Dynamic Prediction of Rectal Cancer Relapse and Mortality Using a Landmarking-Based Machine Learning Model: A Multicenter Retrospective Study from the Italian Society of Surgical Oncology—Colorectal Cancer Network Collaborative Group
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
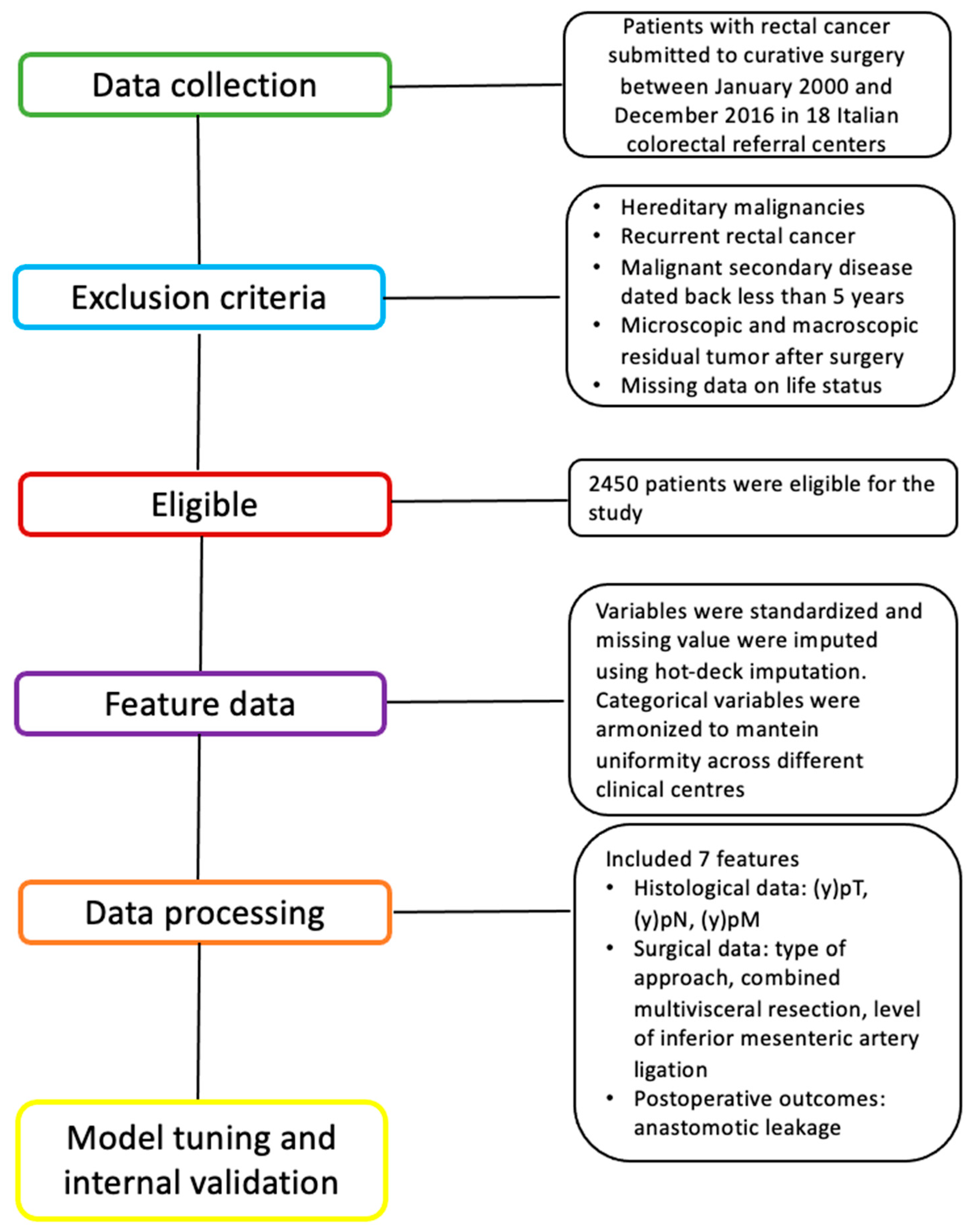
2.1. Patient Selection and Dataset
2.2. Statistical Analysis
- (1)
- Model A: the classical landmark analysis based on the cause-specific Cox model.
- (2)
- Model B: the proposed application of landmarking-based RFS competing risk algorithm.
2.2.1. Data Preparation and Preprocessing
2.2.2. Data Description
2.2.3. Landmarking Approach for Time-Varying Risk Estimation
2.2.4. Model A: Survival Analysis Using Landmark Cause-Specific Cox Models
2.2.5. Model B Machine Learning-Based Competing Risk Survival Analysis: Landmarking with Random Survival Forests
- ✓
- Input Layer—Clinical Data and Landmark Time Points: The model starts with structured clinical data, including baseline and time-varying covariates such as tumor staging, surgical approach, perioperative complications, and comorbidities. For each predefined landmark time point, a subset of patients still at risk is selected to form a “landmark dataset.”
- ✓
- Landmark Dataset Construction: Separate datasets are generated for each landmark time, capturing the patient profile up to that specific time. These are then stacked together into a “super dataset” while preserving the longitudinal structure of the data. Competing events—cancer-specific death, death from other causes, and relapse—are all considered.
- ✓
- RSF Model Training: A random survival forest model is trained using the combined landmark datasets. The RSF, an ensemble of decision trees, learns from the data without assuming proportional hazards or linear relationships. The model captures complex, non-linear associations and interactions among predictors. Before model fitting, hyperparameter tuning was performed to determine the optimal mtry (number of randomly drawn candidate variables) and node size tuning parameters for a random forest, using out-of-sample error as the evaluation metric. Hyperparameter tuning for RSF was conducted using a grid search strategy. The number of candidate variables considered at each split (mtry) and the minimum node size were tuned by evaluating combinations across predefined ranges: mtry (5 to 15) and node size (5 to 50). The number of trees was fixed at 100 to ensure computational consistency. Model performance for each parameter configuration was assessed using out-of-bag (OOB) error estimates, which serve as internal out-of-sample error measures in ensemble tree models. For each tree, approximately one-third of the samples not included in the bootstrap sample were used to compute the prediction error. The optimal parameter set was defined as the one that minimized the average out-of-bag (OOB) error, measured as the OOB concordance error, across all trees.Given the longitudinal nature of the landmarking approach, where the same patient may contribute data at multiple time points, cluster-based resampling was employed during bootstrapping. This ensured that repeated measures for a single patient were sampled as a unit, preserving intra-subject correlation and avoiding data leakage between training and validation sets.
