Germline DNA Repair Gene Mutations and Clonal Hematopoiesis (CH) in 24,849 Patients with BRCA-Associated Cancers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Sequencing Methods
2.3. Statistical Methods
3. Results
3.1. Breast Cancer
3.2. Ovarian Cancer
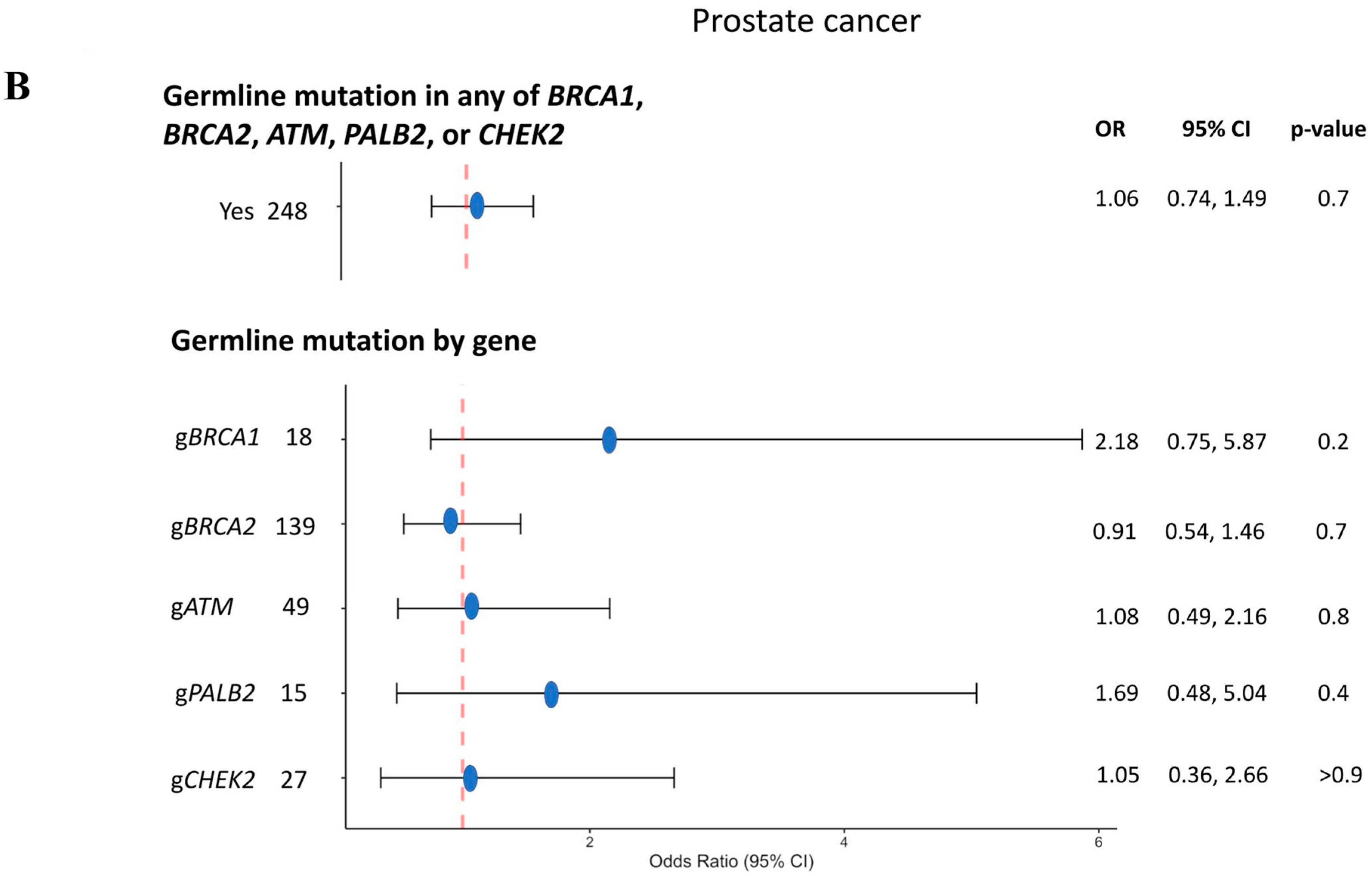
3.3. Prostate Cancer
3.4. Pancreatic Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CH | Clonal hematopoiesis |
| gHRR | Germline Homologous recombination repair |
| HRR | Homologous recombination repair |
| IQR | Interquartile range |
| OR | Odds ratio |
| PARP | poly-ADP ribose polymerase |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Sonkin, D.; Thomas, A.; Teicher, B.A. Cancer treatments: Past, present, and future. Cancer Genet. 2024, 286–287, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Ebert, B.L. Clonal hematopoiesis in human aging and disease. Science 2019, 366, 6465. [Google Scholar] [CrossRef]
- Xie, M.; Lu, C.; Wang, J.; McLellan, M.D.; Johnson, K.J.; Wendl, M.C.; McMichael, J.F.; Schmidt, H.K.; Yellapantula, V.; A Miller, C.; et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014, 20, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.H.; Gondek, L.P.; Luo, J.; Antonarakis, E.S. Clonal Hematopoiesis of Indeterminate Potential in Patients with Solid Tumor Malignancies. Cancer Res. 2022, 82, 4107–4113. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Hong, W.; Li, A.; Liu, Y.; Xiao, X.; Christiani, D.C.; Hung, R.J.; McKay, J.; Field, J.; Amos, C.I.; Cheng, C. Clonal Hematopoiesis Mutations in Patients with Lung Cancer Are Associated with Lung Cancer Risk Factors. Cancer Res. 2021, 82, 199–209. [Google Scholar] [CrossRef]
