derived quantitative risk maps: A feasibility study. Am. J. Roentgenol. 2019, 213, W66–W75. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; He, D. Multiparametric MRI-based artificial intelligence analysis to predict hormone therapy effectiveness for prostate cancer: A clustering-driven ensemble-learning approach. In Proceedings of the China Biomedical Engineering Conference & Innovative Healthcare Summit, Shenzhen, China, 20–22 September 2024; p. 2825. [Google Scholar]
- Sun, C.; Chatterjee, A.; Yousuf, A.; Antic, T.; Eggener, S.; Karczmar, G.S.; Oto, A. Comparison of T2-weighted imaging, DWI, and dynamic contrast-enhanced MRI for calculation of prostate cancer index lesion volume: Correlation with whole-mount pathology. Am. J. Roentgenol. 2019, 212, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Heerkens, H.; Hall, W.; Li, X.; Knechtges, P.; Dalah, E.; Paulson, E.; van den Berg, C.; Meijer, G.; Koay, E.; Crane, C.; et al. Recommendations for MRI-based contouring of gross tumor volume and organs at risk for radiation therapy of pancreatic cancer. Pract. Radiat. Oncol. 2017, 7, 126–136. [Google Scholar]
- Dalah, E.; Moraru, I.; Paulson, E.; Erickson, B.; Li, X.A. Variability of target and normal structure delineation using multimodality imaging for radiation therapy of pancreatic cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 633–640. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Q.; Li, Y.; Yang, Z.; Shen, L. Medaugment: Universal Automatic Data Augmentation Plug-in for Medical Image Analysis. arXiv 2023, arXiv:2306.17466. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Zhuang, H.; Li, B.; Ma, J.; Monkam, P.; Qian, W.; He, D. An attention-based deep learning network for predicting Platinum resistance in ovarian cancer. IEEE Access 2024, 12, 41000–41008. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Xu, X.; Feng, Z.; Cao, C.; Li, M.; Wu, J.; Wu, Z.; Shang, Y.; Ye, S. An improved swin transformer-based model for remote sensing object detection and instance segmentation. Remote Sens. 2021, 13, 4779. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Zheng, X.; Yao, Z.; Huang, Y.; Yu, Y.; Wang, Y.; Liu, Y.; Mao, R.; Li, F.; Xiao, Y.; Wang, Y.; et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat. Commun. 2020, 11, 1236. [Google Scholar] [CrossRef]
- Ali, M.; Benfante, V.; Cutaia, G.; Salvaggio, L.; Rubino, S.; Portoghese, M.; Ferraro, M.; Corso, R.; Piraino, G.; Ingrassia, T.; et al. Prostate Cancer Detection: Performance of Radiomics Analysis in Multiparametric MRI. In Image Analysis and Processing-ICIAP 2023 Workshops; Springer: Berlin/Heidelberg, Germany, 2023; pp. 83–92. [Google Scholar]