- ✓
- Model Outputs—Cumulative Incidence Functions (CIFs): For each patient, the RSF produces predicted cumulative incidence functions for each competing event. These CIFs quantify the probability of experiencing relapse or death at future time points, conditional on survival until the landmark. The RSF framework handled competing risks directly by estimating CIF for each event type (cancer-specific death, other-cause death, and relapse), following the methodology proposed by Ishwaran et al. [22]. This non-parametric ensemble method does not rely on Fine–Gray regression but computes CIFs by aggregating over multiple decision trees. Right-censoring was addressed using inverse probability of censoring weighting (IPCW) during tree construction, ensuring unbiased estimation of survival probabilities. Ties in survival times were resolved using standard RSF splitting rules based on ranked survival times.
2.2.6. Validation and Model Performance Assessment
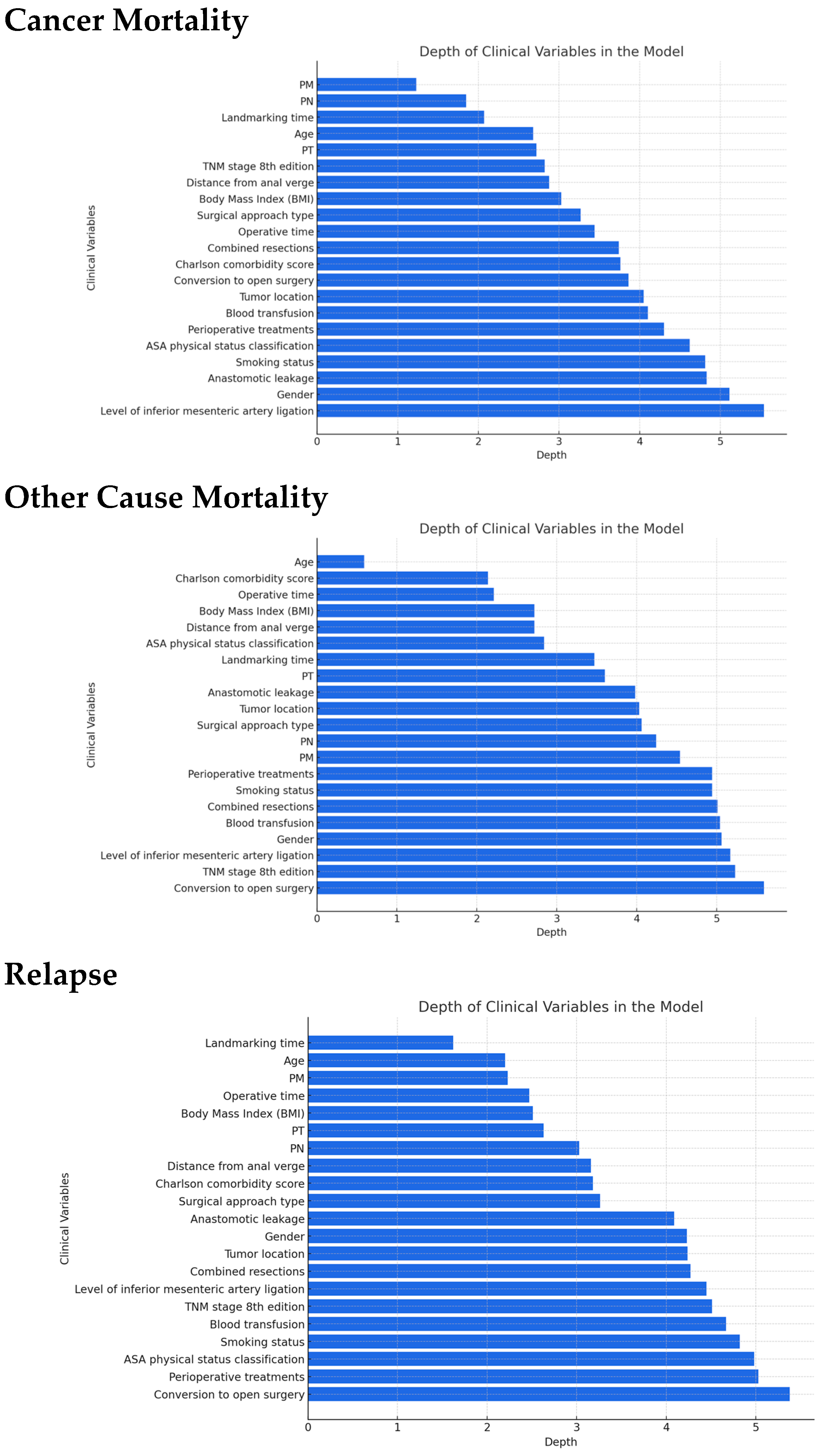
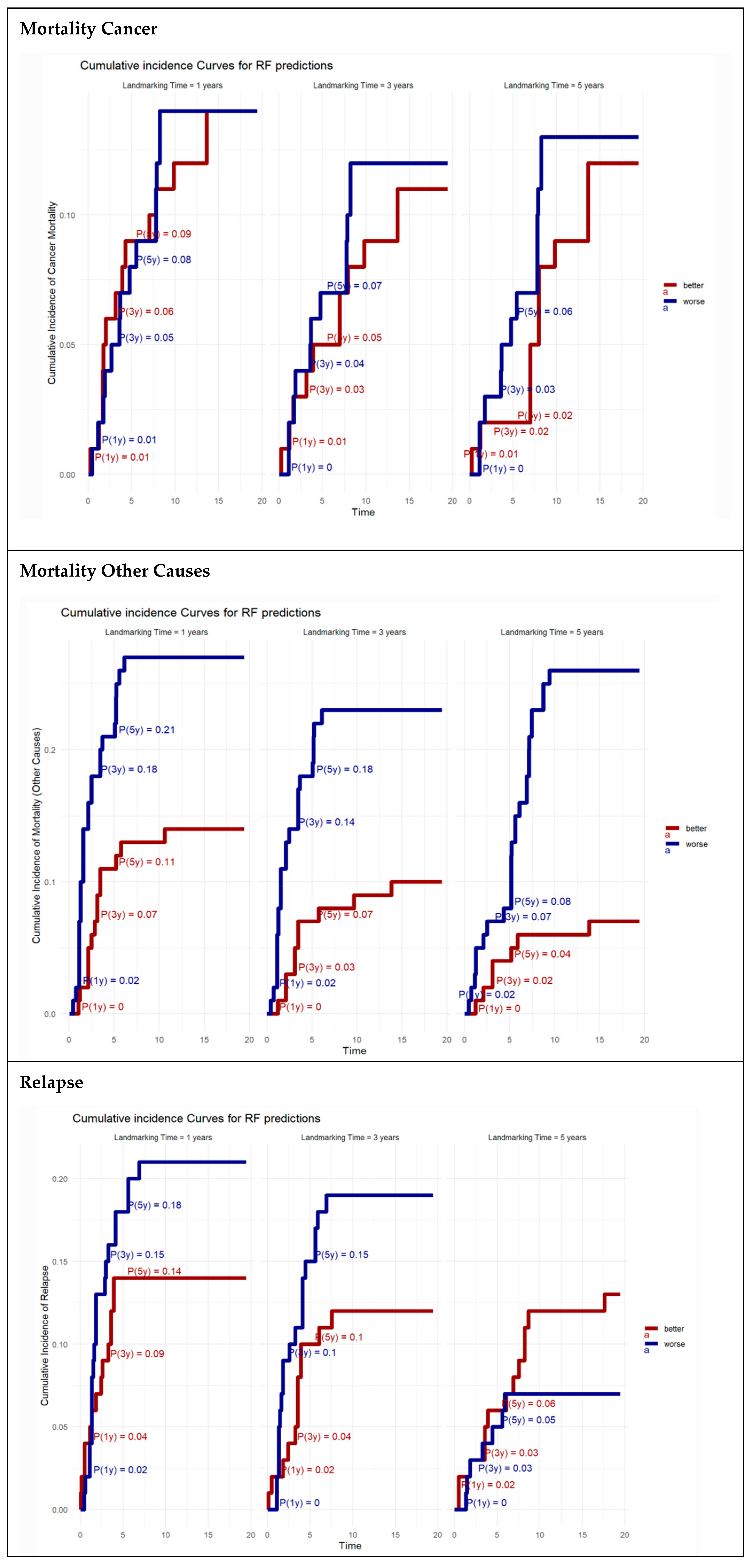
2.2.7. Patient Profiling
3. Results
3.1. Descriptive Statistics
3.2. Model A
3.3. Model B
3.4. Patient Profiling
3.5. Models Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RC | rectal cancer |
| PME | partial mesorectal excision |
| TME | total mesorectal excision |
| CRT | chemoradio therapy |
| CT | chemotherapy |
| RCT | randomized controlled trial |
| CS | conditional survival |
| AI | artificial intelligence |
| ML | machine learning |
| BMI | body mass index |
| MVR | multivisceral resection |
| CRC | colorectal cancer |
| HR | hazard ratio |
| RF | random forest |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- I Numeri del Cancro in Italia 2024 P As S I Progressi nelle Aziende Sanitarie per la Salute in Italia. Available online: https://www.cro.sanita.fvg.it/it/news/2024/numeri-cancro-2024.html (accessed on 25 March 2025).
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up ☆. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Yu, Y.; Wang, Z.; Hong, L.; Shao, L.; Wu, J. Developing Individualized Follow-Up Strategies Based on High-Risk Recurrence Factors and Dynamic Risk Assessment for Locally Advanced Rectal Cancer. Cancer Med. 2024, 13, e70323. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.; Zwaal, C.; Asmis, T.; Cho, C.; Galica, J.; Ginty, A.; Govindarajan, A. An Evidence-Based Guideline for Surveillance of Patients after Curative Treatment for Colon and Rectal Cancer. Curr. Oncol. 2022, 29, 724–740. [Google Scholar] [CrossRef]
- Adam, M.; Chang, G.J.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.A.; Deming, D.; Garrido-Laguna, I.; Grem, J.L.; Buffett Cancer Center Carla Harmath, P.; Randolph Hecht, J.; et al. NCCN Guidelines Version 1.2025 Rectal Cancer Continue NCCN Guidelines Panel Disclosures. 2025. Available online: https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf (accessed on 25 March 2025).
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rödel, C.; Cervantes, A.; Arnold, D. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv22–iv40. [Google Scholar] [CrossRef]
- Hashiguchi, Y.; Muro, K.; Saito, Y.; Ito, Y.; Ajioka, Y.; Hamaguchi, T.; Hasegawa, K.; Hotta, K.; Ishida, H.; Ishiguro, M.; et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int. J. Clin. Oncol. 2020, 25, 1–42. [Google Scholar] [CrossRef]
- Lauretta, A.; Montori, G.; Guerrini, G.P. Surveillance strategies following curative resection and non-operative approach of rectal cancer: How and how long? Review of current recommendations. World J. Gastrointest. Surg. 2023, 15, 177–192. [Google Scholar] [CrossRef]