- Dawoud, A.A.Z.; Tapper, W.J.; Cross, N.C.P. Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia 2020, 34, 2660–2672. [Google Scholar] [CrossRef]
- Miller, P.G.; Qiao, D.; Rojas-Quintero, J.; Honigberg, M.C.; Sperling, A.S.; Gibson, C.J.; Bick, A.G.; Niroula, A.; McConkey, M.E.; Sandoval, B.; et al. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood 2021, 139, 357–368. [Google Scholar] [CrossRef]
- Bolton, K.L.; Ptashkin, R.N.; Gao, T.; Braunstein, L.; Devlin, S.M.; Kelly, D.; Patel, M.; Berthon, A.; Syed, A.; Yabe, M.; et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 2020, 52, 1219–1226. [Google Scholar] [CrossRef]
- Marshall, C.H.; Gondek, L.P.; Daniels, V.; Lu, C.; Pasca, S.; Xie, J.; Markowski, M.C.; Paller, C.J.; Sena, L.A.; Denmeade, S.R.; et al. Association of PARP inhibitor treatment on the prevalence and progression of clonal hematopoiesis in patients with advanced prostate cancer. Prostate 2024, 84, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.P.; Quiros, P.M.; Gu, M.; Jiang, T.; Mitchell, J.; Langdon, R.; Iyer, V.; Barcena, C.; Vijayabaskar, M.S.; Fabre, M.A.; et al. Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat. Genet. 2022, 54, 1155–1166. [Google Scholar] [CrossRef]
- DeBoy, E.A.; Tassia, M.G.; Schratz, K.E.; Yan, S.M.; Cosner, Z.L.; McNally, E.J.; Gable, D.L.; Xiang, Z.; Lombard, D.B.; Antonarakis, E.S.; et al. Familial Clonal Hematopoiesis in a Long Telomere Syndrome. N. Engl. J. Med. 2023, 388, 2422–2433. [Google Scholar] [CrossRef] [PubMed]
