Deciphering Anticancer Mechanisms of Calycosin in Lung Adenocarcinoma Through Multi-Omics: Targeting SMAD3-Mediated NOTCH Signaling in the Tumor Microenvironment
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data Collection
2.2. Liquid Chromatography–Mass Spectrometry
2.3. Network Pharmacology
2.4. Immunohistochemical Staining
2.5. Prognostic Model Construction
2.6. Molecular Docking Analysis for SMAD3-Drug Interactions
2.7. Pearson’s Correlation Analysis to Assess Gene–Gene Relationships in LUAD
2.8. Weight Correlation Network Analysis for SMAD3-Related Genes in LUAD
2.9. Enrichment Analysis
2.10. Differential Immune Cell Infiltration Analysis of SMAD3
2.11. Single-Cell RNA Sequencing Analysis and Cell–Cell Communication
2.12. Statistical Analysis
3. Results
3.1. High-Resolution LC-MS Analysis for Astragalus membranaceus Water Extract
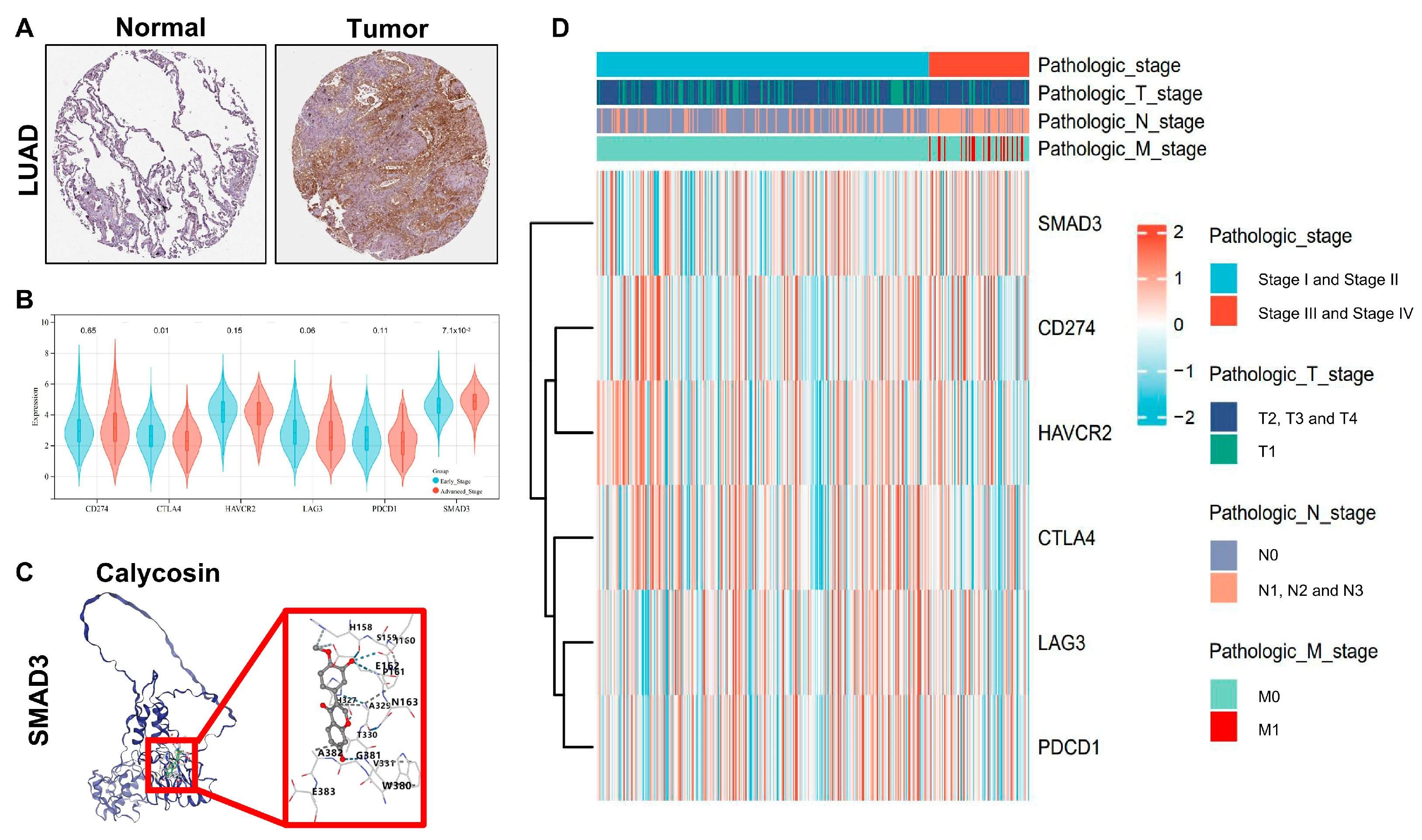
3.2. SMAD3 Identified as a Key Prognostic Biomarker for LUAD Staging and Survival Prediction
3.3. SMAD3 as a Novel Clinical Stage Biomarker in LUAD
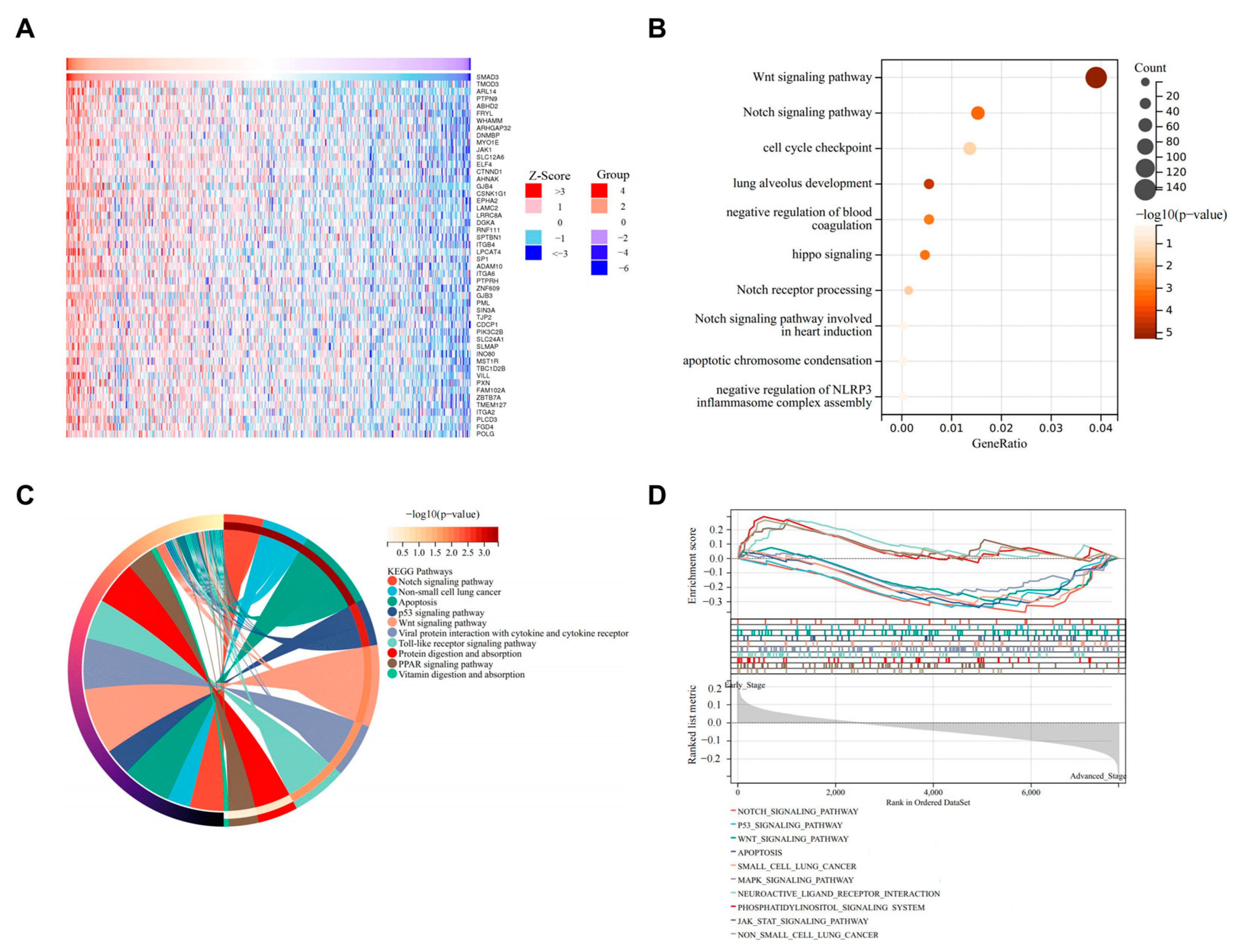
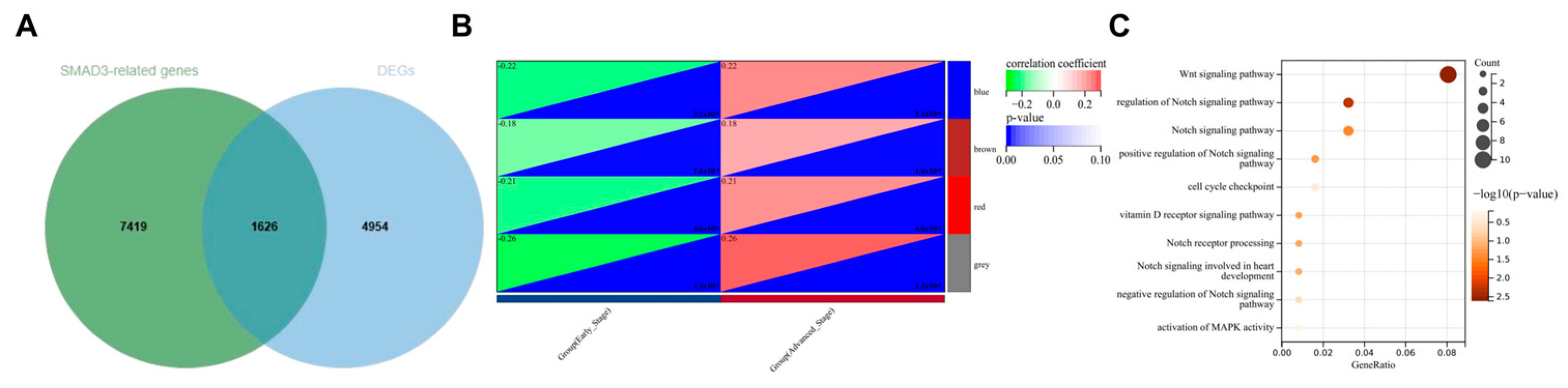
3.4. SMAD3-Mediated LUAD via NOTCH Signaling Pathway
3.5. WGCNA to Confirm NOTCH Signaling Pathway as Primary Mechanism in SMAD3-Mediated LUAD
3.6. Immune Infiltration Analysis of SMAD3 in Common Immune Cell Types
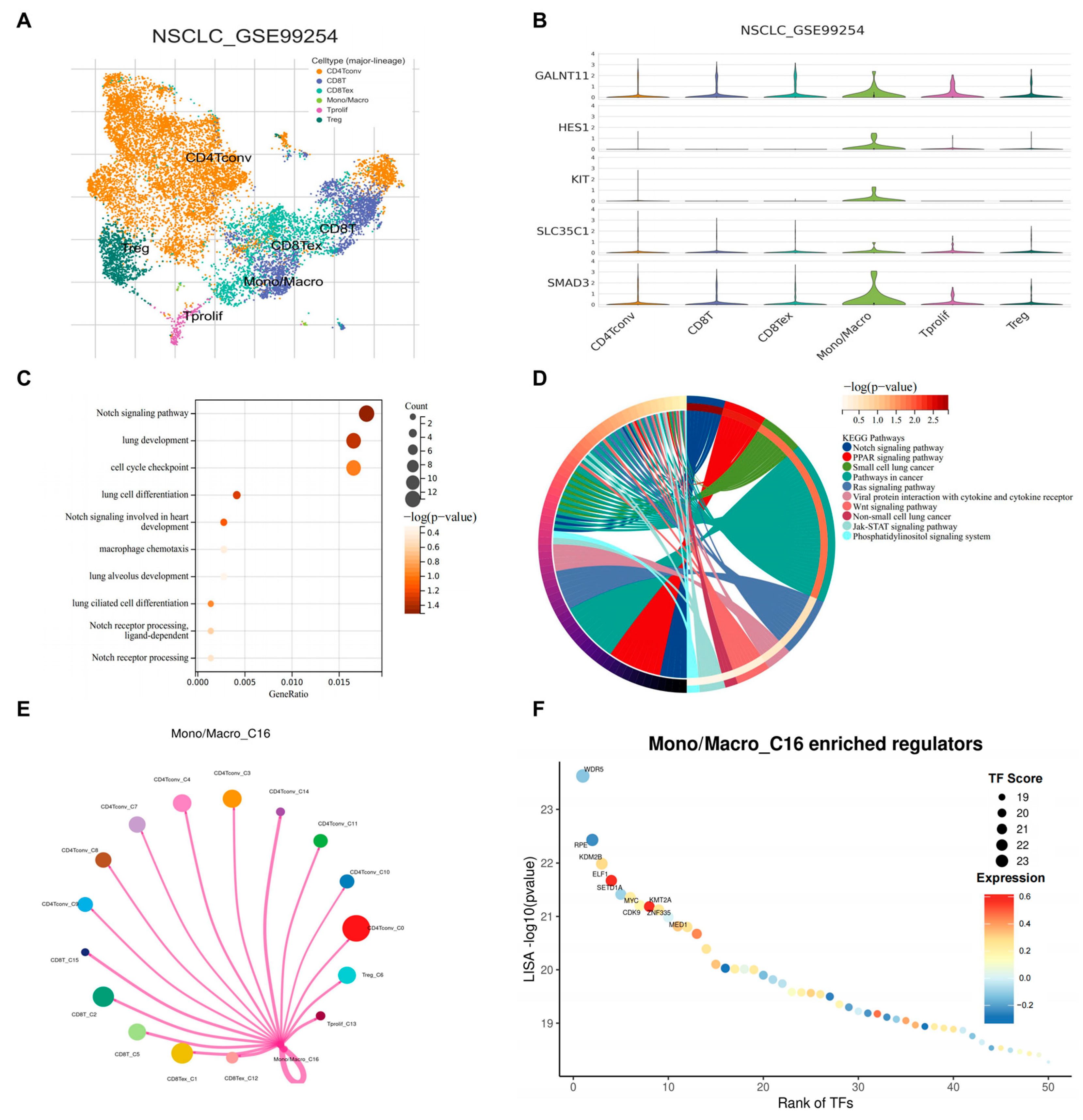
3.7. scRNA-Seq Identified SMAD3 Mediated NOTCH Signaling in Monocytes/Macrophages
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BATMAN-TCM | Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine |
| CD4Tconv | CD4+ T cells conventional |
| CD8T | CD8+ T cells |
| CD8Tex | CD8+ T cells exhausted |
| CTLA4 | Cytotoxic T-Lymphocyte-Associated Protein 4 |
| EMT | Epithelial–mesenchymal transition |