- Silberfein, E.J.; Kattepogu, K.M.; Hu, C.Y.; Skibber, J.M.; Rodriguez-Bigas, M.A.; Feig, B.; Das, P.; Krishnan, S.; Crane, C.; Kopetz, S.; et al. Long-term survival and recurrence outcomes following surgery for distal rectal cancer. Ann. Surg. Oncol. 2010, 17, 2863–2869. [Google Scholar] [CrossRef]
- Fokas, E.; Fietkau, R.; Hartmann, A.; Hohenberger, W.; Grützmann, R.; Ghadimi, M.; Liersch, T.; Ströbel, P.; Grabenbauer, G.G.; Graeven, U.; et al. Neoadjuvant rectal score as individual-level surrogate for disease-free survival in rectal cancer in the CAO/ ARO/AIO-04 randomized phase III trial. Ann. Oncol. 2018, 29, 1521–1527. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, X.; Liu, Z.; Lu, X.; Huang, Y.; Chi, P. Individualized conditional survival nomograms for patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy and radical surgery. Eur. J. Surg. Oncol. 2021, 47, 3175–3181. [Google Scholar] [CrossRef]
- Karagkounis, G.; Liska, D.; Kalady, M.F. Conditional Probability of Survival after Neoadjuvant Chemoradiation and Proctectomy for Rectal Cancer: What Matters and When. In Diseases of the Colon and Rectum; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2019; Volume 62, pp. 33–39. [Google Scholar]
- Van Houwelingen, H.C. Dynamic prediction by landmarking in event history analysis. Scand. J. Stat. 2007, 34, 70–85. [Google Scholar] [CrossRef]
- Fontein, D.B.Y.; Klinten Grand, M.; Nortier, J.W.R.; Seynaeve, C.; Meershoek-Klein Kranenbarg, E.; Dirix, L.Y.; van de Velde, C.J.H.; Putter, H. Dynamic prediction in breast cancer: Proving feasibility in clinical practice using the TEAM trial. Ann. Oncol. 2015, 26, 1254–1262. [Google Scholar] [CrossRef]
- Spolverato, G.; Azzolina, D.; Paro, A.; Lorenzoni, G.; Gregori, D.; Poultsides, G.; Fields, R.C.; Weber, S.M.; Votanopoulos, K.; Maithel, S.K.; et al. Dynamic Prediction of Survival after Curative Resection of Gastric Adenocarcinoma: A landmarking-based analysis. Eur. J. Surg. Oncol. 2022, 48, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- Jeon, Y.; Kim, Y.J.; Jeon, J.; Nam, K.H.; Hwang, T.S.; Kim, K.G.; Baek, J.H. Machine learning based prediction of recurrence after curative resection for rectal cancer. PLoS ONE 2023, 18, e0290141. [Google Scholar] [CrossRef]
- Lou, S.J.; Hou, M.F.; Chang, H.T.; Chiu, C.C.; Lee, H.H.; Yeh, S.C.J.; Shi, H.Y. Machine learning algorithms to predict recurrence within 10 years after breast cancer surgery: A prospective cohort study. Cancers 2020, 12, 3817. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, J.; Wang, Y.; Ji, M.H.; Tong, J.; Yang, J.J.; Xia, H. A machine learning-based predictor for the identification of the recurrence of patients with gastric cancer after operation. Sci. Rep. 2021, 11, 1571. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.D.; Ngo, L.; Samelson, E.J.; Kiel, D.P. Competing Risk of Death: An Important Consideration in Studies of Older Adults. J. Am. Geriatr. Soc. 2010, 58, 783–787. [Google Scholar] [CrossRef]
- Ishwaran, H.; Gerds, T.A.; Kogalur, U.B.; Moore, R.D.; Gange, S.J.; Lau, B.M. Random survival forests for competing risks. Biostatistics 2014, 15, 757–773. [Google Scholar] [CrossRef]
- Degiuli, M.; Elmore, U.; De Luca, R.; De Nardi, P.; Tomatis, M.; Biondi, A.; Persiani, R.; Solaini, L.; Rizzo, G.; Soriero, D.; et al. Risk factors for anastomotic leakage after anterior resection for rectal cancer (RALAR study): A nationwide retrospective study of the Italian Society of Surgical Oncology Colorectal Cancer Network Collaborative Group. Color. Dis. 2022, 24, 264–276. [Google Scholar] [CrossRef]
- White, H. A Heteroskedasticity-Consistent Covariance Matrix Estimator and a Direct Test for Heteroskedasticity. Econom. J. Econom. Soc. 1980, 48, 817. [Google Scholar] [CrossRef]
