Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation
Simple Summary
Abstract
1. Introduction
1.1. Short Open Reading Frames and Translational Potential
1.2. Methods to Detect Translating ncRNAs and the Peptides They Encode
1.3. Translational Mechanisms Among ncRNA ORFs
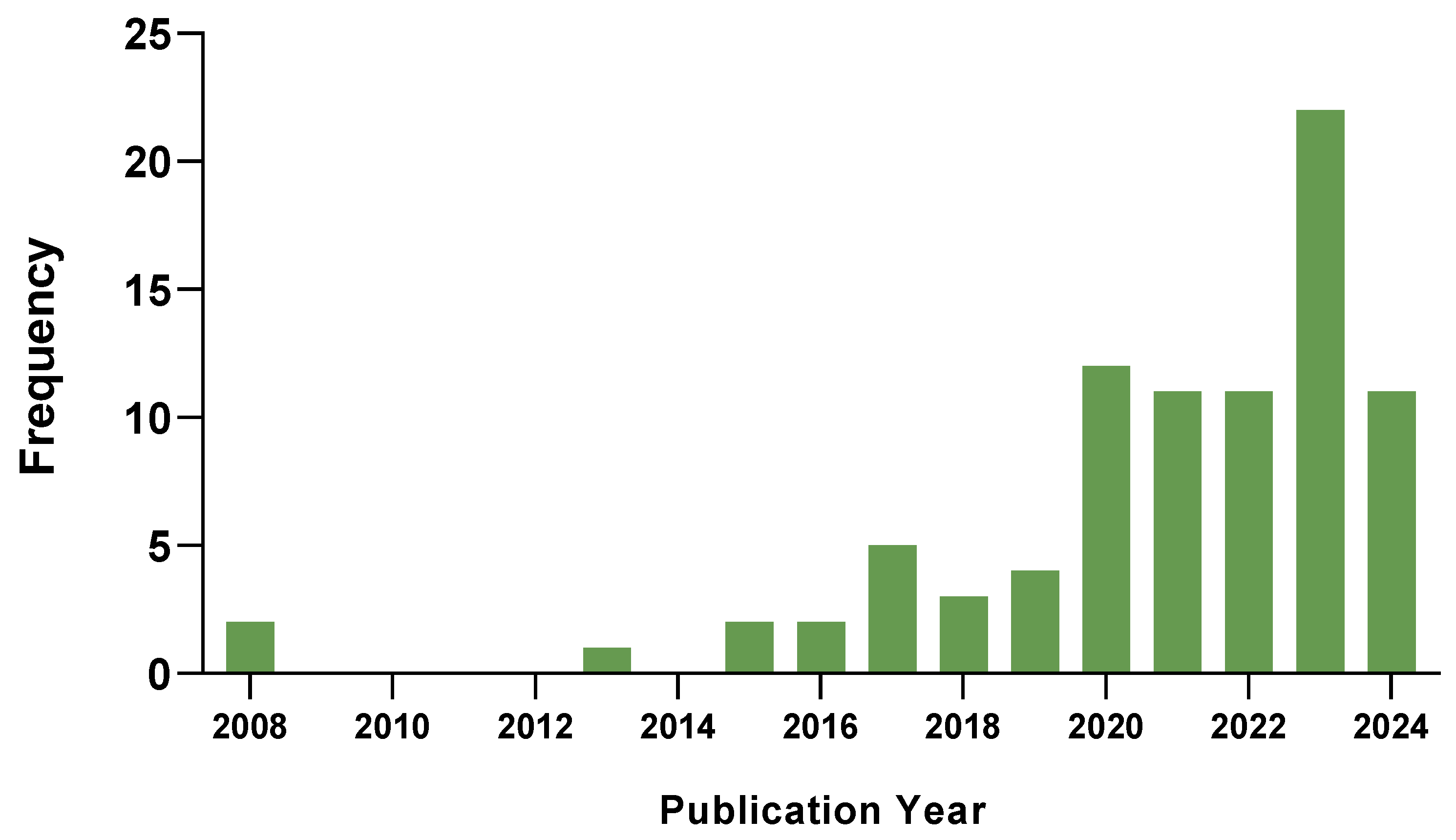
2. Search Approaches and Data Statistics
2.1. Workflow and Data Collection
2.2. Classification of ncRNA-PEP Encoding ncRNAs
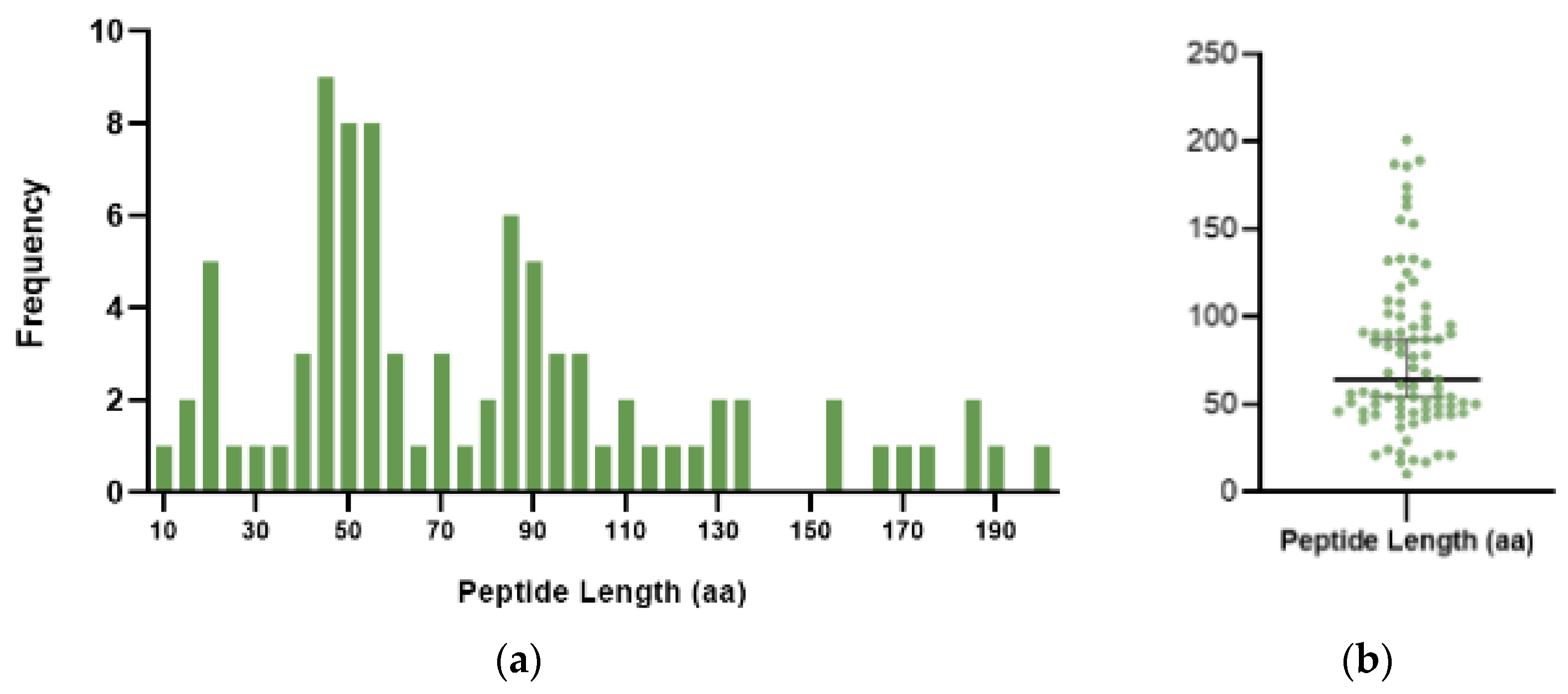
2.3. Size Distribution of ncRNA-PEPs
| Peptide | Transcript | Length (aa) | Function | Sources | |
|---|---|---|---|---|---|
| 1 | AC115619-22aa | AC115619 | 22 | Prevents assembly of the m6A methyltransferase complex and inhibits HCC progression | [41] |
| 2 | AF127577.4-ORF | lncRNA AF127577.4 | 29 | Anti-proliferating function in GBM by reducing both METTL3’s protein stability interaction with ERK2 | [42] |
| 3 | ASRPS | LINC00908 | 60 | Inhibits STAT3 activation, lowering VEGF expression and suppressing tumor angiogenesis | [43] |
| 4 | BVES-AS1-201-50aa | BVES-AS1, C6orf112 | 50 | Enhances CRC invasion ability via Src/mTOR pathway | [44] |
| 5 | CIP2A-BP | LINC00665 | 52 | Directly binds to the oncogene CIP2A and inhibits the migration and invasion of TNBC | [45] |
| 6 | LINC00665_18aa | LINC00665 | 18 | Modulates the CREB1/RPS6KA3 axis to suppresses the proliferation and migration of osteosarcoma cells | [46] |
| 7 | Dleu2-17aa | Dleu2 | 17 | Maintains immune homeostasis by promoting SMAD3 mediated Treg induction | [47] |
| 8 | HOXB-AS3 peptide | HOXB-AS3 | 53 | Competitively binds hnRNP A1 and suppresses glucose metabolism reprogramming in CRC cells | [48] |
| 9 | IMP | VLDLR | 44 | Regulates inflammatory gene expression | [49] |
| 10 | LINC00339-205-49aa | LINC00339 | 49 | Promotes trophoblast adhesion to endometrial cells and contributes to endometrial receptivity (ER) | [50] |
| 11 | LINC00954-ORF polypeptide | LINC00954 | 49 | Promotes pemetrexed sensitivity in drug resistant adenocarcinoma cells | [51] |
| 12 | LINC01013-ORF encoded polypeptide | LINC01013 | 56 | Involved in TGFβ1-mediated fibroblast activation | [52] |
| 13 | M1 peptide | MALAT1 | 43 | Involved in synaptic function | [53] |
| 14 | MAGI2-AS3-ORF5 | MAGI2-AS3 | 45 | Anti-tumor role by inhibiting BC cell viability, possibly though interaction with ECM proteins | [54] |
| 15 | MELOE-1 | meloe | 39 | Involved in T cell immune surveillance; optimal T cell target for melanoma immunotherapy | [55] |
| 16 | MELOE-2 | meloe | 46 | Involved in T cell immune surveillance; optimal T cell target for melanoma immunotherapy | [55] |
| 17 | MELOE-3 | meloe | 54 | Expressed in normal melanocytes—not immunogenic like MELOE-1 and MELOE-2 | [28,55] |
| 18 | MIAC | AQP5-AS1/lncRNA AC025154.2 | 51 | Interacts with AQP2 and inhibits tumor growth and metastasis in HNSCC | [56,57] |
| 19 | miPEP-200b | pri-miR-200b | 54 | Inhibit the EMT of prostate cancer cells by regulating the vimentin mediated pathway | [58] |
| 20 | miPEP155 | MIR155HG | 17 | Suppresses autoimmune inflammation by controlling antigen presentation on cells | [7] |
| 21 | miPEP31 | pri-miRNA-31 | 44 | Represses autoimmunity by promoting Treg differentiation | [40] |
| 22 | miPEP497 | pri-miR-497 | 21 | miPEP497 does not regulate the levels of its own pri-miRNAs, functions not charecterised | [38] |
| 23 | MLN/myoregulin | LINC-RAM/LINC00948 | 46 | Interacts directly with SERCA and regulates muscle performance | [59] |
| 24 | MTLN/LEMP/MOXI/SMIM37/MPM/NCRNA00116 | LINC00116/1500011K16Rik | 56 | Multifunctional protein with roles in respiratory efficiency, beta oxidation of fatty acids and myogenesis | [60,61,62] |
| 25 | N1DARP | LINC00261 | 41 | Acts as a tumor suppressor and chemosensitizer by regulating USP10-Notch1 oncogenic signaling | [63] |
| 26 | PACMP | MARCHF6-DT/CTD-2256P15.2 | 44 | Mediates drug resistance and cancer progression via promoting HR, MMEJ, and PARylation. | [64] |
| 27 | pep-AP | lnc-AP | 37 | Modulates the pentose phosphate pathway and sensitizes CRC cells to Oxaliplatin | [65] |
| 28 | PIGBOS | Antisense to PIGB | 54 | Regulates the unfolded protein response (UPR) in the endoplasmic reticulum and cell death | [66] |
| 29 | pTUNAR | TUNAR/LINC00617 | 48 | Regulates neural differentiation and neurite formation by modulating calcium dynamics | [67] |
| 30 | PVT1 | lncRNA PVT1 | 10 | PVT1 is a tumor antigen recognized by CD8 tumor-infiltrating lymphocytes and mononuclear cells | [68] |
| 31 | RNF217-AS1 ORF3-encoded peptide | RNF217-AS1 | 42 | Inhibits macrophage recruitment, pro-inflammatory responses and stomach cancer tumorigenesis | [69] |
| 32 | SLT | LINC02099 | 24 | Cytoprotective capacity in cardiomyocytes | [29] |
| 33 | SMIM30 | LINC00998 | 59 | Promotes the G1/S transition of cell cycle by regulating cytosolic calcium level | [70,71] |
| 34 | SNHG6 ORF#2 | SNHG6 | 45 | Regulates TGF-β/SMAD pathway, influencing endometrial cell development and potentially, gynecological diseases | [72] |
| 35 | sPEP1 | HNF4A-AS1 | 51 | Facilitates the transcriptional upregulation of stem cell genes related to tumor progression in NB | [73] |
| 36 | STMP1/MM47 | lncRNA 1810058I24Rik in mice | 47 | Regulates mitochondrial function to drive Nlrp3 inflammasome activation, cell differentiation, and metastasis. | [74,75,76] |
| 37 | STORM | LINC00689 | 50 | Molecular mimic of SRP19, with which it competes for 7S RNA binding (involved in translational regulation) | [77] |
| 38 | ELABELA/Toddler | APELA/Gm10664 | 54 | Second endogenous ligand for the Apelin receptor with cardiovascular and developmental roles | [78,79] |
| 39 | XBP1SBM | MLLT4-AS1 | 21 | Promotes the expression of VEGF, angiogenesis and metastasis through XBP1s pathway | [80] |
| 40 | YY1BM | LINC00278 (Y-linked) | 21 | Promotes the apoptosis of ESCC cells, m6a modification changes peptide expression in smokers | [33] |
| 41 | ZFAS1 | Antisense to ZNFX1 | 57 | Promotes higher ROS and HCC cell migration | [81] |
| Microprotein | Transcript | Length (aa) | Function | Sources | |
|---|---|---|---|---|---|
| 1 | 115127-microprotein | NONHSAT115127.2 | 77 | Redox stress in extracellular vesicles derived from glioma cancer cells | [20] |
| 2 | ACLY-BP | LINC00887 | 91 | Promotes lipid deposition through ACLY stabilization and promotes clear cell renal cell carcinoma | [82] |