| GALNT11 | Polypeptide N-Acetylgalactosaminyltransferase 11 |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HAVCR2 | Hepatitis A Virus Cellular Receptor 2 |
| HES1 | Hes Family BHLH Transcription Factor 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KIT | KIT Proto-Oncogene, Receptor Tyrosine Kinase |
| KMT2A | Lysine Methyltransferase 2A |
| LAG3 | Lymphocyte Activation Gene 3 |
| LASSO | Least Absolute Shrinkage and Selection Operator [38] |
| LUAD | Lung adenocarcinoma |
| NOTCH | Notch Signaling Pathway |
| PDCD1 | Programmed Cell Death 1 |
| PD-1 | Programmed Death 1 |
| PD-L1 | Programmed Death-Ligand 1 |
| ROC | Receiver operating characteristic |
| SLC35C1 | Solute Carrier Family 35 Member C1 |
| SMAD2 | SMAD Family Member 2 |
| SMAD3 | SMAD Family Member 3 |
| TCGA | The Cancer Genome Atlas |
| TGF-β | Transforming Growth Factor Beta |
| TIGIT | T-cell Immunoreceptor with Ig and ITIM Domains |
| TIM3 | T-Cell Immunoglobulin and Mucin-Domain Containing 3 |
| TME | Tumor microenvironment |
| TNM | Tumor-Node-Metastasis |
| Tprolif | Proliferating T cells |
| Treg | Regulatory T cells |
| TUBB6 | Tubulin Beta 6 Class V |
| WGCNA | Weighted Correlation Network Analysis |
References
- Hua, L.; Wu, J.; Ge, J.; Li, X.; You, B.; Wang, W.; Hu, B. Identification of lung adenocarcinoma subtypes and predictive signature for prognosis, immune features, and immunotherapy based on immune checkpoint genes. Front. Cell Dev. Biol. 2023, 11, 1060086. [Google Scholar] [CrossRef]
- Wang, D.; Chan, B.C.; Zhang, B.; Wong, K.C.; Kan, L.L.; Wong, C.K. Dark under the Lamp: Neglected Biological Pollutants in the Environment Are Closely Linked to Lung Cancer. Int. J. Mol. Sci. 2024, 25, 3081. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, A.; Akter, Z.; Modareszadeh, P.; Modareszadeh, P.; Berisha, E.; Alemi, P.S.; Chacon Castro, M.D.C.; Deese, A.R.; Zhang, L. Current Landscape of Therapeutic Resistance in Lung Cancer and Promising Strategies to Overcome Resistance. Cancers 2022, 14, 4562. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Ma, Y.; Lei, J.; Zhu, J.; Xie, N.; Tian, F.; Lu, Q.; Wen, M.; Zheng, Q.; Han, Y.; et al. Highly proliferating cancer cells function as novel prognostic biomarkers for lung adenocarcinoma with particular usefulness for stage IA risk stratification. BMC Cancer 2025, 25, 25. [Google Scholar] [CrossRef]
- Lamort, A.S.; Kaiser, J.C.; Pepe, M.A.A.; Lilis, I.; Ntaliarda, G.; Somogyi, K.; Spella, M.; Behrend, S.J.; Giotopoulou, G.A.; Kujawa, W.; et al. Prognostic phenotypes of early-stage lung adenocarcinoma. Eur. Respir. J. 2022, 60, 2101674. [Google Scholar] [CrossRef]
- Roberts, A.B.; Russo, A.; Felici, A.; Flanders, K.C. Smad3: A key player in pathogenetic mechanisms dependent on TGF-beta. Ann. N. Y. Acad. Sci. 2003, 995, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, Y.; Wu, X.; Bo, J.; Zhang, L.; Zhang, J.; Hu, Y.; Chen, Y.; Zeng, Y.; Wei, X.; et al. POH1 induces Smad3 deubiquitination and promotes lung cancer metastasis. Cancer Lett. 2024, 582, 216526. [Google Scholar] [CrossRef]
- Chung, J.Y.-F.; Tang, P.C.-T.; Chan, M.K.-K.; Xue, V.W.; Huang, X.-R.; Ng, C.S.-H.; Zhang, D.; Leung, K.-T.; Wong, C.-K.; Lee, T.-L.; et al. Smad3 is essential for polarization of tumor-associated neutrophils in non-small cell lung carcinoma. Nat. Commun. 2023, 14, 1794. [Google Scholar] [CrossRef]
- Yan, Z.; Lai, Z.; Lin, J. Anticancer Properties of Traditional Chinese Medicine. Comb. Chem. High Throughput Screen. 2017, 20, 423–429. [Google Scholar] [CrossRef]
- Auyeung, K.K.; Han, Q.B.; Ko, J.K. Astragalus membranaceus: A Review of its Protection Against Inflammation and Gastrointestinal Cancers. Am. J. Chin. Med. 2016, 44, 1–22. [Google Scholar] [CrossRef]