- Brouwer, N.P.M.; Bos, A.C.R.K.; Lemmens, V.E.P.P.; Tanis, P.J.; Hugen, N.; Nagtegaal, I.D.; de Wilt, J.H.W.; Verhoeven, R.H.A. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int. J. Cancer 2018, 143, 2758–2766. [Google Scholar] [CrossRef]
- Jiang, W.; Xu, J.; Cui, M.; Qiu, H.; Wang, Z.; Kang, L.; Deng, H.; Chen, W.; Zhang, Q.; Du, X.; et al. Laparoscopy-assisted versus open surgery for low rectal cancer (LASRE): 3-year survival outcomes of a multicentre, randomised, controlled, non-inferiority trial. Lancet Gastroenterol. Hepatol. 2024, 10, 34–43. [Google Scholar] [CrossRef]
- Waldenstedt, S.; Bock, D.; Haglind, E.; Sjöberg, B.; Angenete, E. Intraoperative adverse events as a risk factor for local recurrence of rectal cancer after resection surgery. Color. Dis. 2022, 24, 449–460. [Google Scholar] [CrossRef]
- Ozaki, K.; Kawai, K.; Nozawa, H.; Sasaki, K.; Murono, K.; Emoto, S.; Iida, Y.; Ishii, H.; Yokoyama, Y.; Anzai, H.; et al. Therapeutic effects and limitations of chemoradiotherapy in advanced lower rectal cancer focusing on T4b. Int. J. Color. Dis. 2021, 36, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Fretwell, V.L.; Ang, C.W.; Tweedle, E.M.; Rooney, P.S. The impact of lymph node yield on Duke’s B and C colorectal cancer survival. Color. Dis. 2010, 12, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Sofia, S.; Degiuli, M.; Anania, G.; Baiocchi, G.L.; Baldari, L.; Baldazzi, G.; Bianco, F.; Borghi, F.; Cavaliere, D.; Coco, C.; et al. Textbook Outcome in Colorectal Surgery for Cancer: An Italian Version. J. Clin. Med. 2024, 13, 4687. [Google Scholar] [CrossRef]
- Degiuli, M.; Arolfo, S.; Evangelista, A.; Lorenzon, L.; Reddavid, R.; Staudacher, C.; De Nardi, P.; Rosati, R.; Elmore, U.; Coco, C.; et al. Number of lymph nodes assessed has no prognostic impact in node-negative rectal cancers after neoadjuvant therapy. Results of the “Italian Society of Surgical Oncology (S.I.C.O.) Colorectal Cancer Network” (SICO-CCN) multicentre collaborative study. Eur. J. Surg. Oncol. 2018, 44, 1233–1240. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamaura, T.; Kinjo, Y.; Harada, K.; Kawase, M.; Kawabata, Y.; Kanto, S.; Ogo, Y.; Kuroda, N. Level of Inferior Mesenteric Artery Ligation in Sigmoid Colon and Rectal Cancer Surgery: Analysis of Apical Lymph Node Metastasis and Recurrence. Dig. Surg. 2023, 40, 167–177. [Google Scholar] [CrossRef]
- Yoshida, D.; Sugiyama, M.; Nakazono, K.; Oyama, T.; Hasegawa, T.; Kai, S.; Yamamoto, M.; Matsumoto, T.; Kawanaka, H.; Morita, M.; et al. Oncological Impact of the Level of Inferior Mesenteric Artery Ligation in Low Rectal Cancer Surgery. Anticancer Res. 2023, 47, 3225–3233. [Google Scholar] [CrossRef]
- Baik, H.J. To go high, or to go low: The never-ending debate of inferior mesenteric artery ligation. Ann. Coloproctol. 2024, 40, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Van Der Stok, E.P.; Spaander, M.C.W.; Grünhagen, D.J.; Verhoef, C.; Kuipers, E.J. Surveillance after curative treatment for colorectal cancer. Nat. Rev. Clin. Oncol. 2017, 14, 297–315. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Pang, X.; Ji, G.; Sun, H.; Fan, Q.; Ma, C.; Koniaris, L.G. The impact of anastomotic leakage on oncology after curative anterior resection for rectal cancer: A systematic review and meta-analysis. Medicine 2020, 99, E22139. [Google Scholar] [CrossRef]
- Zhu, S.; Zheng, Z.; Hu, W.; Lei, C. Conditional Cancer-Specific Survival for Inflammatory Breast Cancer: Analysis of SEER, 2010 to 2016. Clin. Breast Cancer 2023, 23, 628–639.e2. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, N.H.; Smith, A.N.; Geyer, S.; Binder, M.; Greipp, P.T.; Kapoor, P.; Dispenzieri, A.; Gertz, M.A.; Baughn, L.B.; Lacy, M.Q.; et al. Conditional survival in multiple myeloma and impact of prognostic factors over time. Blood Cancer J. 2023, 13, 78. [Google Scholar] [CrossRef]