- Franch-Expósito, S.; Mehine, M.; Ptashkin, R.N.; Bolton, K.L.; Bandlamudi, C.; Srinivasan, P.; Zhang, L.; Goodell, M.A.; Gedvilaite, E.; Menghrajani, K.; et al. Associations Between Cancer Predisposition Mutations and Clonal Hematopoiesis in Patients With Solid Tumors. JCO Precis. Oncol. 2023, 7, e2300070. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.H.; Antonarakis, E.S.; Patnaik, M.M. Radiotherapeutics, clonal hematopoiesis, and risk of hematologic malignancies: The good, the bad, the ugly. Blood Rev. 2025, 70, 101269. [Google Scholar] [CrossRef]
- Beaubier, N.; Bontrager, M.; Huether, R.; Igartua, C.; Lau, D.; Tell, R.; Bobe, A.M.; Bush, S.; Chang, A.L.; Hoskinson, D.C.; et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat. Biotechnol. 2019, 37, 1351–1360. [Google Scholar] [CrossRef]
- Beaubier, N.; Tell, R.; Lau, D.; Parsons, J.R.; Bush, S.; Perera, J.; Sorrells, S.; Baker, T.; Chang, A.; Michuda, J.; et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 2019, 10, 2384–2396. [Google Scholar] [CrossRef]
- Arafa, A.T.; Yadav, S.; Marshall, C.H.; Mauer, E.; Huang, M.; Yilma, B.; van der Pol, Y.; Fragkogianni, S.; Teslow, E.A.; Kellen, S.; et al. Germline-Somatic Interactions in BRCA-Associated Cancers: Unique Molecular Profiles and Clinical Outcomes Linking ATM to TP53 Synthetic Essentiality. Clin. Cancer Res. 2025. [Google Scholar] [CrossRef]
- Coombs, C.C.; Zehir, A.; Devlin, S.M.; Kishtagari, A.; Syed, A.; Jonsson, P.; Hyman, D.M.; Solit, D.B.; Robson, M.E.; Baselga, J.; et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017, 21, 374–382.e4. [Google Scholar] [CrossRef]