- Zhou, C.; Zhang, Y.-F.; Guo, S.; Huang, Y.-Q.; Qiao, X.-N.; Wang, R.; Zhao, L.-P.; Chang, D.-H.; Zhao, L.-M.; Da, M.-X.; et al. Multimodal data integration for predicting progression risk in castration-resistant prostate cancer using deep learning: A multicenter retrospective study. Front. Oncol. 2024, 14, 1287995. [Google Scholar] [CrossRef]
- Jin, T.; An, J.; Wu, W.; Zhou, F. Development and validation of a machine learning model for bone metastasis in prostate cancer: Based on inflammatory and nutritional indicators. Urology 2024, 190, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-F.; Zhou, C.; Guo, S.; Wang, C.; Yang, J.; Yang, Z.-J.; Wang, R.; Zhang, X.; Zhou, F.-H. Deep learning algorithm-based multimodal MRI radiomics and pathomics data improve prediction of bone metastases in primary prostate cancer. J. Cancer Res. Clin. Oncol. 2024, 150, 78. [Google Scholar] [CrossRef] [PubMed]
- Cairone, L.; Benfante, V.; Bignardi, S.; Marinozzi, F.; Yezzi, A.; Tuttolomondo, A.; Salvaggio, G.; Bini, F.; Comelli, A. Robustness of radiomics features to varying segmentation algorithms in magnetic resonance images. In Image Analysis and Processing-ICIAP 2023 Workshops; Springer: Berlin/Heidelberg, Germany, 2022; pp. 462–472. [Google Scholar]