- Yang, Y.P.; Guo, C.J.; Gu, Z.X.; Hua, J.J.; Zhang, J.X.; Shi, J. Conditional survival probability of distant-metastatic hepatocellular carcinoma: A population-based study. World J. Gastrointest. Oncol. 2023, 15, 1874–1890. [Google Scholar] [CrossRef]
- Xie, S.H.; Santoni, G.; Bottai, M.; Gottlieb-Vedi, E.; Lagergren, P.; Lagergren, J. Prediction of conditional survival in esophageal cancer in a population-based cohort study. Int. J. Surg. 2023, 109, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Daly, C.; Urbach, D.R.; Stukel, T.A.; Nathan, P.C.; Deitel, W.; Paszat, L.F.; Wilton, A.S.; Baxter, N.N. Patterns of diagnostic imaging and associated radiation exposure among long-term survivors of young adult cancer: A population-based cohort study. BMC Cancer 2015, 15, 1–10. [Google Scholar] [CrossRef]
- Khatri, R.; Quinn, P.L.; Wells-Di Gregorio, S.; Pawlik, T.M.; Cloyd, J.M. Surveillance-Associated Anxiety After Curative-Intent Cancer Surgery: A Systematic Review. Ann. Surg. Oncol. 2024, 32, 47–62. [Google Scholar] [CrossRef]
- Waqas, M.; Humphries, U.W. A critical review of RNN and LSTM variants in hydrological time series predictions. MethodsX 2024, 13, 102946. [Google Scholar] [CrossRef]
- Pedarzani, E.; Fogangolo, A.; Baldi, I.; Berchialla, P.; Panzini, I.; Khan, M.R.; Valpiani, G.; Spadaro, S.; Gregori, D.; Azzolina, D. Prioritizing Patient Selection in Clinical Trials: A Machine Learning Algorithm for Dynamic Prediction of In-Hospital Mortality for ICU Admitted Patients Using Repeated Measurement Data. J. Clin. Med. 2025, 14, 612. [Google Scholar] [CrossRef] [PubMed]
| Death for Cancer | Death for Other Causes | Relapse | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Overall | No Events | Event | HR | P | No Events | Event | HR | P | No Events | Event | HR | P |
| N = 2405 | N = 2218 | N = 187 | N = 1931 | N = 474 | N = 2203 | N = 202 | |||||||
| Age (years) | 66.6 [58.2;74.0] | 66.4 [58.0;73.9] | 69.3 [61.3;75.8] | 1.03 [1.01;1.04] | <0.001 | 64.4 [56.3;70.8] | 74.6 [68.5;79.6] | 1.09 [1.08;1.10] | <0.001 | 66.7 [58.2;74.1] | 66.4 [56.4;73.9] | 1.00 [0.99;1.01] | 0.737 |
| Gender: | 0.171 | 0.001 | 0.096 | ||||||||||
| Female | 921 (38.3%) | 856 (38.6%) | 65 (34.8%) | Ref. | 768 (39.8%) | 153 (32.3%) | Ref. | 852 (38.7%) | 69 (34.2%) | Ref. | |||
| Male | 1484 (61.7%) | 1362 (61.4%) | 122 (65.2%) | 1.23 [0.91;1.67] | 1163 (60.2%) | 321 (67.7%) | 1.40 [1.16;1.70] | 1351 (61.3%) | 133 (65.8%) | 1.28 [0.96;1.71] | |||
| BMI | 25.3 [22.7;27.6] | 25.4 [22.8;27.7] | 24.9 [22.0;27.2] | 0.96 [0.92;1.00] | 0.081 | 25.3 [22.7;27.6] | 25.4 [22.9;27.5] | 1.01 [0.97;1.04] | 0.726 | 25.2 [22.6;27.5] | 26.1 [24.0;29.0] | 1.06 [1.01;1.11] | 0.012 |
| Charlson Comorbidity Score | 2.00 [2.00;3.00] | 2.00 [2.00;3.00] | 2.00 [2.00;3.00] | 1.20 [1.05;1.37] | 0.008 | 2.00 [2.00;3.00] | 3.00 [2.00;3.00] | 1.42 [1.32;1.54] | <0.001 | 2.00 [2.00;3.00] | 2.00 [2.00;3.00] | 1.16 [0.99;1.35] | 0.058 |
| ASA: | 0.011 | <0.001 | 0.42 | ||||||||||
| I–II | 1515 (74.4%) | 1410 (75.0%) | 105 (67.3%) | Ref. | 1283 (78.1%) | 232 (58.9%) | Ref. | 1383 (74.5%) | 132 (73.7%) | Ref. | |||
| III–IV | 521 (25.6%) | 470 (25.0%) | 51 (32.7%) | 1.54 [1.10;2.15] | 359 (21.9%) | 162 (41.1%) | 2.19 [1.79;2.68] | 474 (25.5%) | 47 (26.3%) | 1.15 [0.82;1.60] | |||
| Perioperative treatments: | 0.158 | 0.001 | 0.01 | ||||||||||
| No | 287 (19.7%) | 258 (19.4%) | 29 (22.0%) | Ref. | 227 (18.9%) | 60 (23.3%) | Ref. | 276 (20.8%) | 11 (8.33%) | Ref. | |||
| Yes | 1172 (80.3%) | 1069 (80.6%) | 103 (78.0%) | 0.74 [0.49;1.12] | 974 (81.1%) | 198 (76.7%) | 0.61 [0.46;0.82] | 1051 (79.2%) | 121 (91.7%) | 2.20 [1.19;4.09] | |||