- Mateo, J.; Cheng, H.H.; Beltran, H.; Dolling, D.; Xu, W.; Pritchard, C.C.; Mossop, H.; Rescigno, P.; Perez-Lopez, R.; Sailer, V.; et al. Clinical Outcome of Prostate Cancer Patients with Germline DNA Repair Mutations: Retrospective Analysis from an International Study. Eur. Urol. 2018, 73, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Watson, P.; Conway, T.A.; Lynch, J.F. Clinical/genetic features in hereditary breast cancer. Breast Cancer Res. Treat. 1990, 15, 63–71. [Google Scholar] [CrossRef]
- Takahashi, K.; Nakada, D.; Goodell, M. Distinct landscape and clinical implications of therapy-related clonal hematopoiesis. J. Clin. Investig. 2024, 134, e180069. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713.e6. [Google Scholar] [CrossRef]
- Wong, T.N.; Ramsingh, G.; Young, A.L.; Miller, C.A.; Touma, W.; Welch, J.S.; Lamprecht, T.L.; Shen, D.; Hundal, J.; Fulton, R.S.; et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015, 518, 552–555. [Google Scholar] [CrossRef]
- Brown, T.J.; Reiss, K.A. PARP Inhibitors in Pancreatic Cancer. Cancer J. 2021, 27, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ditonno, F.; Bianchi, A.; Malandra, S.; Porcaro, A.B.; Fantinel, E.; Negrelli, R.; Ferro, M.; Milella, M.; Brunelli, M.; Autorino, R.; et al. PARP Inhibitors in Metastatic Prostate Cancer: A Comprehensive Systematic Review and Meta-analysis of Existing Evidence. Clin. Genitourin. Cancer 2023, 22, 402–412.e17. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.Y.; Jie, Y.E.; Cheng, S.W.; Ling, G.L.; Ming, H.V.Y. PARP Inhibitors in Breast and Ovarian Cancer. Cancers 2023, 15, 2357. [Google Scholar] [CrossRef]
- Arends, C.M.; Kopp, K.; Hablesreiter, R.; Estrada, N.; Christen, F.; Moll, U.M.; Zeillinger, R.; Schmitt, W.D.; Sehouli, J.; Kulbe, H.; et al. Dynamics of clonal hematopoiesis under DNA-damaging treatment in patients with ovarian cancer. Leukemia 2024, 38, 1378–1389. [Google Scholar] [CrossRef]