| 3 | APPLE | ASH1L-AS1 | 90 | Controls eIF4F complex assembly and translation initiation to promote the production of oncoproteins | [83] |
| 4 | Arteridin | LncRNA PSR | 106 | Promotes pathogenic phenotype switching of vascular smooth muscle cells through YBX1 interaction | [84] |
| 5 | ASAP | LINC00467 | 94 | Enhances mitochondrial ATP production by stimulating ATP synthase activity and oxygen consumption | [85] |
| 6 | ATMLP | AFAP1-AS1 | 90 | Suppresses autolysosome formation and mitophagy to increase tumor cell viability | [32] |
| 7 | Aw112010 | lncRNA Aw112010 | 78 | Drives IL-12 production and mediates innate immune response | [86] |
| 8 | C20orf204-189AA | Linc00176 | 189 | Stabilizes nucleolin and promotes HCC | [87] |
| 9 | CASIMO1/SIM22 | lncRNA CASIMO1 | 83 | Interacts with squalene epoxidase to influence cell proliferation and cycle progression | [18] |
| 10 | CRNDEP | CRNDE | 84 | Promotes resistance of OvCa cells to treatment with microtubule-targeted cytostatics | [19,88] |
| 11 | DDUP | lncRNA CTBP1-DT | 186 | Contributes to resistance by maintaining Rad18 at damage sites and promoting DNA damage repair | [36] |
| 12 | FORCP | LINC00675 | 79 | Regulates apoptosis in response to ER stress to prevent CRC progression | [5] |
| 13 | GT3-INCP | LINC00992 | 120 | interacted with GATA3 to coregulate the expression of genes key to the proliferation of ER+ BC cells | [89] |
| 14 | HBVPTPAP | lncRNA HBVPTPAP | 94 | Induces the apoptosis of HCC cells by modulating JAK/STAT signaling pathways | [90] |
| 15 | HCP5-132aa | HCP5 | 132 | contributes to adriamycin resistance and regulates ferroptosis to promote BC | [91,92] |
| 16 | IGF2-AS-168aa | IGF2-AS | 168 | Induces trophoblast cell cycle arrest | [93] |
| 17 | JunBP | LINC02551 | 174 | Promotes HCC metastasis through c-Jun activation | [94] |
| 18 | KRASIM | NCBP2-AS2 | 99 | Decreases KRAS protein levels and the downstream ERK signaling in HCC cells | [95] |
| 19 | LINC00511-133aa | LINC00511 | 133 | Promotes the stemness of BC cells via activation of the Wnt/β-catenin pathway | [96] |
| 20 | LINC01128-MP | LINC01128 | 91 | Role in intracellular trafficking and endocytosis | [97] |
| 21 | Linc013026-68AA | Linc013026/NONHSAT013026.2 | 68 | Role in the proliferation of HCC cells | [98] |
| 22 | Lnc-DLX6-AS1 peptide | DLX6-AS1 | - | May facilitate NSCLC cell growth by activating the Wnt/β-catenin pathway | [99] |
| 23 | MBOP | LINC01234 | 85 | Promoted the expression of MEK1 and activated the MEK1/pERK/MMP2/MMP9 signaling pathway in CRC | [100] |
| 24 | MFRLP | LncRNA-MFRL/MSTRG109 | 64 | Inhibits the pathogenic phenotype switch of vascular smooth muscle cells and improves arterial remodeling | [101] |
| 25 | miPEP-200a | pri-miR-200a | 187 | Inhibit the EMT of prostate cancer cells by regulating the vimentin mediated pathway | [58] |
| 26 | miPEP133 | pri-miR-34a | 133 | Leads to mitochondrial energy loss and reduced ATP production, inhibiting cancer progression | [39] |
| 27 | MRP | LY6E-DT | 153 | Increases EGFR mRNA stability and transcriptionally activates ZEB1 to promote BC | [31] |
| 28 | NBASP/F201A | FAM201A | 155 | Inhibits neuroblastoma progression by reducing FABP5 expression and inactivating the MAPK pathway | [102] |
| 29 | NoBody | LINC01420 | 68 | Regulator of mRNA stability through P-body interaction | [103] |
| 30 | pep-AKR1C2 | lncAKR1C2 | 163 | Promotes gastric cancer lymph node metastasis by regulating fatty acid metabolism | [104] |
| 31 | Pep-KDM4A-AS1 | KDM4A-AS1 | 61 | Function in the fatty acid metabolism pathway and reduces ESCC cell viability and migratory capacity | [105] |
| 32 | PINT87aa | LINC-PINT (circPINT-exon2) | 87 | Induces cell cycle arrest and cellular senescence in HCC cells | [106,107] |
| 33 | pTINCR/TUBL | TINCR | 87 | Promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation | [108,109,110] |
| 34 | RASON | LINC00673 | 108 | Maintains KRAS in the GTP-bound hyperactive state | [111] |
| 35 | RBRP | LINC00266-1 | 71 | Regulates m6A recognition by the m6A reader IGF2BP1 and promotes tumorigenesis | [112] |
| 36 | RIP | LncRNA DGCR5 | 102 | Aggravates steroid-induced osteonecrosis of the femoral head (SONFH) in bone marrow mesenchymal stem cells | [113] |
| 37 | SMIM26 | LINC00493 | 95 | Controls AGK localization and AKT phosphorylation to suppresses CRC metastatic potential | [34] |
| 38 | SP0495 | KIAA0495, TP73-AS1 | 201 | Tumor suppressive protein that binds to phosphoinositides, promotes autophagy and represses oncogenic signaling | [114] |
| 39 | SPAR | LINC00961 | 90 | Regulates Skeletal Muscle Regeneration by inhibiting mTORC1 | [6] |
| 40 | SRSP | LOC90024 | 130 | Promotes oncogenic mRNA spicing through interaction with splicing regulators | [115] |
| 41 | TAT-MIR7-3HG-ORF | MIR7-3HG | 125 | Protects pancreatic β-cells from dexamethasone induced dysfunction by activating the PI3K/AKT pathway | [116] |
| 42 | TP53LC02 | lncRNA AC022075.1, FAM169A-AS1 | 109 | Suppresses cell proliferation | [117] |
| 43 | TP53LC04 | AC022075.1, KLRK1-AS1 | 100 | Suppresses cell proliferation and regulates the cell cycle in response to DNA damage | [117] |
| 44 | U9-ORF protein | U90926 | 87 | Secreted by activated myeloid cells, function not fully characterized | [118] |
| 45 | UBAP1-AST6 | LncRNA UBAP1-AST6 | 117 | Cell proliferation associated function in lung cancer | [22] |
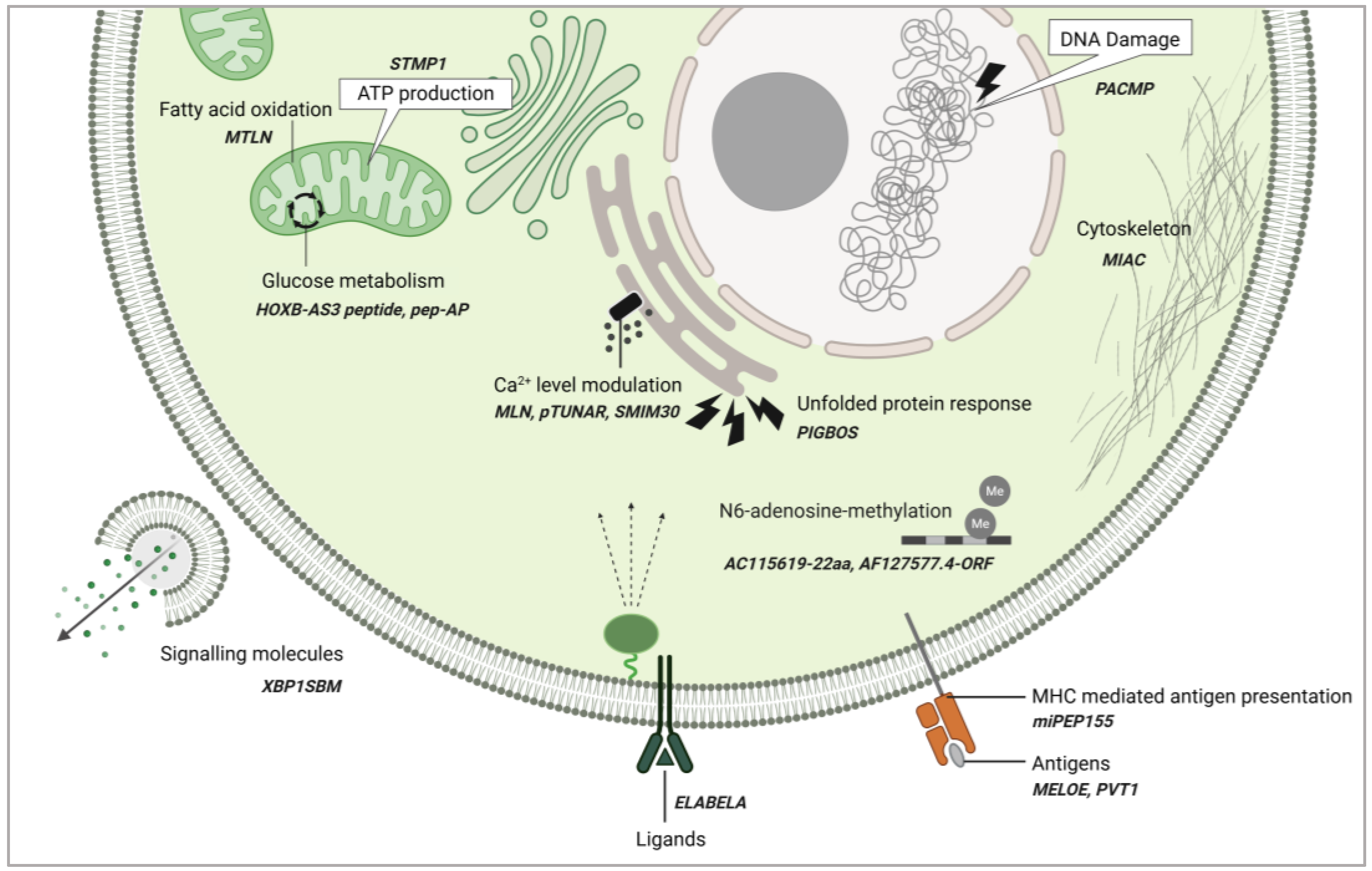
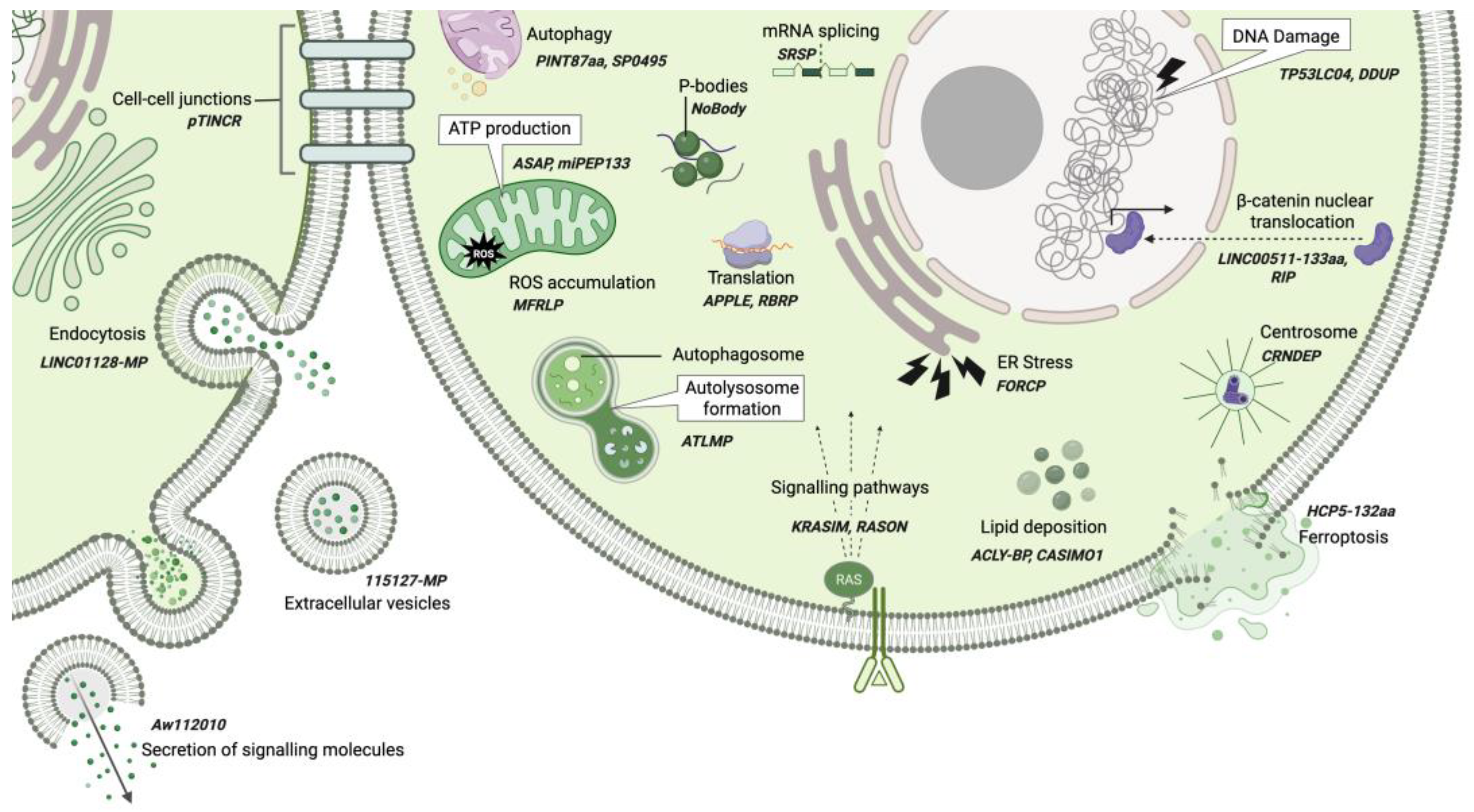
3. Broad Functional Roles of ncRNA-PEPs and ncRNA-MPs
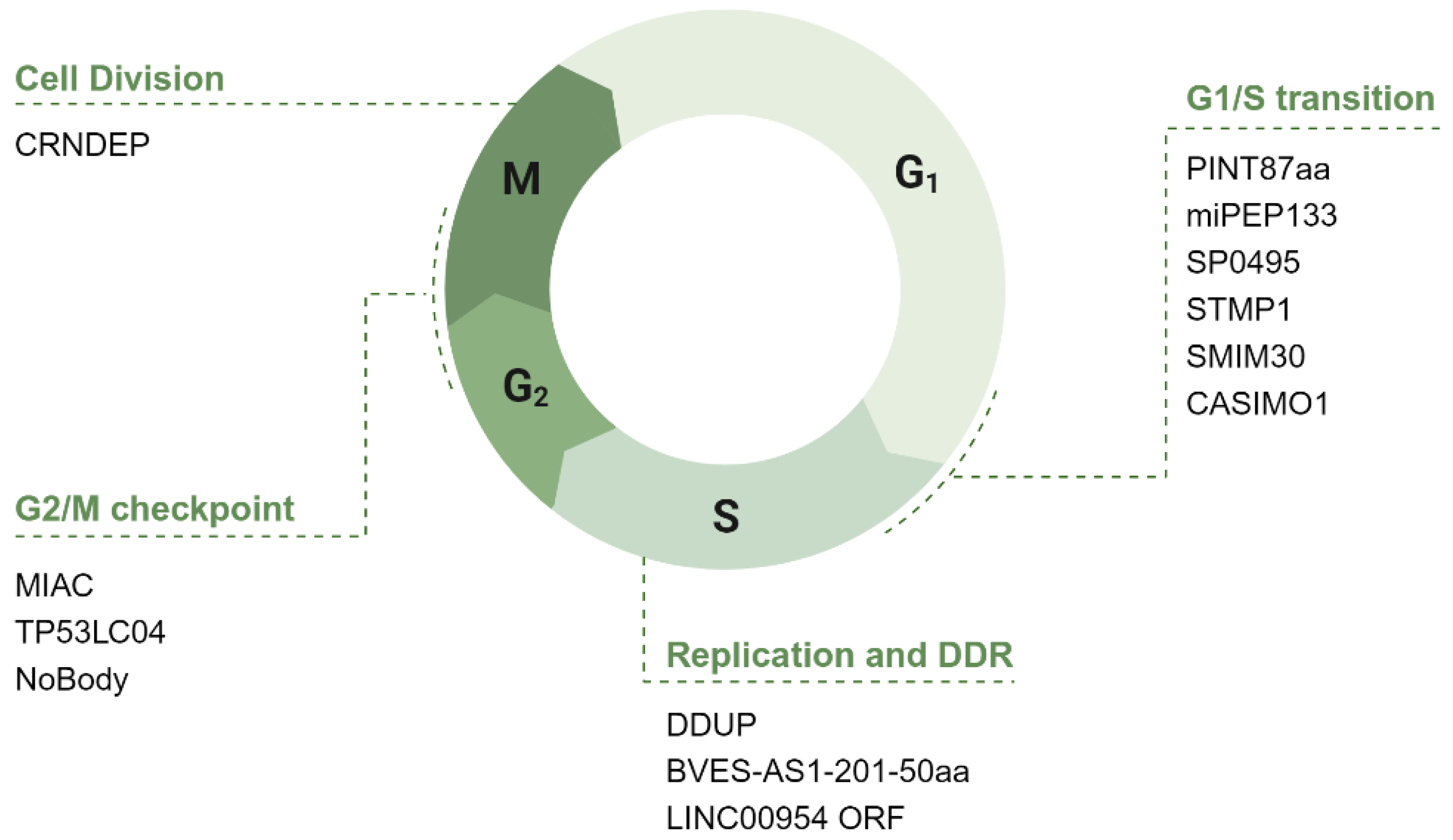
4. ncRNA-PEP/MPs in Cell Cycle Regulation
4.1. The Potential Role of ncRNA-PEP/MPs in G1 Phase of the Cell Cycle
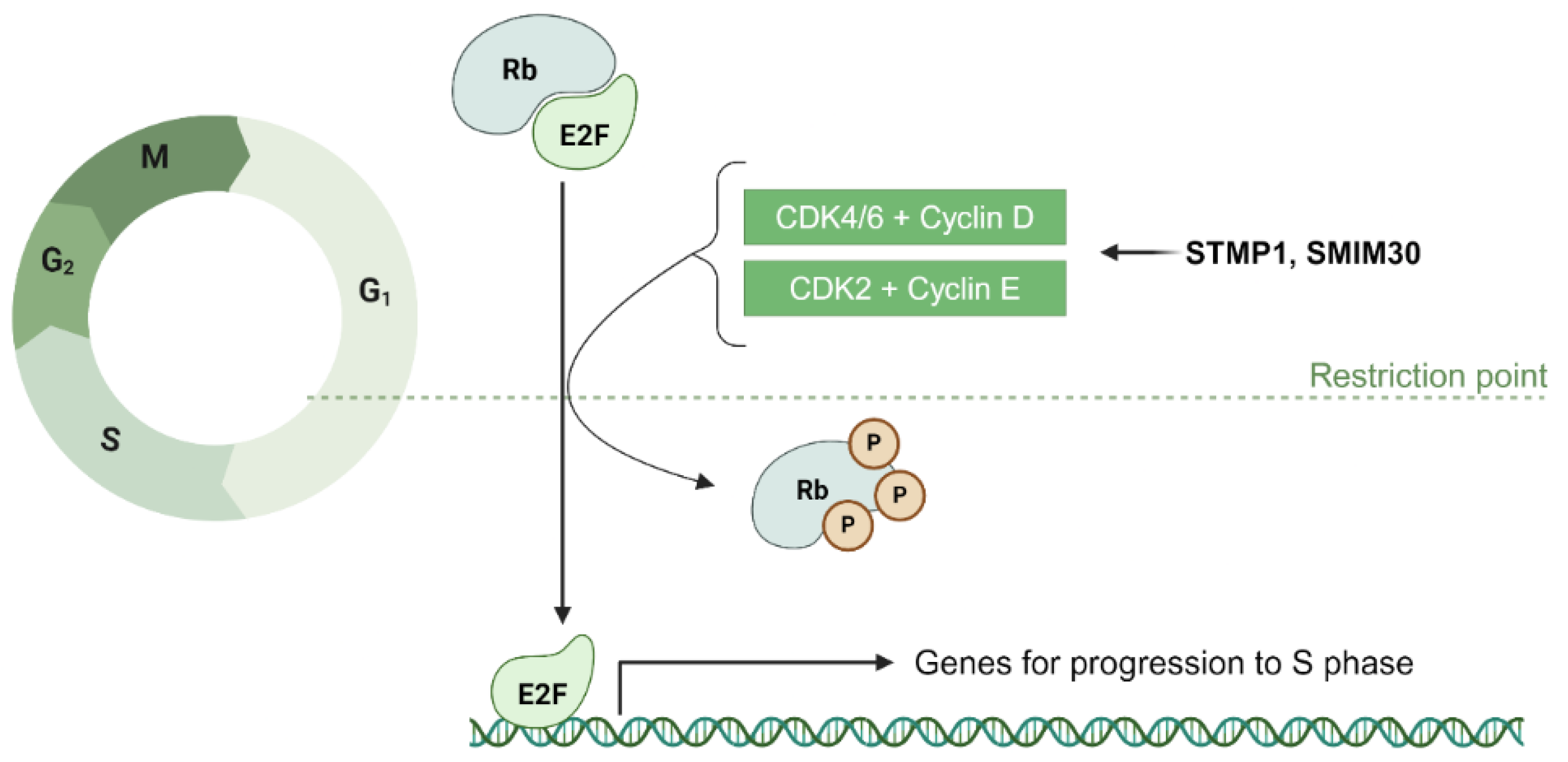
4.1.1. ncRNA-PEP/MPs in G1/S Transition: Modulation of Transcription Factors, Cyclins, and CDKs
4.1.2. Potential Role of ncRNA-PEPs and ncRNA-MPs in G1/S Transition via Metabolic Homeostasis
4.1.3. Linking ncRNA-MPs to G1/S Progression via Autophagy
4.2. The Potential Role of ncRNA-PEP/MPs in S Phase
4.2.1. ncRNA-PEPs in S Phase: PCNA Modulation
4.2.2. ncRNA-PEPs in S Phase: DNA Damage Repair
4.3. The Potential Role of ncRNA-PEP/MPs in G2/M
4.4. The Potential Role of ncRNA-PEP/MPs in Cell Division
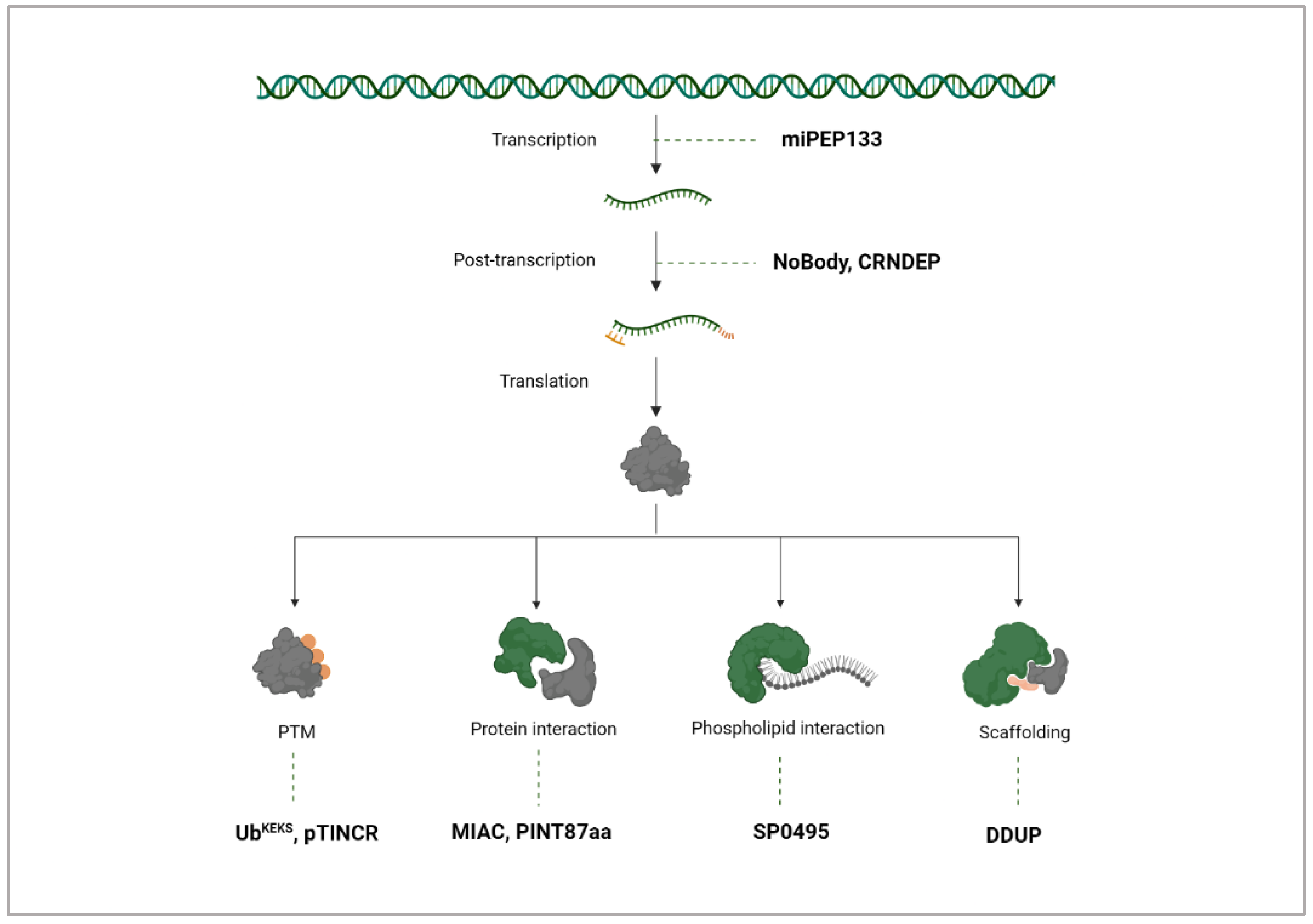
5. Mechanisms of ncRNA-PEP/MPs
Layers of Regulation
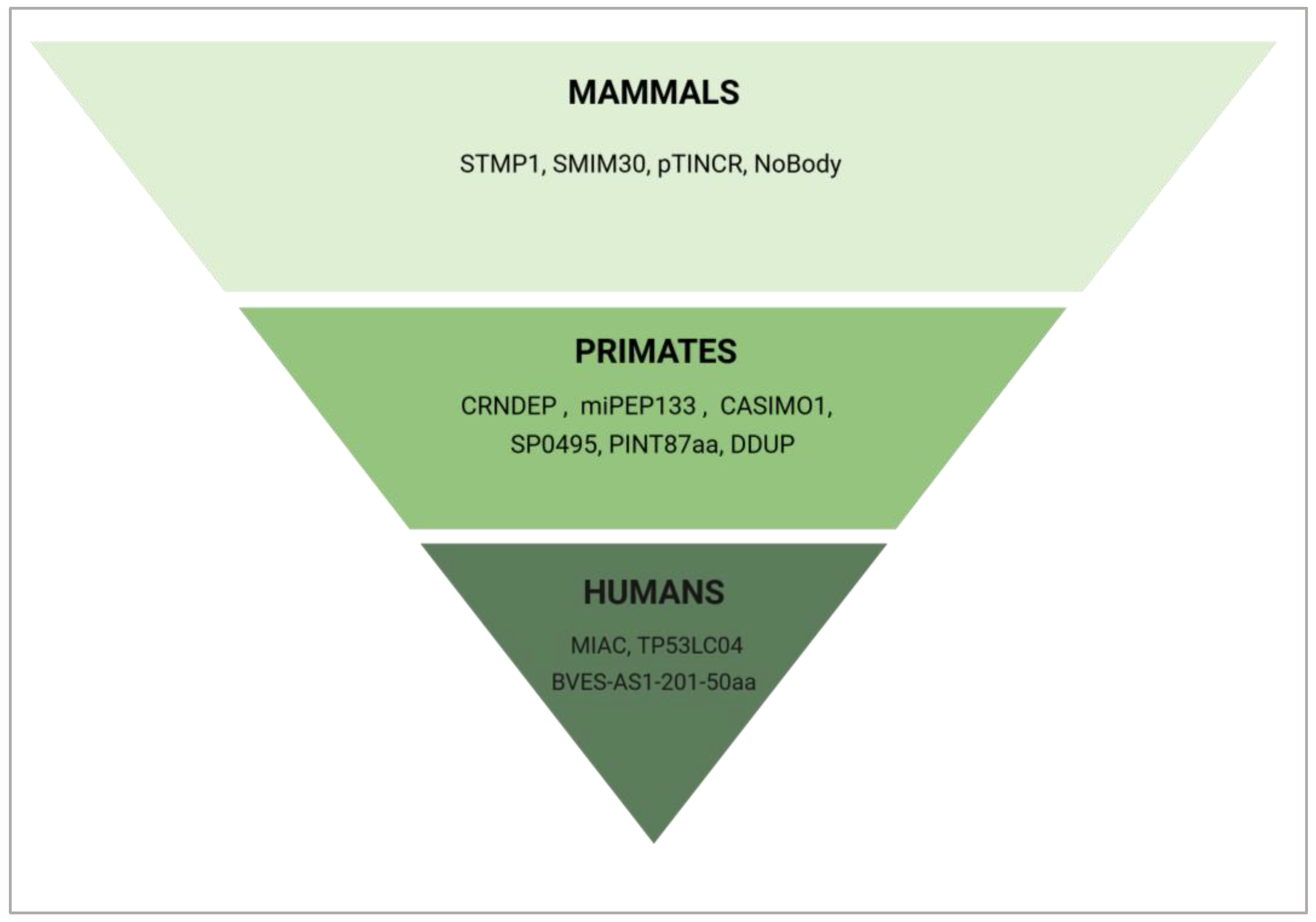
6. Evolutionary Conservation of Cell Cycle Regulating ncRNA-PEP/MPs
7. NcRNA-PEP/MPs in Cancer Biology