| Smoke: | 0.886 | 0.425 | 0.903 | ||||||||||
| No | 1156 (75.8%) | 1053 (75.9%) | 103 (74.6%) | Ref. | 975 (76.1%) | 181 (74.2%) | Ref. | 1060 (75.7%) | 96 (76.2%) | Ref. | |||
| Yes | 370 (24.2%) | 335 (24.1%) | 35 (25.4%) | 1.03 [0.70;1.51] | 307 (23.9%) | 63 (25.8%) | 1.12 [0.84;1.50] | 340 (24.3%) | 30 (23.8%) | 0.97 [0.65;1.47] | |||
| Surgical approach: | <0.001 | 0.026 | 0.682 | ||||||||||
| Laparoscopic | 1312 (56.5%) | 1242 (58.1%) | 70 (38.5%) | Ref. | 1078 (57.5%) | 234 (52.6%) | Ref. | 1208 (56.7%) | 104 (54.7%) | Ref. | |||
| Open | 828 (35.7%) | 734 (34.3%) | 94 (51.6%) | 1.93 [1.41;2.63] | 633 (33.7%) | 195 (43.8%) | 1.02 [0.84;1.24] | 754 (35.4%) | 74 (38.9%) | 0.95 [0.70;1.29] | |||
| Robotic | 181 (7.80%) | 163 (7.62%) | 18 (9.89%) | 1.65 [0.98;2.77] | 165 (8.80%) | 16 (3.60%) | 0.51 [0.31;0.85] | 169 (7.93%) | 12 (6.32%) | 0.77 [0.42;1.40] | |||
| Inferior mesenteric artery ligation level: | 0.085 | 0.647 | 0.373 | ||||||||||
| High tie | 1752 (85.3%) | 1583 (84.8%) | 169 (90.4%) | Ref. | 1450 (85.2%) | 302 (85.8%) | Ref. | 1617 (85.1%) | 135 (88.2%) | Ref. | |||
| Low tie | 301 (14.7%) | 283 (15.2%) | 18 (9.63%) | 0.65 [0.40;1.06] | 251 (14.8%) | 50 (14.2%) | 0.93 [0.69;1.26] | 283 (14.9%) | 18 (11.8%) | 0.80 [0.49;1.31] | |||
| Anastomotic dehiscence: | 0.906 | <0.001 | 0.001 | ||||||||||
| No | 2121 (88.2%) | 1954 (88.1%) | 167 (89.3%) | Ref. | 1723 (89.2%) | 398 (84.0%) | Ref. | 1955 (88.7%) | 166 (82.2%) | Ref. | |||
| Yes | 284 (11.8%) | 264 (11.9%) | 20 (10.7%) | 0.97 [0.61;1.55] | 208 (10.8%) | 76 (16.0%) | 1.67 [1.31;2.13] | 248 (11.3%) | 36 (17.8%) | 1.79 [1.25;2.57] | |||
| Combined multivisceral resections: | <0.001 | 0.037 | 0.006 | ||||||||||
| No | 1676 (80.3%) | 1555 (81.7%) | 121 (65.4%) | Ref. | 1390 (80.8%) | 286 (77.9%) | Ref. | 1559 (80.8%) | 117 (73.6%) | Ref. | |||
| Yes | 412 (19.7%) | 348 (18.3%) | 64 (34.6%) | 2.40 [1.77;3.25] | 331 (19.2%) | 81 (22.1%) | 1.30 [1.02;1.67] | 370 (19.2%) | 42 (26.4%) | 1.64 [1.15;2.34] | |||
| Operative time (min): | 240 [180;300] | 240 [180;300] | 240 [195;300] | 1.00 [1.00;1.00] | 0.809 | 240 [180;300] | 240 [180;300] | 1.00 [1.00;1.00] | 0.033 | 240 [180;300] | 250 [192;312] | 1.00 [1.00;1.00] | 0.029 |
| Trasfusion: | 0.066 | <0.001 | 0.009 | ||||||||||
| No | 1949 (91.6%) | 1805 (91.8%) | 144 (89.4%) | Ref. | 1575 (92.5%) | 374 (88.0%) | Ref. | 1782 (91.9%) | 167 (88.8%) | Ref. | |||
| Yes | 179 (8.41%) | 162 (8.24%) | 17 (10.6%) | 1.60 [0.97;2.64] | 128 (7.52%) | 51 (12.0%) | 2.24 [1.67;3.01] | 158 (8.14%) | 21 (11.2%) | 1.81 [1.15;2.86] | |||
| Conversion: | 0.862 | 0.167 | 0.617 | ||||||||||
| No | 1835 (94.2%) | 1707 (94.3%) | 128 (93.4%) | Ref. | 1452 (94.0%) | 383 (95.0%) | Ref. | 1684 (94.3%) | 151 (92.6%) | Ref. | |||
| Yes | 113 (5.80%) | 104 (5.74%) | 9 (6.57%) | 1.06 [0.54;2.09] | 93 (6.02%) | 20 (4.96%) | 0.73 [0.47;1.14] | 101 (5.66%) | 12 (7.36%) | 1.16 [0.64;2.09] | |||
| AV distance: | 8.00 [6.00;11.0] | 8.80 [6.00;11.0] | 8.00 [6.00;11.0] | 1.00 [0.96;1.04] | 0.987 | 8.00 [6.00;11.0] | 10.0 [6.00;12.0] | 1.05 [1.03;1.08] | <0.001 | 8.00 [6.00;11.0] | 9.00 [6.00;11.0] | 1.03 [0.99;1.07] | 0.161 |
| Localization: | 0.558 | 0.027 | 0.883 | ||||||||||
| High | 682 (29.1%) | 632 (29.2%) | 50 (27.9%) | Ref. | 532 (28.0%) | 150 (33.6%) | Ref. | 627 (29.2%) | 55 (27.8%) | Ref. | |||
| Middle | 1100 (46.9%) | 1011 (46.7%) | 89 (49.7%) | 1.09 [0.77;1.55] | 903 (47.5%) | 197 (44.2%) | 0.80 [0.65;0.99] | 1005 (46.8%) | 95 (48.0%) | 1.05 [0.76;1.47] | |||
| Low | 564 (24.0%) | 524 (24.2%) | 40 (22.3%) | 0.89 [0.59;1.35] | 465 (24.5%) | 99 (22.2%) | 0.72 [0.56;0.93] | 516 (24.0%) | 48 (24.2%) | 0.97 [0.66;1.43] | Low | ||