- Buttigieg, M.M.; Vlasschaert, C.; Bick, A.G.; Vanner, R.J.; Rauh, M.J. Inflammatory reprogramming of the solid tumor microenvironment by infiltrating clonal hematopoiesis is associated with adverse outcomes. Cell Rep. Med. 2025, 6, 101989. [Google Scholar] [CrossRef]
- Lappalainen, T.; Scott, A.J.; Brandt, M.; Hall, I.M. Genomic Analysis in the Age of Human Genome Sequencing. Cell 2019, 177, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Malapelle, U.; André, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients With Cancer. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Marshall, C.H.; Brown, D.W.; Decker, B.J.; Fine, A.D.; Xu, C.; Johnson, K.; Sun, D.; McDevitt, M.; Hughes, J.D.; et al. Equal-depth sequencing of white blood cells (WBC) and plasma from prostate cancer (PCa) liquid biopsies (LBx) and association with clonal hematopoiesis (CH) confounders in clinically relevant genes. J. Clin. Oncol. 2025, 43, 221. [Google Scholar] [CrossRef]
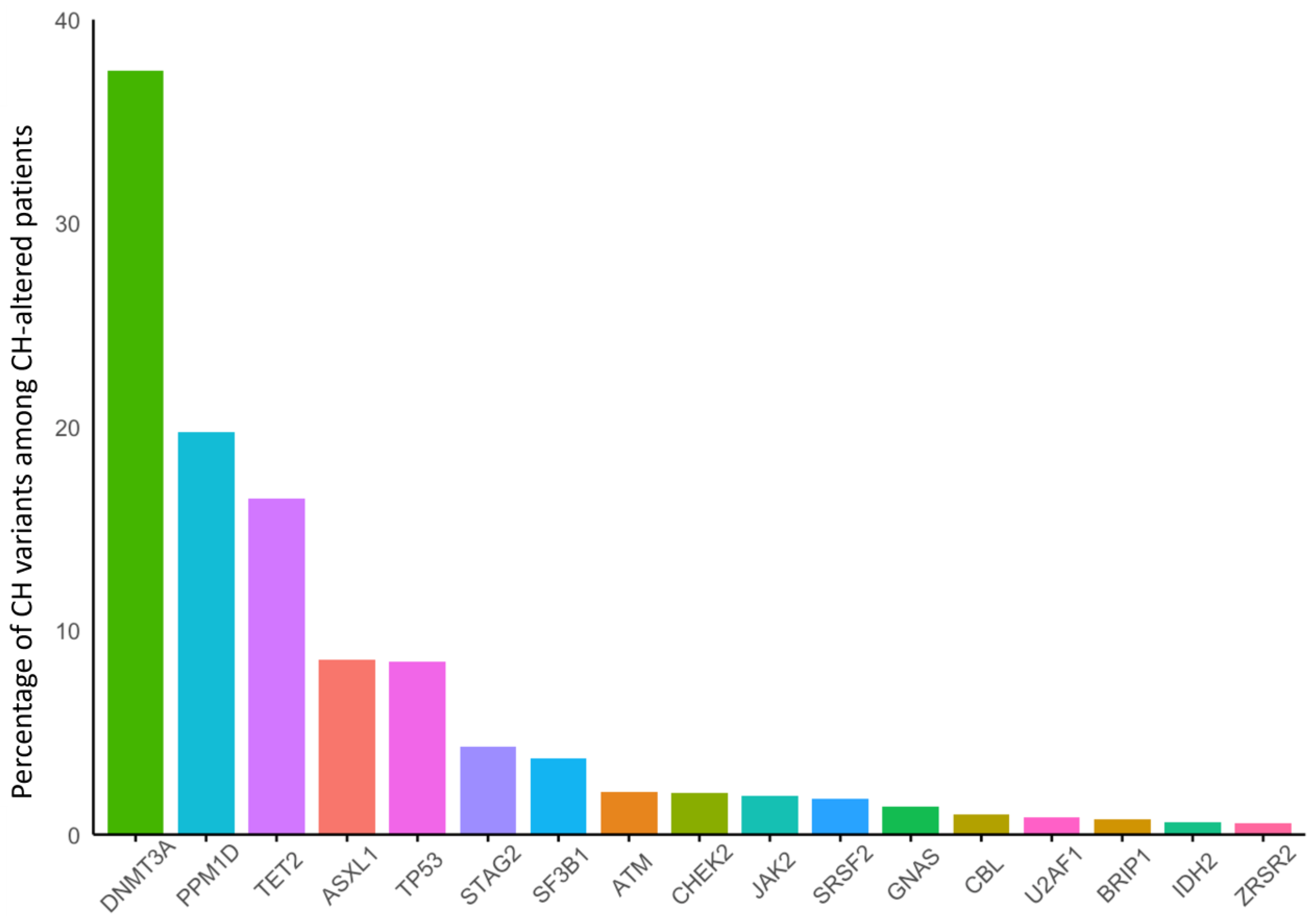
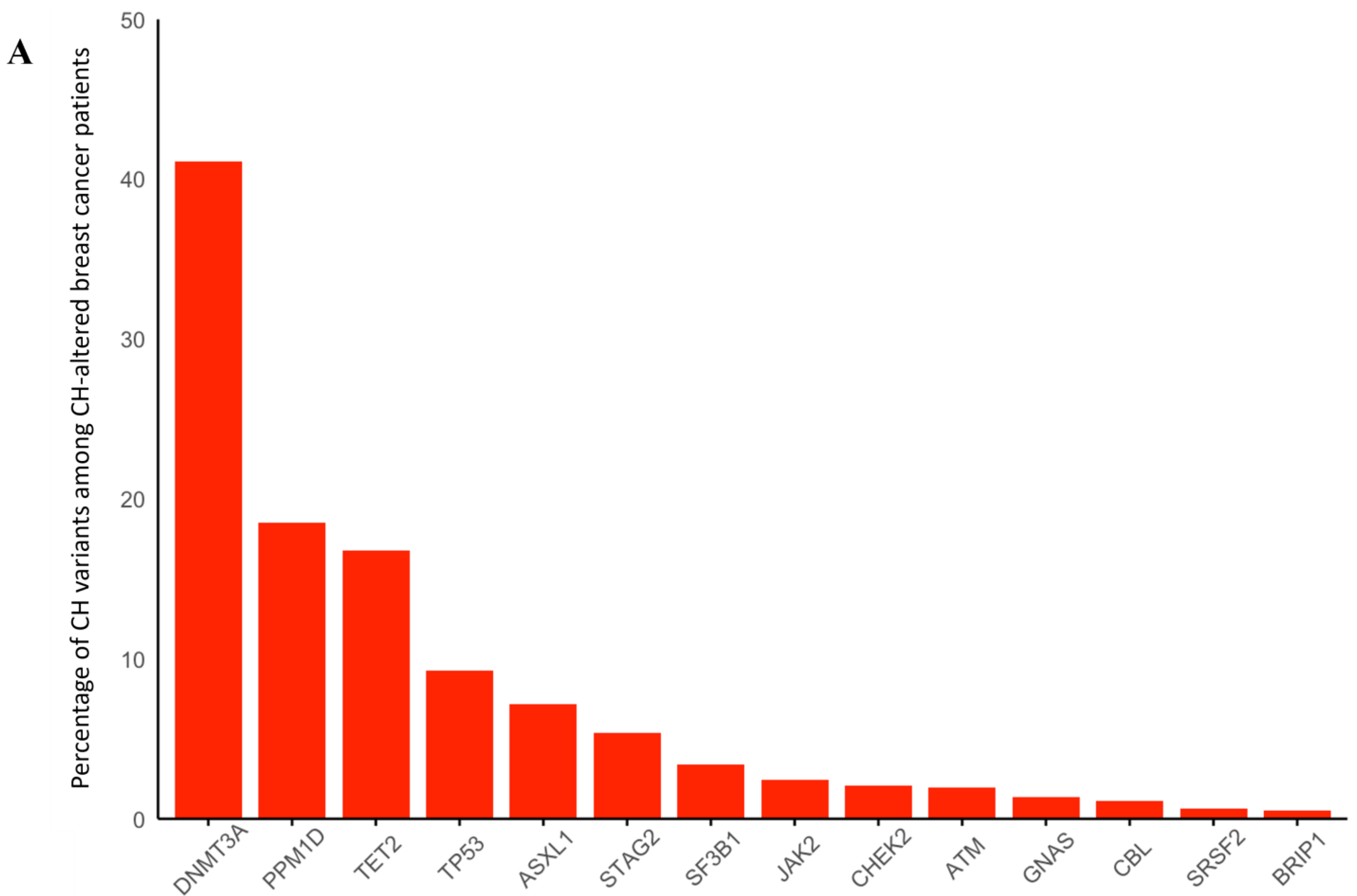
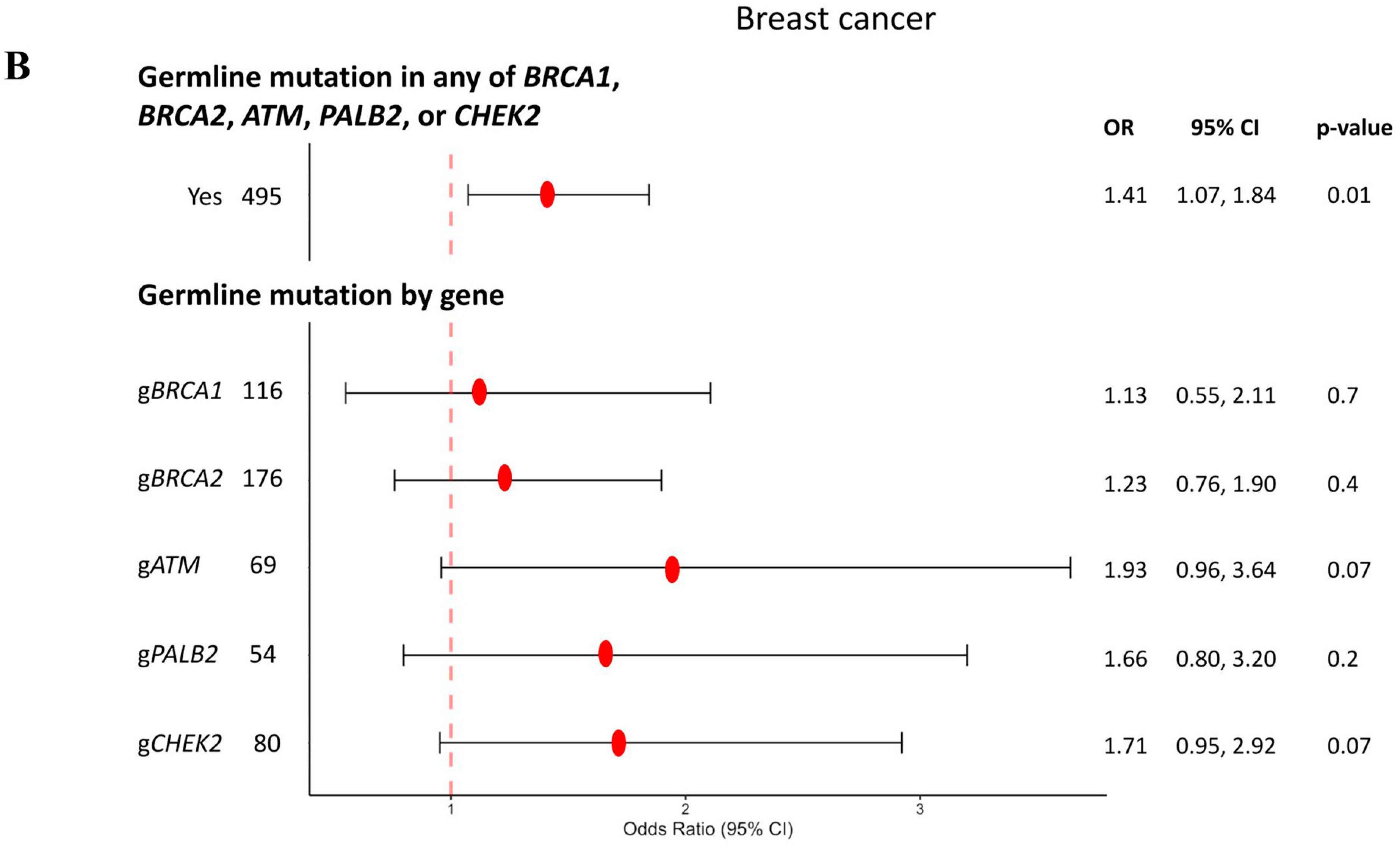


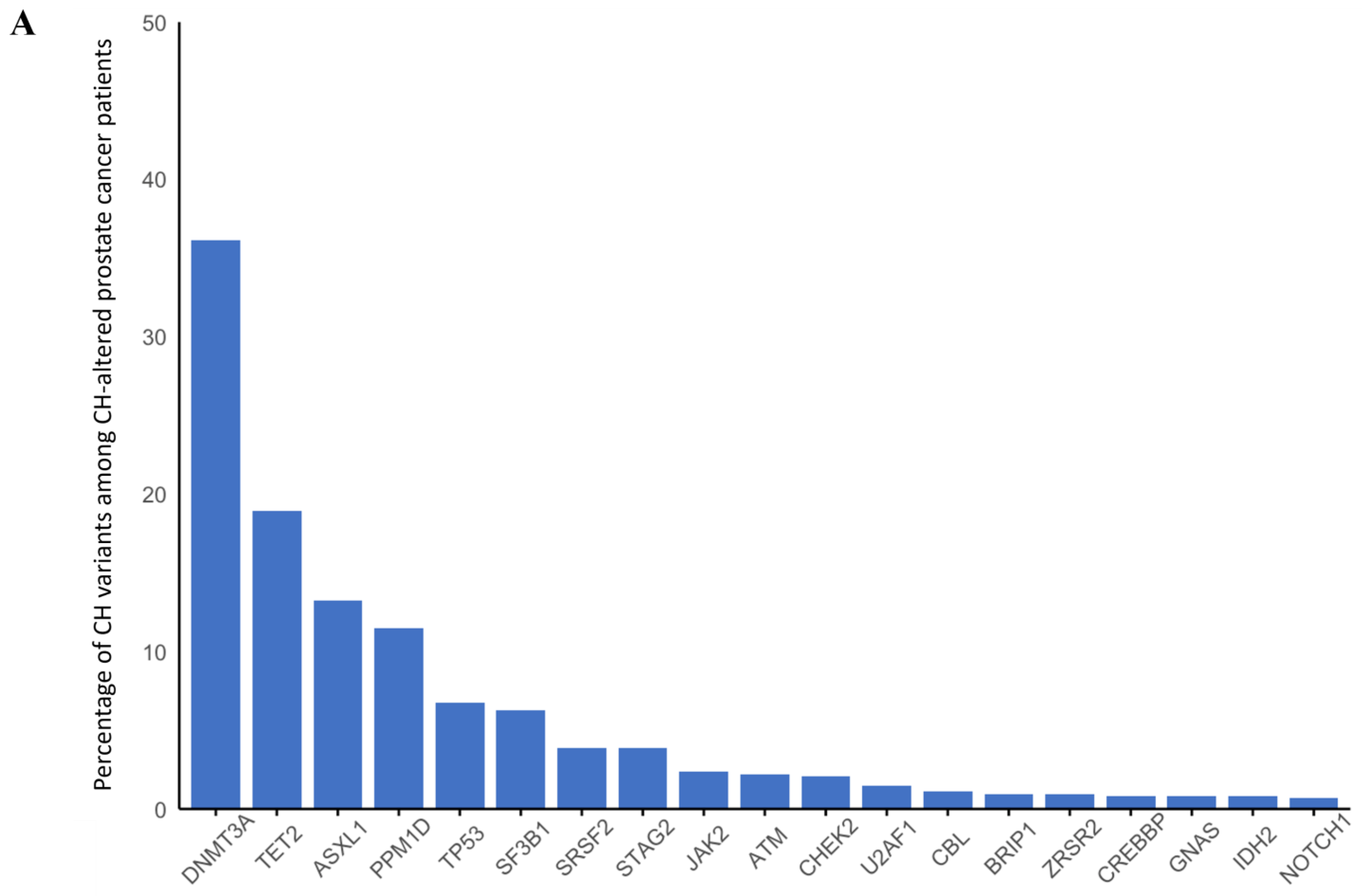


| Sporadic | gBRCA1 | gBRCA2 | gATM | gPALB2 | gCHEK2 | |
|---|---|---|---|---|---|---|
| N (%) | 7591 (94) | 116 (1) | 176 (2) | 69 (1) | 54 (1) | 80 (1) |
| Age at testing, mean | 60 | 48 | 56 | 59 | 58 | 63 |
| IQR | 50, 69 | 38, 59 | 45, 64 | 44, 69 | 50, 74 | 55, 70 |
| N (%) Female | 7511 (99) | 116 (100) | 164 (93) | 68 (99) | 54 (100) | 78 (98) |
| Race, N (%) | ||||||
| White | 3761 (50) | 46 (40) | 72 (41) | 35 (51) | 26 (48) | 45 (56) |