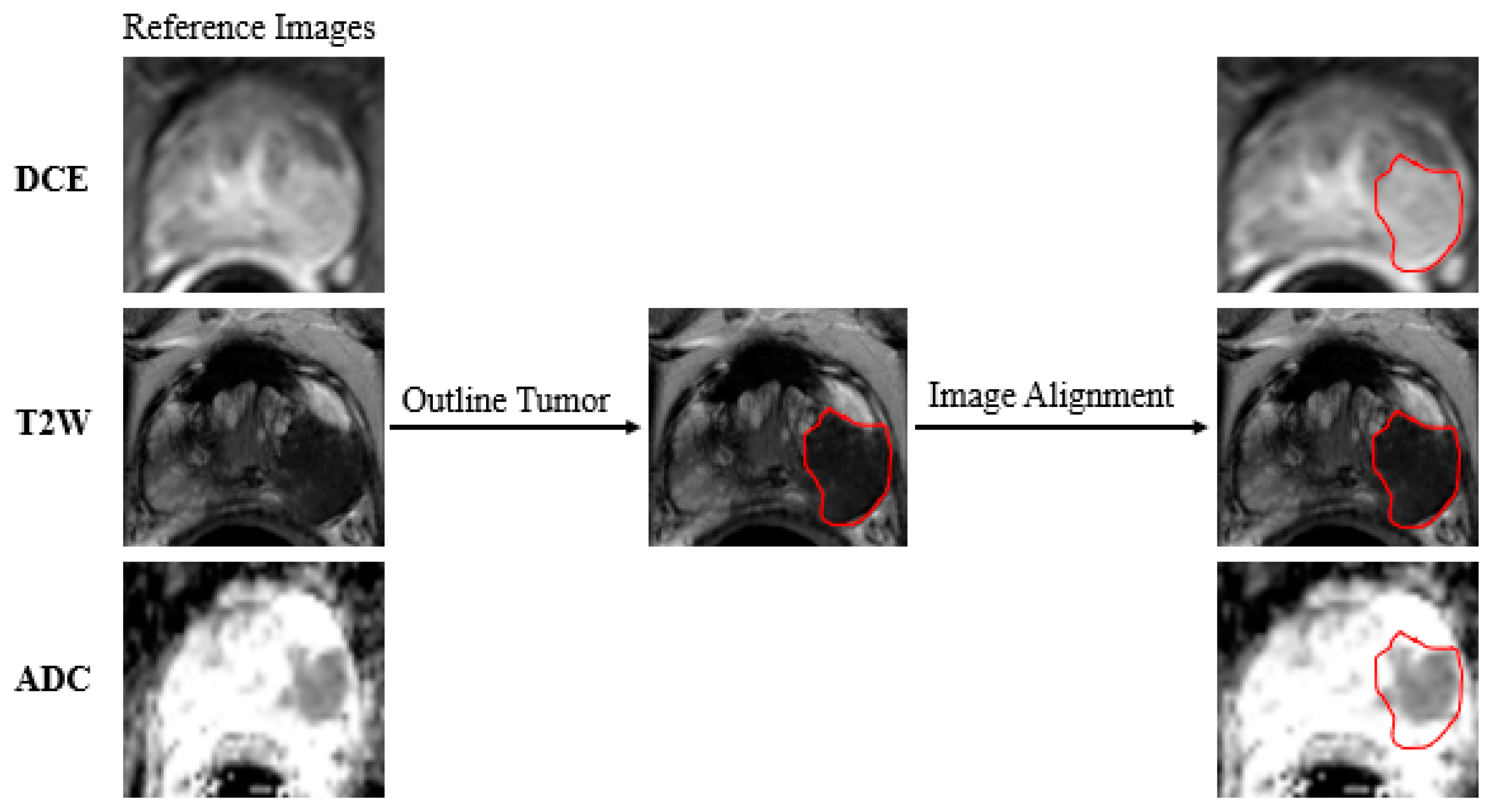

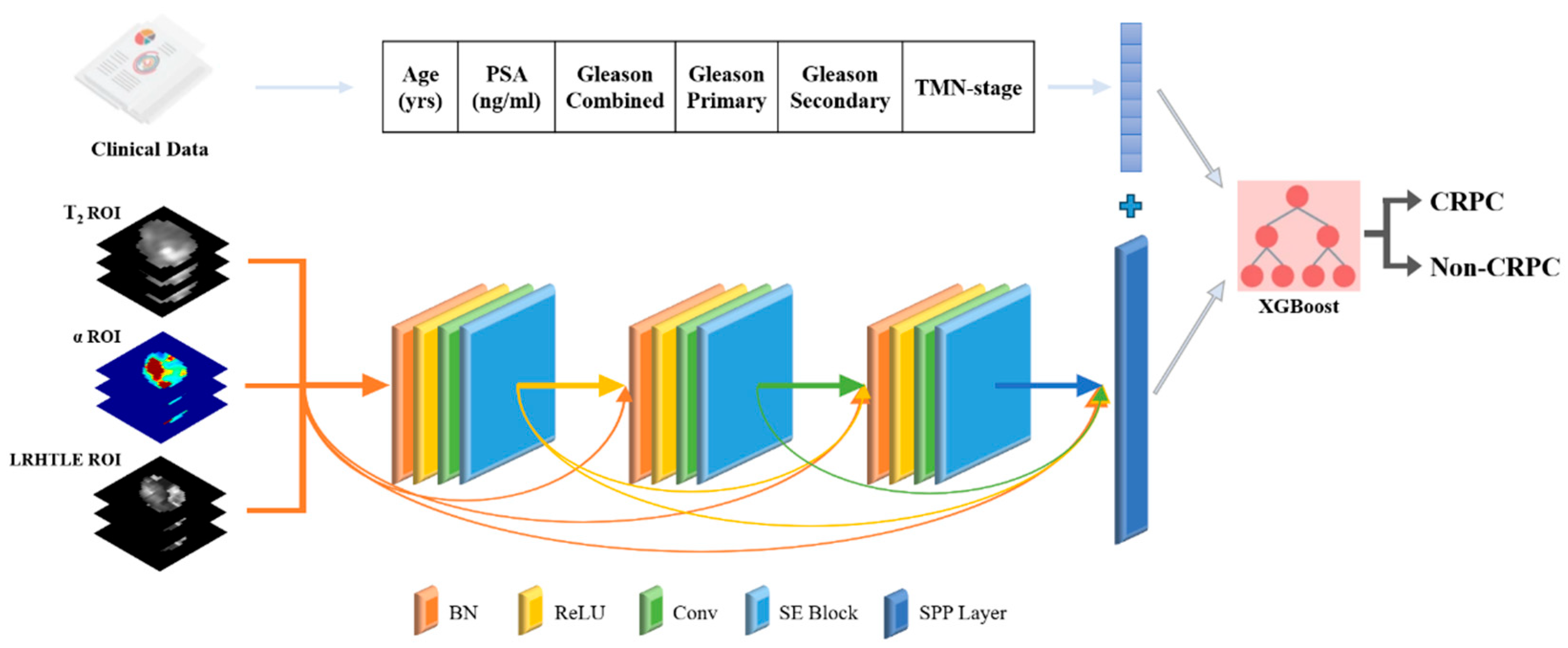
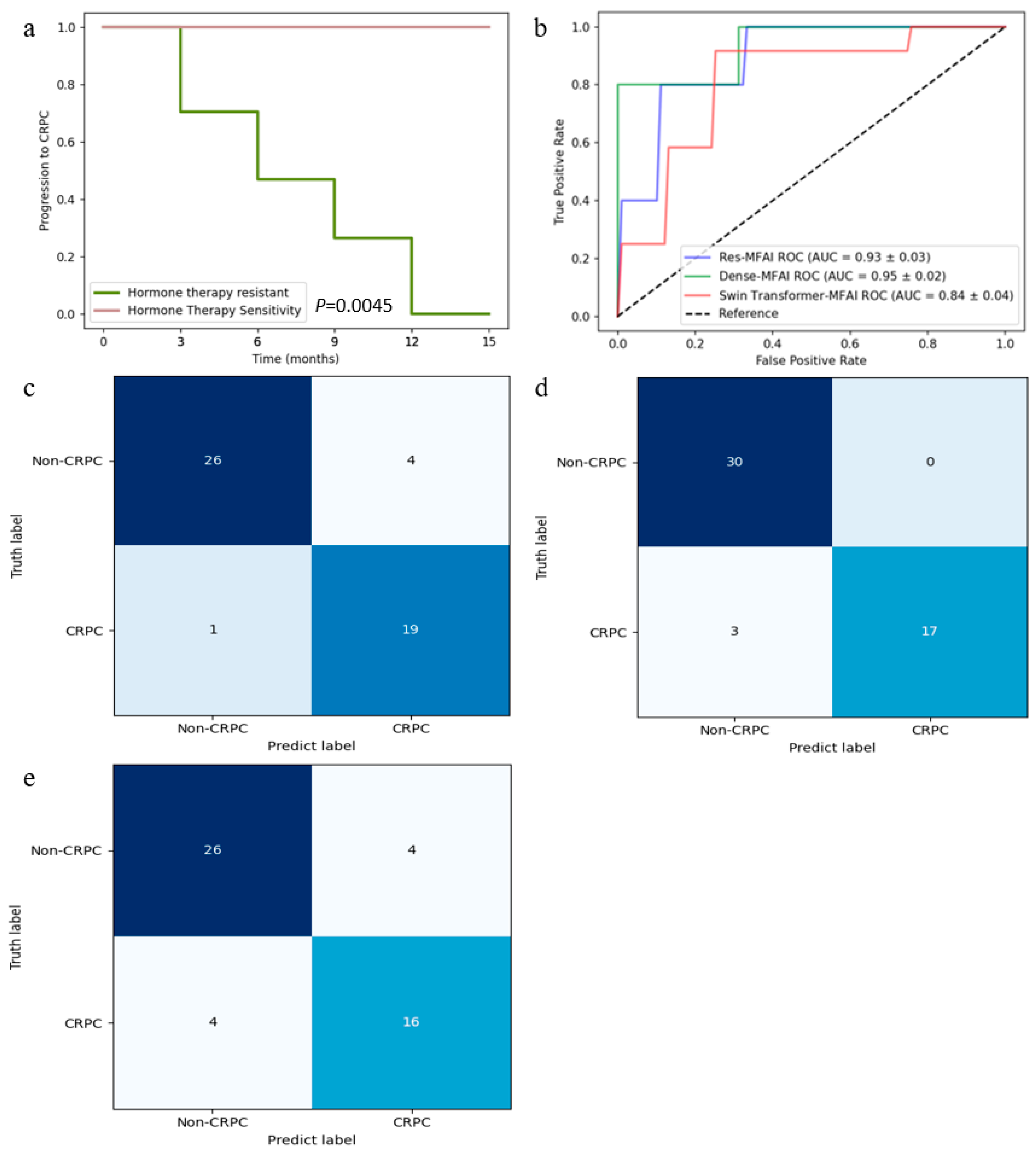
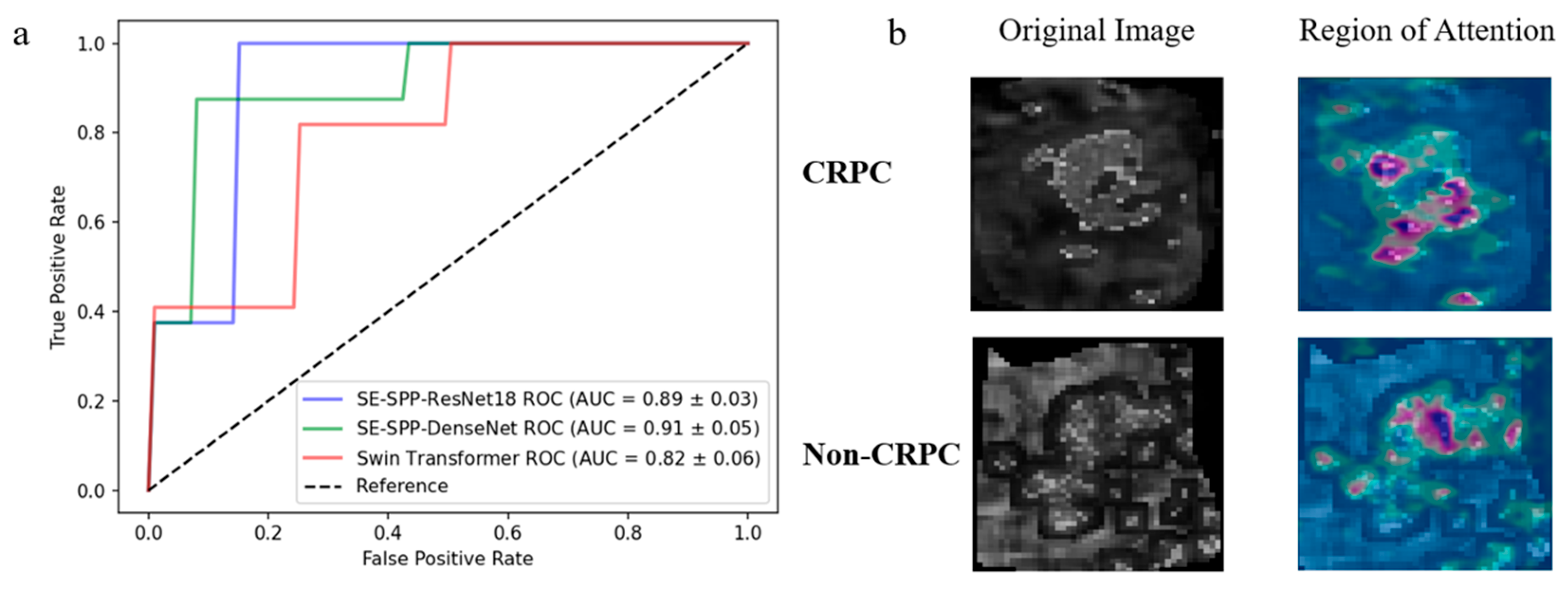
| Imaging Sequence | FOV (mm) | Scan Matrix Size | TE (ms) | TR (ms) | Slice Thickness (mm) | Interval (mm) | Flip Angle (°) |
|---|---|---|---|---|---|---|---|
| T2WI | 200 | 512 × 512 | 128 | 500 | 3 | 0.6 | 90 |
| DWI | 260 | 120 × 96 | 72 | 400 | 3 | 0.6 | / |
| DCE | 340 | 203 × 320 | 1.3 | 3.4 | 3.5 | 0 | 9 |
| Parameter | Epoch | Learning Rate | Batch Size | Momentum | Weight Decay | Optimizer |
|---|---|---|---|---|---|---|
| ResNet18 | 50 | 0.001 | 48 | 0.9 | 0.0001 | SGD |
| DenseNet | 50 | 0.0001 | 48 | 0.9 | 0.0001 | SGD |
| Swin Transformer | 50 | 0.001 | 64 | 0.9 | 0.0001 | SGD |
| Accuracy | Sensitivity | Specificity | PPV | NPV | ||
|---|---|---|---|---|---|---|
| SE-SPP-ResNet18 | T2-ADC | 76.3% | 74.6% | 80.3% | 78.1% | 75.7% |
| T2-α | 83.5% | 80.3% | 87.7% | 87.3% | 80.5% | |
| T2-ADC-α | 85.2% | 83.6% | 87.0% | 87.5% | 83.6% | |
| T2-ADC-LRHGLE | 80.4% | 80.7% | 86.4% | 86.7% | 81.8% | |
| T2-α-LRHGLE | 90.3% | 83.3% | 100.0% | 100.0% | 81.6% | |
| SE-SPP-DenseNet | T2-ADC | 77.6% | 80.5% | 75.1% | 77.2% | 78.5% |