7.1. Role of Cell Cycle Related ncRNA-PEP/MPs in Physiology and Disease Physiology
7.2. Cell Cycle Related ncRNA-PEP/MPs as Oncogenes and Tumor Suppressors
7.3. ncRNA-PEP/MPs in Drug Resistance
7.4. Clinical Relevance of Cell Cycle Related ncRNA-PEP/MPs
8. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Pooresmaeil, F.; Azadi, S.; Hasannejad-Asl, B.; Takamoli, S.; Bolhassani, A. Pivotal Role of miRNA-lncRNA Interactions in Human Diseases. Mol. Biotechnol. 2024. [Google Scholar] [CrossRef]
- Ge, A.; Chan, C.; Yang, X. Exploring the Dark Matter of Human Proteome: The Emerging Role of Non-Canonical Open Reading Frame (ncORF) in Cancer Diagnosis, Biology, and Therapy. Cancers 2024, 16, 2660. [Google Scholar] [CrossRef]
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, E.A.; Hartford, C.C.; Grammatikakis, I.; et al. A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. eLife 2020, 9, e53734. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, S. lncRNA-Encoded Polypeptide SPAR(s) with mTORC1 to Regulate Skeletal Muscle Regeneration. Cell Stem Cell 2017, 20, 428–430. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, P.; Pan, W.; Wu, F.; Qiu, J.; Ma, Z. The role of polypeptides encoded by ncRNAs in cancer. Gene 2024, 928, 148817. [Google Scholar] [CrossRef]
- Zhang, Y. LncRNA-encoded peptides in cancer. J. Hematol. Oncol. 2024, 17, 66. [Google Scholar] [CrossRef]
- Tian, H.; Tang, L.; Yang, Z.; Xiang, Y.; Min, Q.; Yin, M.; You, H.; Xiao, Z.; Shen, J. Current understanding of functional peptides encoded by lncRNA in cancer. Cancer Cell Int. 2024, 24, 252. [Google Scholar] [CrossRef]
- Wen, K.; Chen, X.; Gu, J.; Chen, Z.; Wang, Z. Beyond traditional translation: ncRNA derived peptides as modulators of tumor behaviors. J. Biomed. Sci. 2024, 31, 63. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Basrai, M.A.; Hieter, P.; Boeke, J.D. Small open reading frames: Beautiful needles in the haystack. Genome Res. 1997, 7, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, D.; Elsässer, S.J. Revisiting sORFs: Overcoming challenges to identify and characterize functional microproteins. FEBS J. 2022, 289, 53–74. [Google Scholar] [CrossRef]
- Wright, B.W.; Yi, Z.; Weissman, J.S.; Chen, J. The dark proteome: Translation from noncanonical open reading frames. Trends Cell Biol. 2022, 32, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Gadella, T.W.J.; van Weeren, L.; Stouthamer, J.; Hink, M.A.; Wolters, A.H.G.; Giepmans, B.N.G.; Aumonier, S.; Dupuy, J.; Royant, A. mScarlet3: A brilliant and fast-maturing red fluorescent protein. Nat. Methods 2023, 20, 541–545. [Google Scholar] [CrossRef]
- Brizzard, B. Epitope tagging. Biotechniques 2008, 44, 693–695. [Google Scholar] [CrossRef]
- Polycarpou-Schwarz, M.; Groß, M.; Mestdagh, P.; Schott, J.; Grund, S.E.; Hildenbrand, C.; Rom, J.; Aulmann, S.; Sinn, H.P.; Vandesompele, J.; et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018, 37, 4750–4768. [Google Scholar] [CrossRef]
- Balcerak, A.; Szafron, L.A.; Rubel, T.; Swiderska, B.; Bonna, A.M.; Konarzewska, M.; Sołtyszewski, I.; Kupryjanczyk, J.; Szafron, L.M. A Multi-Faceted Analysis Showing CRNDE Transcripts and a Recently Confirmed Micropeptide as Important Players in Ovarian Carcinogenesis. Int. J. Mol. Sci. 2024, 25, 4381. [Google Scholar] [CrossRef]
- Cai, T.; Zhang, Q.; Wu, B.; Wang, J.; Li, N.; Zhang, T.; Wang, Z.; Luo, J.; Guo, X.; Ding, X.; et al. LncRNA-encoded microproteins: A new form of cargo in cell culture-derived and circulating extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12123. [Google Scholar] [CrossRef]
- Li, W.; Yu, Y.; Zhou, G.; Hu, G.; Li, B.; Ma, H.; Yan, W.; Pei, H. Large-scale ORF screening based on LC-MS to discover novel lncRNA-encoded peptides responding to ionizing radiation and microgravity. Comput. Struct. Biotechnol. J. 2023, 21, 5201–5211. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhang, J.; Lian, X.; Sun, L.; Meng, K.; Chen, Y.; Sun, Z.; Yin, X.; Li, Y.; Zhao, J.; et al. A hidden human proteome encoded by ‘non-coding’ genes. Nucleic Acids Res. 2019, 47, 8111–8125. [Google Scholar] [CrossRef] [PubMed]
- Chothani, S.P.; Adami, E.; Widjaja, A.A.; Langley, S.R.; Viswanathan, S.; Pua, C.J.; Zhihao, N.T.; Harmston, N.; D’Agostino, G.; Whiffin, N.; et al. A high-resolution map of human RNA translation. Mol. Cell 2022, 82, 2885–2899.e2888. [Google Scholar] [CrossRef]
- Zhao, S.; Meng, J.; Kang, Q.; Luan, Y. Identifying LncRNA-Encoded Short Peptides Using Optimized Hybrid Features and Ensemble Learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 2873–2881. [Google Scholar] [CrossRef]
- Prensner, J.R.; Abelin, J.G.; Kok, L.W.; Clauser, K.R.; Mudge, J.M.; Ruiz-Orera, J.; Bassani-Sternberg, M.; Moritz, R.L.; Deutsch, E.W.; van Heesch, S. What Can Ribo-Seq, Immunopeptidomics, and Proteomics Tell Us About the Noncanonical Proteome? Mol. Cell Proteom. 2023, 22, 100631. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kim, S.-H.; Yang, E.; Kang, M.; Joo, J.-Y. Molecular insights into regulatory RNAs in the cellular machinery. Exp. Mol. Med. 2024, 56, 1235–1249. [Google Scholar] [CrossRef]
- Walters, B.; Thompson, S.R. Cap-Independent Translational Control of Carcinogenesis. Front. Oncol. 2016, 6, 128. [Google Scholar] [CrossRef]
- Charpentier, M.; Croyal, M.; Carbonnelle, D.; Fortun, A.; Florenceau, L.; Rabu, C.; Krempf, M.; Labarrière, N.; Lang, F. IRES-dependent translation of the long non coding RNA meloe in melanoma cells produces the most immunogenic MELOE antigens. Oncotarget 2016, 7, 59704–59713. [Google Scholar] [CrossRef]
- Ibrahim, A.G.E.; Ciullo, A.; Yamaguchi, S.; Li, C.; Antes, T.; Jones, X.; Li, L.; Murali, R.; Maslennikov, I.; Sundararaman, N.; et al. A novel micropeptide, Slitharin, exerts cardioprotective effects in myocardial infarction. Proteom. Clin. Appl. 2024, 18, 2300128. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, J.; Zhang, H.; Ge, L.; Li, J.; Wang, H. RNA m(6) A methylation in cancer. Mol. Oncol. 2023, 17, 195–229. [Google Scholar] [CrossRef]
- Liu, H.T.; Gao, Z.X.; Li, F.; Guo, X.Y.; Li, C.L.; Zhang, H.; Zhao, R.N.; Liu, Y.; Shi, D.B.; Zhu, W.J.; et al. LncRNA LY6E-DT and its encoded metastatic-related protein play oncogenic roles via different pathways and promote breast cancer progression. Cell Death Differ. 2024, 31, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Dai, Y.; Yu, Y.; Tang, J.; Cao, Z.; Zhang, Y.; Li, B.; Nie, J.; Hei, T.K.; Zhou, G. The Tumorigenic Effect of lncRNA AFAP1-AS1 is Mediated by Translated Peptide ATMLP Under the Control of m(6) A Methylation. Adv. Sci. 2023, 10, e2300314. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef]