| Black | 725 (10) | 9 (8) | 19 (11) | 4 (6) | 3 (6) | 1 (1) |
| Asian | 232 (3) | 5 (4) | 7 (4) | 2 (3) | 3 (6) | 1 (1) |
| Other | 409 (5) | 8 (7) | 14 (8) | 1 (1) | 5 (9) | 5 (6) |
| Hispanic, N (%) | 489 (6) | 9 (8) | 17 (10) | 4 (6) | 6 (11) | 6 (8) |
| Stage | ||||||
| I | 127 (2) | 0 (0) | 2 (1) | 0 (0) | 1 (2) | 0 (0) |
| II | 309 (4) | 5 (4) | 9 (5) | 3 (4) | 6 (11) | 2 (3) |
| III | 333 (4) | 5 (4) | 9 (5) | 4 (6) | 0 (0) | 1 (1) |
| IV | 3973 (52) | 49 (42) | 87 (49) | 39 (57) | 30 (56) | 52 (65) |
| N (%) with CH | 899 (12) | 10 (9) | 23 (13) | 12 (17) | 12 (22) | 17 (21) |
| Sporadic | gBRCA1 | gBRCA2 | gATM | gPALB2 | gCHEK2 | |
|---|---|---|---|---|---|---|
| N (%) | 4689 (93) | 175 (4) | 102 (2) | 27 (1) | 12 (<1) | 13 (<1) |
| Age at testing, median | 65 | 56 | 64 | 63 | 70 | 58 |
| IQR | 56, 72 | 49, 64 | 57, 69 | 54, 71 | 64, 73 | 45, 70 |
| Race, N (%) | ||||||
| White | 2576 (55) | 93 (53) | 51 (50) | 14 (52) | 10 (83) | 7 (54) |
| Black | 260 (6) | 8 (5) | 5 (5) | 2 (7) | 0 (0) | 0 (0) |
| Asian | 124 (3) | 6 (3) | 5 (5) | 1 (4) | 0 (0) | 0 (0) |
| Other | 199 (4) | 10 (6) | 2 (2) | 1 (4) | 0 (0) | 0 (0) |
| Hispanic, N (%) | 258 (6) | 15 (9) | 2 (2) | 0 (0) | 0 (0) | 0 (0) |
| Stage | ||||||
| I | 87 (2) | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| II | 96 (2) | 2 (1) | 1 (1) | 0 (0) | 1 (8) | 0 (0) |
| III | 647 (14) | 14 (8) | 10 (10) | 1 (4) | 1 (8) | 2 (15) |
| IV | 855 (18) | 31 (18) | 16 (16) | 5 (19) | 2 (17) | 4 (31) |
| N (%) with CH | 715 (15) | 28 (16) | 19 (19) | 3 (11) | 2 (17) | 3 (23) |
| Sporadic | gBRCA1 | gBRCA2 | gATM | gPALB2 | gCHEK2 | |
|---|---|---|---|---|---|---|