- Tang, Z.; Tian, X. Astragalus membranaceus: A Traditional Chinese Medicine with Multifaceted Impacts on Breast Cancer Treatment. Biomolecules 2024, 14, 1339. [Google Scholar] [CrossRef]
- Gong, G.; Zheng, Y.; Yang, Y.; Sui, Y.; Wen, Z. Pharmaceutical Values of Calycosin: One Type of Flavonoid Isolated from Astragalus. Evid. Based Complement. Altern. Med. 2021, 2021, 9952578. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Ma, G.; Li, X.; Shi, Q.; Li, X.; Zhou, X.; Tang, Y.; Xie, Z.; Liao, S.; Qin, Y.; et al. Clinical case report of patients with osteosarcoma and anticancer benefit of calycosin against human osteosarcoma cells. J. Cell Biochem. 2019, 120, 10697–10706. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Pan, Q.; Wang, L.; Yi, S.; Liu, P.; Huang, W. Anticancer targets and mechanisms of calycosin to treat nasopharyngeal carcinoma. Biofactors 2020, 46, 675–684. [Google Scholar] [CrossRef]
- Liu, Y.; Piao, X.-J.; Xu, W.-T.; Zhang, Y.; Zhang, T.; Xue, H.; Li, Y.-N.; Zuo, W.-B.; Sun, G.; Fu, Z.-R.; et al. Calycosin induces mitochondrial-dependent apoptosis and cell cycle arrest, and inhibits cell migration through a ROS-mediated signaling pathway in HepG2 hepatocellular carcinoma cells. Toxicol. Vitr. 2021, 70, 105052. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Leung, P.C.; Cho, W.C.; Wong, C.K.; Wang, D. Targeting PI3K signaling in Lung Cancer: Advances, challenges and therapeutic opportunities. J. Transl. Med. 2025, 23, 184. [Google Scholar] [CrossRef]
- Ji, M.; Wang, D.; Lin, S.; Wang, C.; Li, L.; Zhang, Z.; Jin, J.; Wu, D.; Dong, Y.; Xu, H.; et al. A novel PI3K inhibitor XH30 suppresses orthotopic glioblastoma and brain metastasis in mice models. Acta Pharm. Sin. B 2022, 12, 774–786. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Ginnebaugh, K.R.; Ahmad, A.; Sarkar, F.H. The therapeutic potential of targeting the epithelial-mesenchymal transition in cancer. Expert Opin. Ther. Targets 2014, 18, 731–745. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Lin, Y.-T.; Lin, L.-J.; Chang, Y.-H.; Chen, H.-Y.; Wang, Y.-P.; Shih, J.-Y.; Yu, C.-J.; Yang, P.-C. Stage Shift Improves Lung Cancer Survival: Real-World Evidence. J. Thorac. Oncol. 2023, 18, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.A.; Patel, V.P.; Bhosle, K.P.; Nagare, S.D.; Thombare, K.C. The tumor microenvironment: Shaping cancer progression and treatment response. J. Chemother. 2025, 37, 15–44. [Google Scholar] [CrossRef]
- Bied, M.; Ho, W.W.; Ginhoux, F.; Blériot, C. Roles of macrophages in tumor development: A spatiotemporal perspective. Cell. Mol. Immunol. 2023, 20, 983–992. [Google Scholar] [CrossRef]
- Ostuni, R.; Kratochvill, F.; Murray, P.J.; Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015, 36, 229–239. [Google Scholar] [CrossRef]
- Zhang, B.T.; Leung, P.C.; Wong, C.K.; Wang, D.J. The Immunomodulatory Effects of Vitamin D on COVID-19 Induced Glioblastoma Recurrence via the PI3K-AKT Signaling Pathway. Int. J. Mol. Sci. 2024, 25, 2952. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Pu, J.; Teng, Z.; Yang, W.; Zhu, P.; Zhang, T.; Zhang, D.; Wang, B.; Hu, Z.; Song, Q. Construction of a prognostic model for lung squamous cell carcinoma based on immune-related genes. Carcinogenesis 2023, 44, 143–152. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, D.; Xu, B. The combination of molecular docking and network pharmacology reveals the molecular mechanism of Danggui Niantong decoction in treating gout. Medicine 2022, 101, e31535. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Zheng, L.; Zheng, C.; Song, J.; Zhang, Q.; Kang, B.; Liu, Z.; Jin, L.; Xing, R.; et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Cha, H.; Kim, J.; Jang, Y.; Son, Y.; Joe, C.Y.; Kim, J.; Kim, J.; Lee, S.H.; Lee, S. Dissecting transcriptome signals of anti-PD-1 response in lung adenocarcinoma. Sci. Rep. 2024, 14, 21096. [Google Scholar] [CrossRef] [PubMed]