- Meng, K.; Lu, S.; Li, Y.Y.; Hu, L.L.; Zhang, J.; Cao, Y.; Wang, Y.; Zhang, C.Z.; He, Q.Y. LINC00493-encoded microprotein SMIM26 exerts anti-metastatic activity in renal cell carcinoma. EMBO Rep. 2023, 24, e56282. [Google Scholar] [CrossRef]
- Yi, Q.; Feng, J.; Lan, W.; shi, H.; Sun, W.; Sun, W. CircRNA and lncRNA-encoded peptide in diseases, an update review. Mol. Cancer 2024, 23, 214. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Hu, Y.; Zhang, S.; Li, X.; Tang, M.; Yang, M.; Wu, X.; Li, Z.; Liao, X.; Xu, Y.; et al. LncRNA CTBP1-DT-encoded microprotein DDUP sustains DNA damage response signalling to trigger dual DNA repair mechanisms. Nucleic Acids Res. 2022, 50, 8060–8079. [Google Scholar] [CrossRef]
- Ormancey, M.; Thuleau, P.; Combier, J.-P.; Plaza, S. The Essentials on microRNA-Encoded Peptides from Plants to Animals. Biomolecules 2023, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Prel, A.; Dozier, C.; Combier, J.-P.; Plaza, S.; Besson, A. Evidence That Regulation of Pri-miRNA/miRNA Expression Is Not a General Rule of miPEPs Function in Humans. Int. J. Mol. Sci. 2021, 22, 3432. [Google Scholar] [CrossRef]
- Kang, M.; Tang, B.; Li, J.; Zhou, Z.; Liu, K.; Wang, R.; Jiang, Z.; Bi, F.; Patrick, D.; Kim, D.; et al. Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol. Cancer 2020, 19, 143. [Google Scholar] [CrossRef]
- Zhou, H.; Lou, F.; Bai, J.; Sun, Y.; Cai, W.; Sun, L.; Xu, Z.; Liu, Z.; Zhang, L.; Yin, Q.; et al. A peptide encoded by pri-miRNA-31 represses autoimmunity by promoting Treg differentiation. EMBO Rep. 2022, 23, e53475. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, T.; Yan, L.; Zhu, S.; Jin, W.; Bai, Y.; Zeng, Y.; Zhang, X.; Yin, Z.; Yang, J.; et al. Hypoxia-Responsive lncRNA AC115619 Encodes a Micropeptide That Suppresses m6A Modifications and Hepatocellular Carcinoma Progression. Cancer Res. 2023, 83, 2496–2512. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Zhang, Z.; Jia, L.; Zhang, H.; Zhang, S.; Wang, H.; Cheng, Z. Micropeptide AF127577.4-ORF hidden in a lncRNA diminishes glioblastoma cell proliferation via the modulation of ERK2/METTL3 interaction. Sci. Rep. 2024, 14, 12090. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020, 217, jem.20190950. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Guo, Y.; Zhang, G.; Bai, J.; Song, Y.; Song, X.; Zhu, Q.; Bao, X.; Wu, G.; Zhang, C. Peptide encoded by lncRNA BVES-AS1 promotes cell viability, migration, and invasion in colorectal cancer cells via the SRC/mTOR signaling pathway. PLoS ONE 2023, 18, e0287133. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020, 39, e102190. [Google Scholar] [CrossRef]
- Pan, J.; Liu, M.; Duan, X.; Wang, D. A short peptide LINC00665_18aa encoded by lncRNA LINC00665 suppresses the proliferation and migration of osteosarcoma cells through the regulation of the CREB1/RPS6KA3 interaction. PLoS ONE 2023, 18, e0286422. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, J.; Lou, F.; Zhou, H.; Cai, X.; Wang, Z.; Sun, L.; Sun, Y.; Li, X.; Fan, L.; et al. A lncRNA Dleu2-encoded peptide relieves autoimmunity by facilitating Smad3-mediated Treg induction. EMBO Rep. 2024, 25, 1208–1232. [Google Scholar] [CrossRef]
- Huang, J.Z.; Chen, M.; Chen, D.; Gao, X.C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184.e176. [Google Scholar] [CrossRef]
- van Solingen, C.; Sharma, M.; Bijkerk, R.; Afonso, M.S.; Koelwyn, G.J.; Scacalossi, K.R.; Holdt, L.M.; Maegdefessel, L.; van Zonneveld, A.J.; Moore, K.J. Abstract 544: A Novel Micropeptide, IMP, Directs Inflammation Through Interaction with Transcriptional Co-activators. Arterioscler. Thromb. Vasc. Biol. 2019, 39, A544. [Google Scholar] [CrossRef]
- Zhou, B.; Yu, G.; Zhao, M.; Li, Y.; Li, J.; Xiang, Y.; Tong, L.; Chu, X.; Wang, C.; Song, Y. The lncRNA LINC00339-encoded peptide promotes trophoblast adhesion to endometrial cells via MAPK and PI3K-Akt signaling pathways. J. Assist. Reprod. Genet. 2024, 41, 493–504. [Google Scholar] [CrossRef]
- Han, X.; Chen, L.; Sun, P.; Wang, X.; Zhao, Q.; Liao, L.; Lou, D.; Zhou, N.; Wang, Y. A novel lncRNA-hidden polypeptide regulates malignant phenotypes and pemetrexed sensitivity in A549 pulmonary adenocarcinoma cells. Amino Acids 2024, 56, 15. [Google Scholar] [CrossRef]
- Quaife, N.M.; Chothani, S.; Schulz, J.F.; Lindberg, E.L.; Vanezis, K.; Adami, E.; O’Fee, K.; Greiner, J.; Litviňuková, M.; van Heesch, S.; et al. LINC01013 Is a Determinant of Fibroblast Activation and Encodes a Novel Fibroblast-Activating Micropeptide. J. Cardiovasc. Transl. Res. 2023, 16, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Halabi, R.; Lin, C.H.; Nazim, M.; Yeom, K.H.; Black, D.L. The lncRNA Malat1 is trafficked to the cytoplasm as a localized mRNA encoding a small peptide in neurons. Genes. Dev. 2024, 38, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yi, Y.; Wang, Z.; Zhang, H.; Zhao, Y.; He, R.; Luo, Y.; Cui, Z. LncRNA MAGI2-AS3-Encoded Polypeptide Restrains the Proliferation and Migration of Breast Cancer Cells. Mol. Biotechnol. 2024, 66, 1409–1423. [Google Scholar] [CrossRef]
- Godet, Y.; Moreau-Aubry, A.; Guilloux, Y.; Vignard, V.; Khammari, A.; Dreno, B.; Jotereau, F.; Labarriere, N. MELOE-1 is a new antigen overexpressed in melanomas and involved in adoptive T cell transfer efficiency. J. Exp. Med. 2008, 205, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Zhang, Y.; Wu, H.; Zhou, H.; Ding, X.; Zhang, X.; Jin, X.; Wang, Y.; Yin, X.; et al. Micropeptide MIAC Inhibits HNSCC Progression by Interacting with Aquaporin 2. J. Am. Chem. Soc. 2020, 142, 6708–6716. [Google Scholar] [CrossRef]
- Li, M.; Liu, G.; Jin, X.; Guo, H.; Setrerrahmane, S.; Xu, X.; Li, T.; Lin, Y.; Xu, H. Micropeptide MIAC inhibits the tumor progression by interacting with AQP2 and inhibiting EREG/EGFR signaling in renal cell carcinoma. Mol. Cancer 2022, 21, 181. [Google Scholar] [CrossRef]