| (y)pT: | <0.001 | <0.001 | <0.001 | ||||||||||
| 0 | 223 (9.55%) | 215 (10.0%) | 8 (4.28%) | Ref. | 187 (9.95%) | 36 (7.89%) | Ref. | 214 (10.0%) | 9 (4.48%) | Ref. | |||
| 1 | 316 (13.5%) | 307 (14.3%) | 9 (4.81%) | 0.74 [0.29;1.93] | 263 (14.0%) | 53 (11.6%) | 0.88 [0.57;1.34] | 304 (14.2%) | 12 (5.97%) | 0.84 [0.36;2.01] | |||
| 2 | 589 (25.2%) | 565 (26.3%) | 24 (12.8%) | 1.14 [0.51;2.54] | 481 (25.6%) | 108 (23.7%) | 1.06 [0.73;1.55] | 553 (25.9%) | 36 (17.9%) | 1.48 [0.71;3.07] | |||
| 3 | 1079 (46.2%) | 957 (44.6%) | 122 (65.2%) | 3.73 [1.82;7.63] | 850 (45.2%) | 229 (50.2%) | 1.62 [1.14;2.31] | 960 (45.0%) | 119 (59.2%) | 3.27 [1.66;6.44] | |||
| 4 | 128 (5.48%) | 104 (4.84%) | 24 (12.8%) | 7.19 [3.23;16.0] | 98 (5.22%) | 30 (6.58%) | 2.05 [1.26;3.33] | 103 (4.83%) | 25 (12.4%) | 6.77 [3.16;14.5] | |||
| (y)pM: | <0.001 | 0.001 | <0.001 | ||||||||||
| 0 | 2188 (92.2%) | 2060 (94.3%) | 128 (68.4%) | Ref. | 1755 (92.0%) | 433 (93.3%) | Ref. | 2030 (93.5%) | 158 (78.6%) | Ref. | |||
| 1 | 184 (7.76%) | 125 (5.72%) | 59 (31.6%) | 10.6 [7.77;14.5] | 153 (8.02%) | 31 (6.68%) | 1.86 [1.29;2.69] | 141 (6.49%) | 43 (21.4%) | 6.86 [4.87;9.65] | |||
| (y)pN: | <0.001 | 0.001 | <0.001 | ||||||||||
| 0 | 1551 (65.4%) | 1489 (68.1%) | 62 (33.2%) | Ref. | 1243 (65.2%) | 308 (66.0%) | Ref. | 1452 (66.8%) | 99 (49.5%) | Ref. | |||
| 1 | 545 (23.0%) | 473 (21.6%) | 72 (38.5%) | 3.81 [2.72;5.36] | 439 (23.0%) | 106 (22.7%) | 1.20 [0.96;1.49] | 489 (22.5%) | 56 (28.0%) | 1.92 [1.38;2.67] | |||
| 2 | 277 (11.7%) | 224 (10.2%) | 53 (28.3%) | 7.19 [4.97;10.4] | 224 (11.8%) | 53 (11.3%) | 1.72 [1.28;2.31] | 232 (10.7%) | 45 (22.5%) | 4.16 [2.91;5.94] | |||
| Stage (y)pTNM 8th: | <0.001 | 0.013 | <0.001 | ||||||||||
| <II | 1541 (64.1%) | 1485 (67.0%) | 56 (29.9%) | Ref. | 1233 (63.9%) | 308 (65.0%) | Ref. | 1447 (65.7%) | 94 (46.5%) | Ref. | |||
| >III | 864 (35.9%) | 733 (33.0%) | 131 (70.1%) | 5.16 [3.77;7.06] | 698 (36.1%) | 166 (35.0%) | 1.27 [1.05;1.54] | 756 (34.3%) | 108 (53.5%) | 2.64 [2.00;3.49] | |||
| Variables | 1 Year HR (95% CI) | 3 Years HR (95% CI) | 5 Years HR (95% CI) | Global Effect | Interaction with Time |
|---|---|---|---|---|---|
| Age | 1.12 (0.89–1.41) | 0.96 (0.76–1.22) | 0.83 (0.64–1.09) | 0.08 | <0.001 |
| BMI | 1.15 (0.96–1.37) | 1.15 (0.96–1.38) | 1.15 (0.94–1.40) | 0.06 | 0.99 |
| Charlson Comorbidity Score | 1.05 (0.91–1.23) | 0.99 (0.83–1.17) | 0.92 (0.74–1.14) | 0.28 | 0.06 |
| Operative Time | 1.10 (0.88–1.37) | 1.05 (0.81–1.35) | 1.00 (0.74–1.35) | 0.74 | 0.31 |
| Transfusion | 1.60 (0.97–2.64) | 1.33 (0.73–2.41) | 1.11 (0.51–2.43) | 0.06 | 0.2 |
| Conversion | 0.78 (0.42–1.42) | 0.92 (0.50–1.71) | 1.09 (0.53–2.23) | 0.42 | 0.16 |
| AV Distance | 1.68 (0.90–3.14) | 1.33 (0.68–2.58) | 1.05 (0.49–2.26) | 0.07 | 0.04 |
| Landmarking Time | 0.40 (0.25–0.63) | 0.02 | |||
| Gender—F:M | 0.84 (0.60–1.18) | 0.85 (0.60–1.22) | 0.86 (0.57–1.31) | 0.32 | 0.85 |
| ASA—III-IV:I-II | 0.97 (0.66–1.43) | 0.91 (0.59–1.40) | 0.85 (0.50–1.43) | 0.93 | 0.42 |
| Perioperative Treatments—No:Yes | 0.77 (0.49–1.20) | 0.78 (0.47–1.31) | 0.80 (0.42–1.53) | 0.47 | 0.84 |
| Smoking—Yes:No | 1.07 (0.74–1.55) | 0.99 (0.66–1.49) | 0.91 (0.56–1.49) | 0.81 | 0.28 |
| Surgical Approach—Open: Laparoscopic | 0.59 (0.41–0.85) | 0.58 (0.40–0.85) | 0.57 (0.37–0.89) | 0.03 | 0.48 |
| Surgical Approach—Robotic: Laparoscopic | 0.91 (0.48–1.75) | 1.07 (0.52–2.19) | 1.25 (0.52–3.00) | 0.91 | 0.4 |
| Lower Mesenteric Artery Ligation—Low:High | 0.85 (0.50–1.47) | 0.47 (0.24–0.91) | 0.26 (0.11–0.61) | <0.001 | <0.001 |
| Anastomotic Dehiscence—Yes:No | 1.53 (1.01–2.31) | 1.56 (0.99–2.48) | 1.60 (0.91–2.81) | 0.02 | 0.13 |