- Guégan, J.P.; Peyraud, F.; Dadone-Montaudie, B.; Teyssonneau, D.; Palmieri, L.J.; Clot, E.; Cousin, S.; Roubaud, G.; Cabart, M.; Leroy, L.; et al. Analysis of PD1, LAG3, TIGIT, and TIM3 expression in human lung adenocarcinoma reveals a 25-gene signature predicting immunotherapy response. Cell Rep. Med. 2024, 5, 101831. [Google Scholar] [CrossRef]
- Song, P.; Li, W.; Wu, X.; Qian, Z.; Ying, J.; Gao, S.; He, J. Integrated analysis of single-cell and bulk RNA-sequencing identifies a signature based on B cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma. Cancer Immunol. Immunother. 2022, 71, 2341–2354. [Google Scholar] [CrossRef] [PubMed]
- Bacolod, M.D.; Barany, F.; Fisher, P.B. Can CpG methylation serve as surrogate markers for immune infiltration in cancer? Adv. Cancer Res. 2019, 143, 351–384. [Google Scholar] [CrossRef]
- Tang, P.C.; Chung, J.Y.; Xue, V.W.; Xiao, J.; Meng, X.M.; Huang, X.R.; Zhou, S.; Chan, A.S.; Tsang, A.C.; Cheng, A.S.; et al. Smad3 Promotes Cancer-Associated Fibroblasts Generation via Macrophage-Myofibroblast Transition. Adv. Sci. 2022, 9, e2101235. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, W.; Yin, T.; Lu, L. Calycosin suppresses TGF-β-induced epithelial-to-mesenchymal transition and migration by upregulating BATF2 to target PAI-1 via the Wnt and PI3K/Akt signaling pathways in colorectal cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 240. [Google Scholar] [CrossRef]
- Tang, P.M.-K.; Zhou, S.; Meng, X.-M.; Wang, Q.-M.; Li, C.-J.; Lian, G.-Y.; Huang, X.-R.; Tang, Y.-J.; Guan, X.-Y.; Yan, B.P.-Y.; et al. Smad3 promotes cancer progression by inhibiting E4BP4-mediated NK cell development. Nat. Commun. 2017, 8, 14677. [Google Scholar] [CrossRef]