- Fang, J.; Morsalin, S.; Rao, V.; Reddy, E.S. Decoding of Non-Coding DNA and Non-Coding RNA: Pri-Micro RNA-Encoded Novel Peptides Regulate Migration of Cancer Cells. J. Pharm. Sci. Pharmacol. 2017, 3, 23–27. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e3718. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Baskin, K.K.; Munir, A.Z.; Bezprozvannaya, S.; Sharma, G.; Khemtong, C.; Shah, A.M.; McAnally, J.R.; Malloy, C.R.; Szweda, L.I.; et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018, 23, 3701–3709. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, J.; Han, L.; Qi, H.; Wang, Y.; Wang, H.; Chen, S.; Du, L.; Li, S.; Zhang, Y.; et al. The micropeptide LEMP plays an evolutionarily conserved role in myogenesis. Cell Death Dis. 2020, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Lin, J.; Ji, Y.; Zhang, R.; Zhang, Z.; Cao, Y.; Liu, Y.; Tang, X.; Liu, J.; Liu, P.; et al. A microprotein N1DARP encoded by LINC00261 promotes Notch1 intracellular domain (N1ICD) degradation via disrupting USP10-N1ICD interaction to inhibit chemoresistance in Notch1-hyperactivated pancreatic cancer. Cell Discov. 2023, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, B.; Gu, F.; Liu, H.; Wu, H.; Yao, F.; Zheng, H.; Fu, H.; Chong, W.; Cai, S.; et al. Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol. Cell 2022, 82, 1297–1312.e1298. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yin, S.; Yang, Y.; Yang, H.; Yang, J.; Zhou, Z.; Li, S.; Ying, G.; Ba, Y. lncRNA-encoded pep-AP attenuates the pentose phosphate pathway and sensitizes colorectal cancer cells to Oxaliplatin. EMBO Rep. 2022, 23, e53140. [Google Scholar] [CrossRef]
- Chu, Q.; Martinez, T.F.; Novak, S.W.; Donaldson, C.J.; Tan, D.; Vaughan, J.M.; Chang, T.; Diedrich, J.K.; Andrade, L.; Kim, A.; et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019, 10, 4883. [Google Scholar] [CrossRef]
- Senís, E.; Esgleas, M.; Najas, S.; Jiménez-Sábado, V.; Bertani, C.; Giménez-Alejandre, M.; Escriche, A.; Ruiz-Orera, J.; Hergueta-Redondo, M.; Jiménez, M.; et al. TUNAR lncRNA Encodes a Microprotein that Regulates Neural Differentiation and Neurite Formation by Modulating Calcium Dynamics. Front. Cell Dev. Biol. 2021, 9, 747667. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Tokita, S.; Hirama, T.; Kochin, V.; Nakatsugawa, M.; Shinkawa, T.; Hirohashi, Y.; Tsukahara, T.; Hata, F.; Takemasa, I.; et al. CD8(+) T-cell Immune Surveillance against a Tumor Antigen Encoded by the Oncogenic Long Noncoding RNA PVT1. Cancer Immunol. Res. 2021, 9, 1342–1353. [Google Scholar] [CrossRef]
- Ma, Q.; Ma, F.; Zhang, B.; Zhang, Y.; Peng, L.; Li, X. The short peptide encoded by long non-coding RNA RNF217-AS1 inhibits stomach cancer tumorigenesis, macrophage recruitment, and pro-inflammatory responses. Amino Acids 2024, 56, 45. [Google Scholar] [CrossRef]
- Yang, J.E.; Zhong, W.J.; Li, J.F.; Lin, Y.Y.; Liu, F.T.; Tian, H.; Chen, Y.J.; Luo, X.Y.; Zhuang, S.M. LINC00998-encoded micropeptide SMIM30 promotes the G1/S transition of cell cycle by regulating cytosolic calcium level. Mol. Oncol. 2023, 17, 901–916. [Google Scholar] [CrossRef]
- Pang, Y.; Liu, Z.; Han, H.; Wang, B.; Li, W.; Mao, C.; Liu, S. Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J. Hepatol. 2020, 73, 1155–1169. [Google Scholar] [CrossRef]
- Zou, Q.; Du, X.; Zhou, L.; Yao, D.; Dong, Y.; Jin, J. A short peptide encoded by long non-coding RNA small nucleolar RNA host gene 6 promotes cell migration and epithelial-mesenchymal transition by activating transforming growth factor-beta/SMAD signaling pathway in human endometrial cells. J. Obs. Gynaecol. Res. 2023, 49, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, J.; Wang, X.; Yuan, B.; Li, D.; Hu, A.; Guo, Y.; Cai, S.; Jin, S.; Zhou, Y.; et al. HNF4A-AS1-encoded small peptide promotes self-renewal and aggressiveness of neuroblastoma stem cells via eEF1A1-repressed SMAD4 transactivation. Oncogene 2022, 41, 2505–2519. [Google Scholar] [CrossRef]
- Zheng, X.; Guo, Y.; Zhang, R.; Chen, H.; Liu, S.; Qiu, S.; Xiang, M. The mitochondrial micropeptide Stmp1 promotes retinal cell differentiation. Biochem. Biophys. Res. Commun. 2022, 636, 79–86. [Google Scholar] [CrossRef]
- Sang, Y.; Liu, J.Y.; Wang, F.Y.; Luo, X.Y.; Chen, Z.Q.; Zhuang, S.M.; Zhu, Y. Mitochondrial micropeptide STMP1 promotes G1/S transition by enhancing mitochondrial complex IV activity. Mol. Ther. 2022, 30, 2844–2855. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, M.; Liu, S.; Chen, H.; Li, Y.; Yuan, F.; Yang, L.; Qiu, S.; Wang, H.; Xie, Z.; et al. A lncRNA-encoded mitochondrial micropeptide exacerbates microglia-mediated neuroinflammation in retinal ischemia/reperfusion injury. Cell Death Dis. 2023, 14, 126. [Google Scholar] [CrossRef] [PubMed]
- Min, K.W.; Davila, S.; Zealy, R.W.; Lloyd, L.T.; Lee, I.Y.; Lee, R.; Roh, K.H.; Jung, A.; Jemielity, J.; Choi, E.J.; et al. eIF4E phosphorylation by MST1 reduces translation of a subset of mRNAs, but increases lncRNA translation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Chng, S.C.; Ho, L.; Tian, J.; Reversade, B. ELABELA: A hormone essential for heart development signals via the apelin receptor. Dev. Cell 2013, 27, 672–680. [Google Scholar] [CrossRef]
- Lu, L.; Cao, J.; Li, L.; Chen, L. Elabela, a new endogenous ligand of APJ, functions in embryos and adults organisms. Acta Biochim. Biophys. Sin. 2017, 49, 378–381. [Google Scholar] [CrossRef]
- Wu, S.; Guo, B.; Zhang, L.; Zhu, X.; Zhao, P.; Deng, J.; Zheng, J.; Li, F.; Wang, Y.; Zhang, S.; et al. A micropeptide XBP1SBM encoded by lncRNA promotes angiogenesis and metastasis of TNBC via XBP1s pathway. Oncogene 2022, 41, 2163–2172. [Google Scholar] [CrossRef]
- Guo, Z.W.; Meng, Y.; Zhai, X.M.; Xie, C.; Zhao, N.; Li, M.; Zhou, C.L.; Li, K.; Liu, T.C.; Yang, X.X.; et al. Translated Long Non-Coding Ribonucleic Acid ZFAS1 Promotes Cancer Cell Migration by Elevating Reactive Oxygen Species Production in Hepatocellular Carcinoma. Front. Genet. 2019, 10, 1111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Z.; Liu, X.; Deng, Y.; Zheng, J.; Deng, J.; Wang, Y.; Guo, B.; Li, F.; Chen, X.; et al. LncRNA-Encoded Micropeptide ACLY-BP Drives Lipid Deposition and Cell Proliferation in Clear Cell Renal Cell Carcinoma via Maintenance of ACLY Acetylation. Mol. Cancer Res. 2023, 21, 1064–1078. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, W.; Han, C.; Huang, W.; Sun, Y.; Fang, K.; Zeng, Z.; Yang, Q.; Pan, Q.; Chen, T.; et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol. Cell 2021, 81, 4493–4508.e4499. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, W.; Yang, J.; Zhang, Y.; Gong, X.; Luo, H.; Cao, N.; Xu, Z.; Tian, M.; Yang, P.; et al. LncRNA PSR Regulates Vascular Remodeling Through Encoding a Novel Protein Arteridin. Circ. Res. 2022, 131, 768–787. [Google Scholar] [CrossRef]