| N (%) | 5023 (95) | 18 (<1) | 139 (3) | 49 (1) | 15 (<1) | 27 (1) |
| Age at testing, median | 66 | 65 | 63 | 66 | 69 | 70 |
| IQR | 61, 74 | 60, 78 | 60, 70 | 62, 74 | 63, 73 | 66, 79 |
| Race, N (%) | ||||||
| White | 2366 (47) | 9 (50) | 74 (53) | 24 (49) | 10 (67) | 16 (59) |
| Black | 555 (11) | 0 (0) | 16 (12) | 4 (8) | 0 (0) | 0 (0) |
| Asian | 89 (2) | 2 (11) | 0 (0) | 1 (2) | 0 (0) | 0 (0) |
| Other | 158 (3) | 0 (0) | 4 (3) | 1 (2) | 1 (7) | 0 (0) |
| Hispanic, N (%) | 239 (5) | 1 (6) | 10 (7) | 3 (6) | 0 (0) | 0 (0) |
| Stage | ||||||
| I | 8 (<1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| II | 64 (1) | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| III | 193 (4) | 1 (6) | 4 (3) | 1 (2) | 0 (0) | 1 (4) |
| IV | 2487 (50) | 4 (22) | 75 (54) | 24 (49) | 10 (67) | 21 (78) |
| N (%) with CH | 834 (17) | 6 (33) | 19 (14) | 9 (18) | 4 (27) | 5 (19) |
| Sporadic | gBRCA1 | gBRCA2 | gATM | gPALB2 | gCHEK2 | |
|---|---|---|---|---|---|---|
| N (%) | 6211 (96) | 27 (<1) | 108 (2) | 76 (1) | 25 (<1) | 27 (<1) |
| Age at testing, median | 67 | 62 | 65 | 65 | 64 | 68 |
| IQR | 60, 74 | 54, 71 | 58, 72 | 61, 72 | 55, 67 | 62, 74 |
| N (%) Female | 2898 (47) | 11 (41) | 48 (44) | 37 (49) | 14 (56) | 11 (41) |
| Race, N (%) | ||||||
| White | 3071 (49) | 10 (37) | 50 (46) | 30 (39) | 22 (88) | 19 (70) |
| Black | 393 (6) | 3 (11) | 4 (4) | 6 (8) | 0 (0) | 0 (0) |
| Asian | 132 (2) | 0 (0) | 6 (6) | 1 (1) | 0 (0) | 0 (0) |
| Other | 164 (3) | 2 (7) | 4 (4) | 4 (5) | 0 (0) | 0 (0) |
| Hispanic, N (%) | 243 (4) | 1 (4) | 8 (7) | 5 (7) | 0 (0) | 0 (0) |
| Stage | ||||||
| I | 203 (3) | 2 (7) | 6 (6) | 2 (3) | 0 (0) | 1 (4) |