- Chen, C.; Du, Y.; Nie, R.; Wang, S.; Wang, H.; Li, P. Notch signaling in cancers: Mechanism and potential therapy. Front. Cell Dev. Biol. 2025, 13, 1542967. [Google Scholar] [CrossRef]
- Akil, A.; Gutiérrez-García, A.K.; Guenter, R.; Rose, J.B.; Beck, A.W.; Chen, H.; Ren, B. Notch Signaling in Vascular Endothelial Cells, Angiogenesis, and Tumor Progression: An Update and Prospective. Front. Cell Dev. Biol. 2021, 9, 642352. [Google Scholar] [CrossRef]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch signaling pathway in cancer: From mechanistic insights to targeted therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, R.; Hernandez, S.C.; Hanna, A.; Su, K.; Shinde, A.V.; Frangogiannis, N.G. Differential effects of Smad2 and Smad3 in regulation of macrophage phenotype and function in the infarcted myocardium. J. Mol. Cell Cardiol. 2022, 171, 1–15. [Google Scholar] [CrossRef]
- Ye, Y.C.; Zhao, J.L.; Lu, Y.T.; Gao, C.C.; Yang, Y.; Liang, S.Q.; Lu, Y.Y.; Wang, L.; Yue, S.Q.; Dou, K.F.; et al. NOTCH Signaling via WNT Regulates the Proliferation of Alternative, CCR2-Independent Tumor-Associated Macrophages in Hepatocellular Carcinoma. Cancer Res. 2019, 79, 4160–4172. [Google Scholar] [CrossRef] [PubMed]
- Palaga, T.; Wongchana, W.; Kueanjinda, P. Notch Signaling in Macrophages in the Context of Cancer Immunity. Front. Immunol. 2018, 9, 652. [Google Scholar] [CrossRef]
- Li, X.; Yan, X.; Wang, Y.; Kaur, B.; Han, H.; Yu, J. The Notch signaling pathway: A potential target for cancer immunotherapy. J. Hematol. Oncol. 2023, 16, 45. [Google Scholar] [CrossRef]
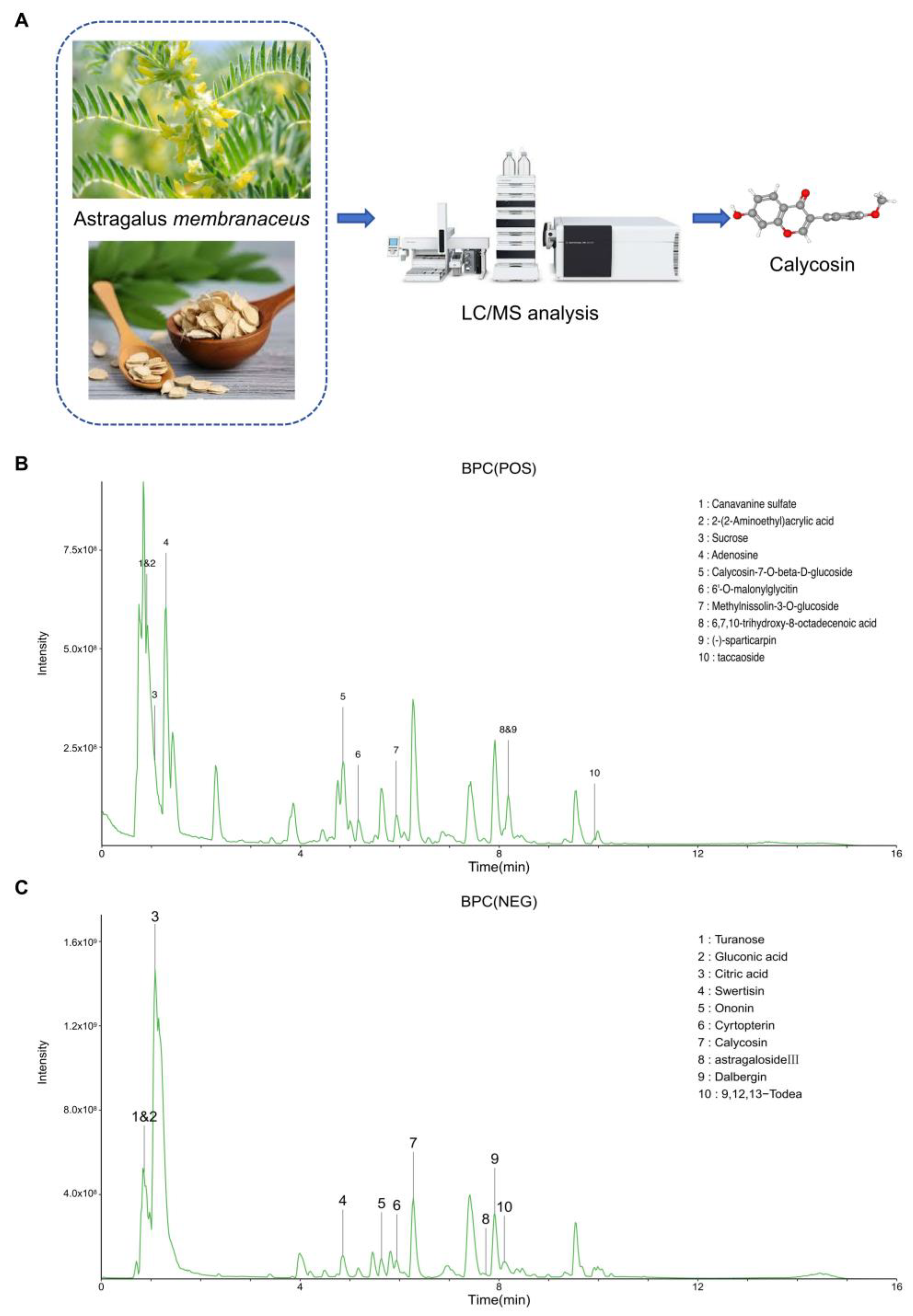


| Category | Components | Percentage% |
|---|---|---|
| POS | Adenosine | 4.640 |
| Canavanine sulfate | 4.366 | |
| Calycosin-7-O-beta-D-glucoside | 3.864 | |
| 2-(2-Aminoethyl)acrylic acid | 2.304 | |