- Ge, Q.; Jia, D.; Cen, D.; Qi, Y.; Shi, C.; Li, J.; Sang, L.; Yang, L.J.; He, J.; Lin, A.; et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J. Clin. Investig. 2021, 131, e152911. [Google Scholar] [CrossRef]
- Jackson, R.; Kroehling, L.; Khitun, A.; Bailis, W.; Jarret, A.; York, A.G.; Khan, O.M.; Brewer, J.R.; Skadow, M.H.; Duizer, C.; et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature 2018, 564, 434–438. [Google Scholar] [CrossRef]
- Burbano De Lara, S.; Tran, D.D.H.; Allister, A.B.; Polenkowski, M.; Nashan, B.; Koch, M.; Tamura, T. C20orf204, a hepatocellular carcinoma-specific protein interacts with nucleolin and promotes cell proliferation. Oncogenesis 2021, 10, 31. [Google Scholar] [CrossRef]
- Szafron, L.M.; Balcerak, A.; Grzybowska, E.A.; Pienkowska-Grela, B.; Felisiak-Golabek, A.; Podgorska, A.; Kulesza, M.; Nowak, N.; Pomorski, P.; Wysocki, J.; et al. The Novel Gene CRNDE Encodes a Nuclear Peptide (CRNDEP) Which Is Overexpressed in Highly Proliferating Tissues. PLoS ONE 2015, 10, e0127475. [Google Scholar] [CrossRef]
- Zheng, C.; Wei, Y.; Zhang, P.; Xu, L.; Zhang, Z.; Lin, K.; Hou, J.; Lv, X.; Ding, Y.; Chiu, Y.; et al. CRISPR/Cas9 screen uncovers functional translation of cryptic lncRNA-encoded open reading frames in human cancer. J. Clin. Investig. 2023, 133, e159940. [Google Scholar] [CrossRef]
- Lun, Y.-Z.; Pan, Z.-P.; Liu, S.-A.; Sun, J.; Han, M.; Liu, B.; Dong, W.; Pan, L.-H.; Cheng, J. The peptide encoded by a novel putative lncRNA HBVPTPAP inducing the apoptosis of hepatocellular carcinoma cells by modulating JAK/STAT signaling pathways. Virus Res. 2020, 287, 198104. [Google Scholar] [CrossRef]
- Tong, X.; Yu, Z.; Xing, J.; Liu, H.; Zhou, S.; Huang, Y.; Lin, J.; Jiang, W.; Wang, L. LncRNA HCP5-Encoded Protein Regulates Ferroptosis to Promote the Progression of Triple-Negative Breast Cancer. Cancers 2023, 15, 1880. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.N.; Shang, Y.N.; Yu, Z.L.; Zhou, S.H.; Chen, W.Y.; Wang, L.H. LncRNA HCP5-encoded protein contributes to adriamycin resistance through ERK/mTOR pathway-mediated autophagy in breast cancer cells. Genes. Dis. 2024, 11, 101024. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Chen, X.L.; Guo, L.Y.; Lu, D.F.; Lu, S.; Wang, A.A.; Liang, X.F. Downregulation of lncRNA IGF2-AS-encoded peptide induces trophoblast—Cycle arrest. Reprod. Biomed. Online 2021, 43, 598–606. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, Z.; Wang, W.; Liu, Y.; Zhu, H.; Liang, H.; Zhang, B.; Chen, X. A micropeptide JunBP regulated by TGF-β promotes hepatocellular carcinoma metastasis. Oncogene 2023, 42, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Deng, B.; Lin, P.; Liu, C.; Li, B.; Huang, Q.; Zhou, H.; Yang, J.; Qu, L. Ribosome profiling analysis identified a KRAS-interacting microprotein that represses oncogenic signaling in hepatocellular carcinoma cells. Sci. China Life Sci. 2020, 63, 529–542. [Google Scholar] [CrossRef]
- Tan, Z.; Zhao, L.; Huang, S.; Jiang, Q.; Wei, Y.; Wu, J.L.; Zhang, Z.; Li, Y. Small peptide LINC00511-133aa encoded by LINC00511 regulates breast cancer cell invasion and stemness through the Wnt/β-catenin pathway. Mol. Cell Probes 2023, 69, 101913. [Google Scholar] [CrossRef]
- Sandmann, C.-L.; Schulz, J.F.; Ruiz-Orera, J.; Kirchner, M.; Ziehm, M.; Adami, E.; Marczenke, M.; Christ, A.; Liebe, N.; Greiner, J.; et al. Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol. Cell 2023, 83, 994–1011.e1018. [Google Scholar] [CrossRef]
- Polenkowski, M.; Burbano de Lara, S.; Allister, A.B.; Nguyen, T.N.Q.; Tamura, T.; Tran, D.D.H. Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA. Int. J. Mol. Sci. 2021, 23, 58. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Wang, M.; Zhang, X.; Jiang, W.; Wu, S.; Ti, X. A Peptide Encoded by a Long Non-Coding RNA DLX6-AS1 Facilitates Cell Proliferation, Migration, and Invasion by Activating the wnt/β-Catenin Signaling Pathway in Non-Small-Cell Lung Cancer Cell. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 43–53. [Google Scholar] [CrossRef]
- Tang, C.; Zhou, Y.; Sun, W.; Hu, H.; Liu, Y.; Chen, L.; Ou, F.; Zeng, S.; Lin, N.; Yu, L. Oncopeptide MBOP Encoded by LINC01234 Promotes Colorectal Cancer through MAPK Signaling Pathway. Cancers 2022, 14, 2338. [Google Scholar] [CrossRef]
- Liu, X.; Chen, S.; Luo, W.; Yu, C.; Yan, S.; Lei, L.; Qiu, S.; Lin, X.; Feng, T.; Shi, J.; et al. LncRNA MFRL regulates the phenotypic switch of vascular smooth muscle cells to attenuate arterial remodeling by encoding a novel micropeptide MFRLP. Transl. Res. 2024, 272, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Gao, R.; Chen, S.; Bai, J.; Chen, J.; Lu, F.; Gu, D.; Shi, X.; Yu, P.; Tian, Y.; et al. FAM201A encodes small protein NBASP to inhibit neuroblastoma progression via inactivating MAPK pathway mediated by FABP5. Commun. Biol. 2023, 6, 714. [Google Scholar] [CrossRef] [PubMed]
- D’Lima, N.G.; Ma, J.; Winkler, L.; Chu, Q.; Loh, K.H.; Corpuz, E.O.; Budnik, B.A.; Lykke-Andersen, J.; Saghatelian, A.; Slavoff, S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017, 13, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.G.; Yang, J.; Zhu, Y.; Zhu, Q.; Pan, W.; Deng, S.; He, Y.; Zuo, D.; Wang, P.; Han, Y.; et al. The microprotein encoded by exosomal lncAKR1C2 promotes gastric cancer lymph node metastasis by regulating fatty acid metabolism. Cell Death Dis. 2023, 14, 708. [Google Scholar] [CrossRef]
- Zhou, B.; Wu, Y.; Cheng, P.; Wu, C. Long noncoding RNAs with peptide-encoding potential identified in esophageal squamous cell carcinoma: KDM4A-AS1-encoded peptide weakens cancer cell viability and migratory capacity. Mol. Oncol. 2023, 17, 1419–1436. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef]
- Xiang, X.; Fu, Y.; Zhao, K.; Miao, R.; Zhang, X.; Ma, X.; Liu, C.; Zhang, N.; Qu, K. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 2021, 11, 4929–4944. [Google Scholar] [CrossRef]
- Nita, A.; Matsumoto, A.; Tang, R.; Shiraishi, C.; Ichihara, K.; Saito, D.; Suyama, M.; Yasuda, T.; Tsuji, G.; Furue, M.; et al. A ubiquitin-like protein encoded by the “noncoding” RNA TINCR promotes keratinocyte proliferation and wound healing. PLoS Genet. 2021, 17, e1009686. [Google Scholar] [CrossRef]