| II | 416 (7) | 2 (7) | 5 (5) | 9 (12) | 1 (4) | 3 (11) |
| III | 321 (5) | 1 (4) | 3 (3) | 6 (8) | 2 (8) | 2 (7) |
| IV | 3372 (54) | 15 (56) | 73 (68) | 28 (37) | 15 (60) | 16 (59) |
| N (%) with CH | 889 (14) | 3 (11) | 13 (12) | 16 (21) | 3 (12) | 4 (15) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marshall, C.H.; Arafa, A.T.; Jaeger, E.; Fragkogianni, S.; Sonnenschein, A.; Mauer, E.; Gondek, L.P.; Chao, C.; Luo, J.; Antonarakis, E.S. Germline DNA Repair Gene Mutations and Clonal Hematopoiesis (CH) in 24,849 Patients with BRCA-Associated Cancers. Cancers 2025, 17, 1432. https://doi.org/10.3390/cancers17091432
Marshall CH, Arafa AT, Jaeger E, Fragkogianni S, Sonnenschein A, Mauer E, Gondek LP, Chao C, Luo J, Antonarakis ES. Germline DNA Repair Gene Mutations and Clonal Hematopoiesis (CH) in 24,849 Patients with BRCA-Associated Cancers. Cancers. 2025; 17(9):1432. https://doi.org/10.3390/cancers17091432
Chicago/Turabian StyleMarshall, Catherine H., Ali T. Arafa, Ellen Jaeger, Stamatina Fragkogianni, Anne Sonnenschein, Elizabeth Mauer, Lukasz P. Gondek, Calvin Chao, Jun Luo, and Emmanuel S. Antonarakis. 2025. "Germline DNA Repair Gene Mutations and Clonal Hematopoiesis (CH) in 24,849 Patients with BRCA-Associated Cancers" Cancers 17, no. 9: 1432. https://doi.org/10.3390/cancers17091432
APA StyleMarshall, C. H., Arafa, A. T., Jaeger, E., Fragkogianni, S., Sonnenschein, A., Mauer, E., Gondek, L. P., Chao, C., Luo, J., & Antonarakis, E. S. (2025). Germline DNA Repair Gene Mutations and Clonal Hematopoiesis (CH) in 24,849 Patients with BRCA-Associated Cancers. Cancers, 17(9), 1432. https://doi.org/10.3390/cancers17091432