| Methylnissolin-3-O-glucoside | 1.702 | |
| Taccaoside | 1.620 | |
| 6′-O-malonylglycitin | 1.503 | |
| 6,7,10-trihydroxy-8-octadecenoic acid | 1.493 | |
| Sucrose | 1.203 | |
| (-)-sparticarpin | 1.001 | |
| NEG | Turanose | 20.302 |
| Citric acid | 12.130 | |
| Calycosin | 3.143 | |
| Dalbergin | 2.981 | |
| 9,12,13-Todea | 2.402 | |
| Swertisin | 2.324 | |
| Gluconic acid | 2.107 | |
| Astragaloside III | 1.599 | |
| Ononin | 1.512 | |
| Cyrtopterin | 1.501 |
| CurPocket ID | Docking Score (kcal/mol) |
|---|---|
| C4 | −7.9 |
| C2 | −7.1 |
| C3 | −7.1 |
| C1 | −7 |
| C5 | −7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.-T.; Song, X.; Cho, C.-S.; Wong, C.-K.; Wang, D. Deciphering Anticancer Mechanisms of Calycosin in Lung Adenocarcinoma Through Multi-Omics: Targeting SMAD3-Mediated NOTCH Signaling in the Tumor Microenvironment. Cancers 2025, 17, 1455. https://doi.org/10.3390/cancers17091455
Zhang B-T, Song X, Cho C-S, Wong C-K, Wang D. Deciphering Anticancer Mechanisms of Calycosin in Lung Adenocarcinoma Through Multi-Omics: Targeting SMAD3-Mediated NOTCH Signaling in the Tumor Microenvironment. Cancers. 2025; 17(9):1455. https://doi.org/10.3390/cancers17091455
Chicago/Turabian StyleZhang, Bi-Tian, Xiaoyu Song, Chi-Shing (William) Cho, Chun-Kwok Wong, and Dongjie Wang. 2025. "Deciphering Anticancer Mechanisms of Calycosin in Lung Adenocarcinoma Through Multi-Omics: Targeting SMAD3-Mediated NOTCH Signaling in the Tumor Microenvironment" Cancers 17, no. 9: 1455. https://doi.org/10.3390/cancers17091455
APA StyleZhang, B.-T., Song, X., Cho, C.-S., Wong, C.-K., & Wang, D. (2025). Deciphering Anticancer Mechanisms of Calycosin in Lung Adenocarcinoma Through Multi-Omics: Targeting SMAD3-Mediated NOTCH Signaling in the Tumor Microenvironment. Cancers, 17(9), 1455. https://doi.org/10.3390/cancers17091455