| T2-α | 85.1% | 90.4% | 75.2% | 80.4% | 91.2% | |
| T2-ADC-α | 87.7% | 89.6% | 87.8% | 88.9% | 88.7% | |
| T2-ADC-LRHGLE | 83.8% | 82.4% | 80.7% | 81.2% | 81.8% | |
| T2-α-LRHGLE | 91.2% | 91.9% | 87.3% | 91.7% | 87.8% | |
| Swin Transformer | T2-ADC | 67.9% | 75.4% | 54.7% | 62.2% | 69.3% |
| T2-α | 73.1% | 71.4% | 75.4% | 75.8% | 70.5% | |
| T2-ADC-α | 75.5% | 82.4% | 68.7% | 70.3% | 76.7% | |
| T2-ADC-LRHGLE | 71.5% | 79.7% | 62.7% | 69.9% | 74.0% | |
| T2-α-LRHGLE | 78.9% | 98.1% | 51.7% | 73.3% | 96.8% |
| Accuracy | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|
| Baseline Clinical Features | 70.4% | 87.1% | 41.6% | 72.5% | 67.7% |
| SE-SPP-ResNet18 | 90.3% | 83.3% | 100.0% | 100.0% | 81.6% |
| SE-SPP-ResNet18 + MFAI | 92.7% | 87.5% | 96.4% | 97.5% | 84.1% |
| SE-SPP-DenseNet | 91.2% | 91.9% | 87.3% | 91.7% | 87.8% |
| SE-SPP-DenseNet + MFAI | 94.2% | 100.0% | 82.1% | 89.6% | 100.0% |
| Swin Transformer | 78.9% | 98.1% | 51.7% | 73.3% | 96.8% |
| Swin Transformer + MFAI | 85.2% | 87.8% | 81.4% | 86.5% | 80.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, D.; Zhuang, H.; Ma, Y.; Xia, B.; Chatterjee, A.; Fan, X.; Qi, S.; Qian, W.; Zhang, Z.; Liu, J. A Multiparametric MRI and Baseline-Clinical-Feature-Based Dense Multimodal Fusion Artificial Intelligence (MFAI) Model to Predict Castration-Resistant Prostate Cancer Progression. Cancers 2025, 17, 1556. https://doi.org/10.3390/cancers17091556