- Morgado-Palacin, L.; Brown, J.A.; Martinez, T.F.; Garcia-Pedrero, J.M.; Forouhar, F.; Quinn, S.A.; Reglero, C.; Vaughan, J.; Heydary, Y.H.; Donaldson, C.; et al. The TINCR ubiquitin-like microprotein is a tumor suppressor in squamous cell carcinoma. Nat. Commun. 2023, 14, 1328. [Google Scholar] [CrossRef]
- Boix, O.; Martinez, M.; Vidal, S.; Giménez-Alejandre, M.; Palenzuela, L.; Lorenzo-Sanz, L.; Quevedo, L.; Moscoso, O.; Ruiz-Orera, J.; Ximénez-Embún, P.; et al. pTINCR microprotein promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation and activation. Nat. Commun. 2022, 13, 6840. [Google Scholar] [CrossRef]
- Cheng, R.; Li, F.; Zhang, M.; Xia, X.; Wu, J.; Gao, X.; Zhou, H.; Zhang, Z.; Huang, N.; Yang, X.; et al. A novel protein RASON encoded by a lncRNA controls oncogenic RAS signaling in KRAS mutant cancers. Cell Res. 2023, 33, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, J.Z.; Chen, D.; He, Y.T.; Meng, N.; Chen, M.; Lu, R.X.; Chen, X.H.; Zhang, X.L.; Yan, G.R. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat. Commun. 2020, 11, 1685. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Chen, Y.; Sun, M.; Huang, X.; Zhang, H.; Fu, Z.; Wang, J.; Zhang, S.; Lian, C.; Tang, B.; et al. LncRNA DGCR5-encoded polypeptide RIP aggravates SONFH by repressing nuclear localization of β-catenin in BMSCs. Cell Rep. 2023, 42, 112969. [Google Scholar] [CrossRef]
- Li, L.; Shu, X.-S.; Geng, H.; Ying, J.; Guo, L.; Luo, J.; Xiang, T.; Wu, L.; Ma, B.B.Y.; Chan, A.T.C.; et al. A novel tumor suppressor encoded by a 1p36.3 lncRNA functions as a phosphoinositide-binding protein repressing AKT phosphorylation/activation and promoting autophagy. Cell Death Differ. 2023, 30, 1166–1183. [Google Scholar] [CrossRef]
- Meng, N.; Chen, M.; Chen, D.; Chen, X.H.; Wang, J.Z.; Zhu, S.; He, Y.T.; Zhang, X.L.; Lu, R.X.; Yan, G.R. Small Protein Hidden in lncRNA LOC90024 Promotes “Cancerous” RNA Splicing and Tumorigenesis. Adv. Sci. 2020, 7, 1903233. [Google Scholar] [CrossRef]
- Mao, X.; Zhou, J.; Kong, L.; Zhu, L.; Yang, D.; Zhang, Z. A peptide encoded by lncRNA MIR7-3 host gene (MIR7-3HG) alleviates dexamethasone-induced dysfunction in pancreatic β-cells through the PI3K/AKT signaling pathway. Biochem. Biophys. Res. Commun. 2023, 647, 62–71. [Google Scholar] [CrossRef]
- Xu, W.; Liu, C.; Deng, B.; Lin, P.; Sun, Z.; Liu, A.; Xuan, J.; Li, Y.; Zhou, K.; Zhang, X.; et al. TP53-inducible putative long noncoding RNAs encode functional polypeptides that suppress cell proliferation. Genome Res. 2022, 32, 1026–1041. [Google Scholar] [CrossRef] [PubMed]
- Sabikunnahar, B.; Caldwell, S.; Varnum, S.; Hogan, T.; Cooper, A.; Lahue, K.G.; Bivona, J.J.; Cousens, P.M.; Symeonides, M.; Ballif, B.A.; et al. Long Noncoding RNA U90926 Is Induced in Activated Macrophages, Is Protective in Endotoxic Shock, and Encodes a Novel Secreted Protein. J. Immunol. 2023, 210, 807–819. [Google Scholar] [CrossRef]
- Chugunova, A.; Loseva, E.; Mazin, P.; Mitina, A.; Navalayeu, T.; Bilan, D.; Vishnyakova, P.; Marey, M.; Golovina, A.; Serebryakova, M.; et al. LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 4940–4945. [Google Scholar] [CrossRef]
- Barczak, W.; Carr, S.M.; Liu, G.; Munro, S.; Nicastri, A.; Lee, L.N.; Hutchings, C.; Ternette, N.; Klenerman, P.; Kanapin, A.; et al. Long non-coding RNA-derived peptides are immunogenic and drive a potent anti-tumour response. Nat. Commun. 2023, 14, 1078. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, M.; Diao, H.; Xiong, L.; Yang, X.; Xing, S. LncRNA-encoded peptides: Unveiling their significance in cardiovascular physiology and pathology—Current research insights. Cardiovasc. Res. 2023, 119, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Frion, J.; Meller, A.; Marbach, G.; Lévesque, D.; Roucou, X.; Boisvert, F.M. CRISPR/Cas9-mediated knockout of the ubiquitin variant UbKEKS reveals a role in regulating nucleolar structures and composition. Biol. Open 2023, 12, bio059984. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef]
- Humeau, J.; Bravo-San Pedro, J.M.; Vitale, I.; Nuñez, L.; Villalobos, C.; Kroemer, G.; Senovilla, L. Calcium signaling and cell cycle: Progression or death. Cell Calcium 2018, 70, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, J.; Zhu, Y.; Wang, L.; Jiang, X.; Liu, B.; He, G. Targeting autophagy and beyond: Deconvoluting the complexity of Beclin-1 from biological function to cancer therapy. Acta Pharm. Sin. B 2023, 13, 4688–4714. [Google Scholar] [CrossRef] [PubMed]
- De Tito, S.; Hervás, J.H.; van Vliet, A.R.; Tooze, S.A. The Golgi as an Assembly Line to the Autophagosome. Trends Biochem. Sci. 2020, 45, 484–496. [Google Scholar] [CrossRef]
- Mathiassen, S.G.; De Zio, D.; Cecconi, F. Autophagy and the Cell Cycle: A Complex Landscape. Front. Oncol. 2017, 7, 51. [Google Scholar] [CrossRef]
- Liu, S.; Yao, S.; Yang, H.; Liu, S.; Wang, Y. Autophagy: Regulator of cell death. Cell Death Dis. 2023, 14, 648. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Park, S.H. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 2020, 52, 1948–1958. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef]
- Peng, M.; Zhou, Y.; Wan, C. Identification of phosphorylated small ORF-encoded peptides in Hep3B cells by LC/MS/MS. J. Proteom. 2024, 303, 105214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Regulation of Cell Cycle Progression by Growth Factor-Induced Cell Signaling. Cells 2021, 10, 3327. [Google Scholar] [CrossRef]
- Diril, M.K.; Ratnacaram, C.K.; Padmakumar, V.C.; Du, T.; Wasser, M.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 3826–3831. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jakymiw, A.; Wood, M.R.; Eystathioy, T.; Rubin, R.L.; Fritzler, M.J.; Chan, E.K.L. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004, 117, 5567–5578. [Google Scholar] [CrossRef]
- Safieddine, A.; Benassy, M.-N.; Bonte, T.; Slimani, F.; Pourcelot, O.; Kress, M.; Ernoult-Lange, M.; Courel, M.; Coleno, E.; Imbert, A.; et al. Cell-cycle-dependent mRNA localization in P-bodies. Mol. Cell 2024, 84, 4191–4208.e4197. [Google Scholar] [CrossRef] [PubMed]
- Standart, N.; Weil, D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef]
- Na, Z.; Luo, Y.; Cui, D.S.; Khitun, A.; Smelyansky, S.; Loria, J.P.; Slavoff, S.A. Phosphorylation of a Human Microprotein Promotes Dissociation of Biomolecular Condensates. J. Am. Chem. Soc. 2021, 143, 12675–12687. [Google Scholar] [CrossRef]