He D, Zhuang H, Ma Y, Xia B, Chatterjee A, Fan X, Qi S, Qian W, Zhang Z, Liu J. A Multiparametric MRI and Baseline-Clinical-Feature-Based Dense Multimodal Fusion Artificial Intelligence (MFAI) Model to Predict Castration-Resistant Prostate Cancer Progression. Cancers. 2025; 17(9):1556. https://doi.org/10.3390/cancers17091556
Chicago/Turabian StyleHe, Dianning, Haoming Zhuang, Ying Ma, Bixuan Xia, Aritrick Chatterjee, Xiaobing Fan, Shouliang Qi, Wei Qian, Zhe Zhang, and Jing Liu. 2025. "A Multiparametric MRI and Baseline-Clinical-Feature-Based Dense Multimodal Fusion Artificial Intelligence (MFAI) Model to Predict Castration-Resistant Prostate Cancer Progression" Cancers 17, no. 9: 1556. https://doi.org/10.3390/cancers17091556
APA StyleHe, D., Zhuang, H., Ma, Y., Xia, B., Chatterjee, A., Fan, X., Qi, S., Qian, W., Zhang, Z., & Liu, J. (2025). A Multiparametric MRI and Baseline-Clinical-Feature-Based Dense Multimodal Fusion Artificial Intelligence (MFAI) Model to Predict Castration-Resistant Prostate Cancer Progression. Cancers, 17(9), 1556. https://doi.org/10.3390/cancers17091556