- Dubois, M.-L.; Meller, A.; Samandi, S.; Brunelle, M.; Frion, J.; Brunet, M.A.; Toupin, A.; Beaudoin, M.C.; Jacques, J.-F.; Lévesque, D.; et al. UBB pseudogene 4 encodes functional ubiquitin variants. Nat. Commun. 2020, 11, 1306. [Google Scholar] [CrossRef]
- Li, L.; Tong, M.; Fu, Y.; Chen, F.; Zhang, S.; Chen, H.; Ma, X.; Li, D.; Liu, X.; Zhong, Q. Lipids and membrane-associated proteins in autophagy. Protein Cell 2021, 12, 520–544. [Google Scholar] [CrossRef]
- Andersson, D.I.; Jerlström-Hultqvist, J.; Näsvall, J. Evolution of new functions de novo and from preexisting genes. Cold Spring Harb. Perspect. Biol. 2015, 7, a017996. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Verdaguer-Grau, P.; Villanueva-Cañas, J.L.; Messeguer, X.; Albà, M.M. Translation of neutrally evolving peptides provides a basis for de novo gene evolution. Nat. Ecol. Evol. 2018, 2, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Heames, B.; Buchel, F.; Aubel, M.; Tretyachenko, V.; Loginov, D.; Novák, P.; Lange, A.; Bornberg-Bauer, E.; Hlouchová, K. Experimental characterization of de novo proteins and their unevolved random-sequence counterparts. Nat. Ecol. Evol. 2023, 7, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs kinases—The lessons from the mouse models: Inhibition ≠ deletion. Cell Biosci. 2020, 10, 8. [Google Scholar] [CrossRef]
- Patraquim, P.; Magny, E.G.; Pueyo, J.I.; Platero, A.I.; Couso, J.P. Translation and natural selection of micropeptides from long non-canonical RNAs. Nat. Commun. 2022, 13, 6515. [Google Scholar] [CrossRef]
- Omote, N.; Sakamoto, K.; Li, Q.; Schupp, J.C.; Adams, T.; Ahangari, F.; Chioccioli, M.; DeIuliis, G.; Hashimoto, N.; Hasegawa, Y.; et al. Long noncoding RNA TINCR is a novel regulator of human bronchial epithelial cell differentiation state. Physiol. Rep. 2021, 9, e14727. [Google Scholar] [CrossRef]
- Koirala, M.; DiPaola, M. Overcoming Cancer Resistance: Strategies and Modalities for Effective Treatment. Biomedicines 2024, 12, 1801. [Google Scholar] [CrossRef] [PubMed]
- Suski, J.M.; Braun, M.; Strmiska, V.; Sicinski, P. Targeting cell-cycle machinery in cancer. Cancer Cell 2021, 39, 759–778. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e229. [Google Scholar] [CrossRef]
- Bonilauri, B.; Holetz, F.B.; Dallagiovanna, B. Long Non-Coding RNAs Associated with Ribosomes in Human Adipose-Derived Stem Cells: From RNAs to Microproteins. Biomolecules 2021, 11, 1673. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, R.; Lee, I.; Zhang, W.; Sun, J.; Meng, X.; Wang, W. Prediction of LncRNA-encoded small peptides in glioma and oligomer channel functional analysis using in silico approaches. PLoS ONE 2021, 16, e0248634. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.H.; Luo, J.L.; Hsu, M.H.; Chen, L.H.; Lu, T.P.; Tsai, M.H.; Chuang, E.Y.; Chuang, L.L.; Lai, L.C. Regulatory mechanisms and function of hypoxia-induced long noncoding RNA NDRG1-OT1 in breast cancer cells. Cell Death Dis. 2022, 13, 807. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Wang, H.; Xie, S.; Wu, Z.; Zhang, C. BVES-AS1 inhibits the malignant behaviors of colon adenocarcinoma cells via regulating BVES. Cell Biol. Int. 2021, 45, 1945–1956. [Google Scholar] [CrossRef]
- Poliseno, L.; Lanza, M.; Pandolfi, P.P. Coding, or non-coding, that is the question. Cell Res. 2024, 34, 609–629. [Google Scholar] [CrossRef]
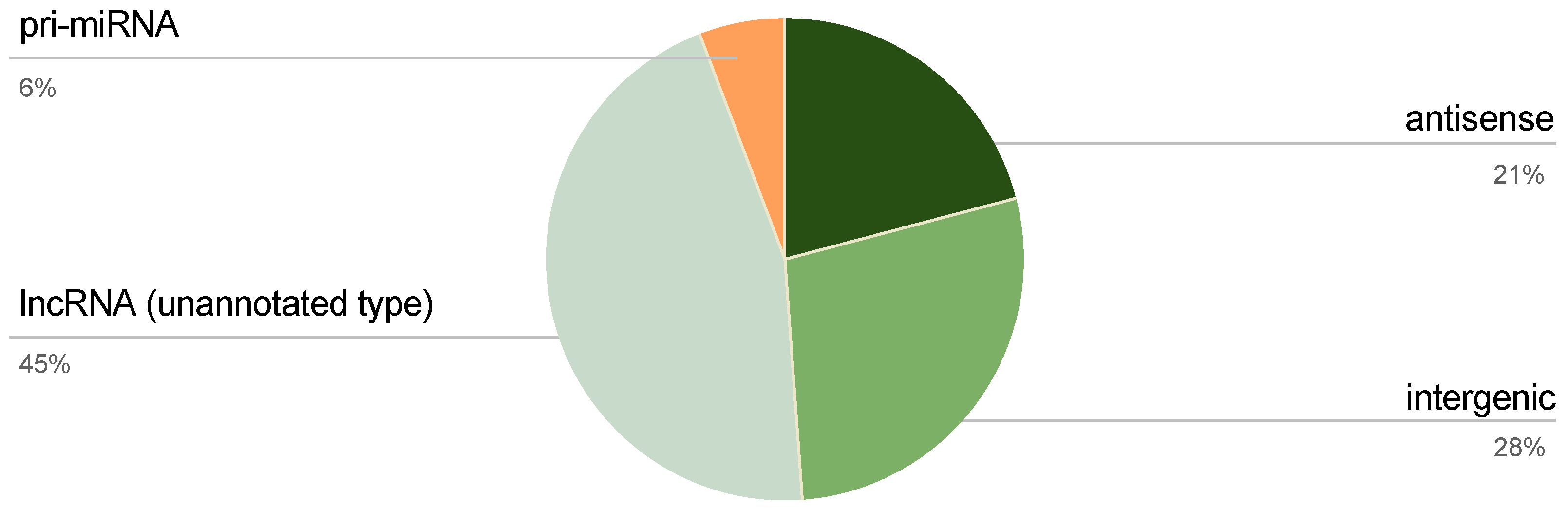
| Method | Strengths | Limitations |
|---|---|---|
| Ribo-seq | Maps ribosome-protected RNA fragments; identifies translation | Cannot distinguish functional vs. non-functional translation |
| Mass spectrometry | Direct peptide identification | Bias toward abundant proteins, missing low-expressed peptides |
| Proteogenomics | Integrates genomics with Mass spectrometry for novel peptide discovery | Computationally intensive |
| Machine learning bioinformatics (e.g., sORFfinder, ORFscore) | Predict sORFs and coding potential | Risk of false positives |
| ncRNA-PEP/MPs | ncRNA Transcript | Length (aa) | Cell Cycle Function | Sources |
|---|---|---|---|---|
| miPEP133 | miR-34a | 133 | Induces mitochondrial dysfunction- p53 dependent and independent cell cycle arrest | [39] |
| PINT87aa | circPINT-exon2 | 87 | G1 arrest, mitophagy inhibition via the PINK1/Parkin axis | [107] |
| SP0495 | TP73-AS1/KIAA0495 | 201 | G1 arrest, binds phosphoinositides, promotes autophagy | [114] |
| CASIMO1 | lncRNA CASIMO1 | 83 | Positively regulates mevalonate pathway, promotes G1/S transition | [18] |
| SMIM30 | LINC00998 | 59 | Controls intracellular calcium levels, modulates CDK4 and cyclin E2 to favor of G1/S transition | [70] |
| STMP1 | lncRNA 1810058I24Rik | 47 | Enhances mitochondrial respiration, modulates cyclin E2, CDK2, and E2F1 to favor of G1/S transition | [75] |
| BVES-AS1-201-50aa | BVES-AS1 | 50 | Promotes PCNA expression | [44] |
| DDUP | CTBP1-DT | 186 | Stabilizes Rad18 on damage sites and promotes homologous recombination and post-replication repair | [36] |
| LINC00954-ORF polypeptide | LINC00954 | 49 | Downregulation of PCNA and CDK1 | [51] |
| MIAC | AQP5-AS1 | 51 | Binds AQP2 and controls EGFR signaling, overexpression causes arrest in S and G2 phases | [56] |
| TP53LC04 | KLRK1-AS1 | 100 | Arrests cells in response to DNA damage, part of the p53 response | [117] |
| NoBody | LINC01420 | 68 | P-body dissolution during mitosis | [103] |
| CRNDEP | LINC00180 | 84 | Centrosome maturation and microtubules | [19] |
| pTINCR | TINCR | 87 | Expression negatively correlated with cell cycle genes | [110] |
| UbKEKS | UBBP4 | 76 | Nuclear protein trafficking | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshpande, A.; Mahale, S.; Kanduri, C. Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation. Cancers 2025, 17, 1555. https://doi.org/10.3390/cancers17091555
Deshpande A, Mahale S, Kanduri C. Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation. Cancers. 2025; 17(9):1555. https://doi.org/10.3390/cancers17091555
Chicago/Turabian StyleDeshpande, Ananya, Sagar Mahale, and Chandrasekhar Kanduri. 2025. "Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation" Cancers 17, no. 9: 1555. https://doi.org/10.3390/cancers17091555
APA StyleDeshpande, A., Mahale, S., & Kanduri, C. (2025). Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation. Cancers, 17(9), 1555. https://doi.org/10.3390/cancers17091555