| Combined Multivisceral Resections—Yes:No | 1.22 (0.80–1.86) | 1.53 (1.00–2.35) | 1.93 (1.20–3.09) | 0.01 | <0.001 |
| Localization—High:Middle | 0.59 (0.33–1.04) | 0.68 (0.37–1.25) | 0.78 (0.39–1.60) | 0.07 | 0.34 |
| Localization—Low:Middle | 1.33 (0.70–2.52) | 1.15 (0.58–2.27) | 0.99 (0.44–2.20) | 0.1 | 0.26 |
| (y)pT—0:3 | 0.35 (0.16–0.77) | 0.50 (0.23–1.08) | 0.71 (0.31–1.66) | <0.001 | 0.002 |
| (y)pT—1:3 | 0.41 (0.22–0.77) | 0.54 (0.28–1.02) | 0.71 (0.35–1.41) | 0.001 | 0.02 |
| (y)pT—2:3 | 0.55 (0.35–0.86) | 0.58 (0.37–0.93) | 0.62 (0.37–1.06) | 0.01 | 0.28 |
| (y)pT—4:3 | 1.41 (0.82–2.42) | 1.24 (0.67–2.29) | 1.09 (0.51–2.36) | 0.12 | 0.37 |
| (y)pM—1:0 | 4.42 (2.60–7.53) | 3.00 (1.52–5.92) | 2.03 (0.81–5.10) | <0.001 | 0.02 |
| (y)pN—1:0 | 1.40 (0.65–3.02) | 1.46 (0.68–3.13) | 1.53 (0.68–3.47) | 0.28 | 0.99 |
| (y)pN—2:0 | 2.53 (1.13–5.68) | 2.85 (1.24–6.55) | 3.22 (1.24–8.32) | 0.03 | 0.53 |
| (y)pTNM Stage—>III:<II | 0.77 (0.35–1.68) | 0.61 (0.28–1.31) | 0.48 (0.21–1.09) | 0.54 | 0.15 |
| Feature | Model A (Cause-Specific Cox + Landmarking) | Model B (Random Survival Forest + Landmarking) |
|---|---|---|
| Model Type | Traditional Cox proportional hazards model | Machine learning random survival forest |
| Time-dependency Handling | Handled via landmarking at fixed time points | Handled via landmarking at fixed time points |
| Assumptions | Proportional hazards, linearity | Non-parametric, no proportional hazards assumption |
| Outcome Type | Relapse, with cancer and other-cause death as competing risks | Relapse, with cancer and other-cause death as competing risks |
| Validation Method | fivefold cross-validation, bootstrap CI | fivefold cross-validation, bootstrap CI |
| Performance Metric | Harrell Concordance Index | Harrell Concordance Index |
| Harrell C-Index | 0.78 [95% CI: 0.75–0.79] | 0.95 [95% CI: 0.82–0.96] |
| Interpretability | High (hazard ratios) | Moderate (feature importance via minimal depth) |
| Ability to Capture Non-linear Effects | Limited | High |
| Competing Risks Framework | Handled indirectly through a cause-specific approach | Explicitly modeled with cumulative incidence functions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reddavid, R.; Elmore, U.; Moro, J.; De Nardi, P.; Biondi, A.; Persiani, R.; Solaini, L.; Pafundi, D.P.; Cianflocca, D.; Sasia, D.; et al. Dynamic Prediction of Rectal Cancer Relapse and Mortality Using a Landmarking-Based Machine Learning Model: A Multicenter Retrospective Study from the Italian Society of Surgical Oncology—Colorectal Cancer Network Collaborative Group. Cancers 2025, 17, 1294. https://doi.org/10.3390/cancers17081294
Reddavid R, Elmore U, Moro J, De Nardi P, Biondi A, Persiani R, Solaini L, Pafundi DP, Cianflocca D, Sasia D, et al. Dynamic Prediction of Rectal Cancer Relapse and Mortality Using a Landmarking-Based Machine Learning Model: A Multicenter Retrospective Study from the Italian Society of Surgical Oncology—Colorectal Cancer Network Collaborative Group. Cancers. 2025; 17(8):1294. https://doi.org/10.3390/cancers17081294
Chicago/Turabian StyleReddavid, Rossella, Ugo Elmore, Jacopo Moro, Paola De Nardi, Alberto Biondi, Roberto Persiani, Leonardo Solaini, Donato P. Pafundi, Desiree Cianflocca, Diego Sasia, and et al. 2025. "Dynamic Prediction of Rectal Cancer Relapse and Mortality Using a Landmarking-Based Machine Learning Model: A Multicenter Retrospective Study from the Italian Society of Surgical Oncology—Colorectal Cancer Network Collaborative Group" Cancers 17, no. 8: 1294. https://doi.org/10.3390/cancers17081294
APA StyleReddavid, R., Elmore, U., Moro, J., De Nardi, P., Biondi, A., Persiani, R., Solaini, L., Pafundi, D. P., Cianflocca, D., Sasia, D., Milone, M., Turri, G., Mineccia, M., Pecchini, F., Gallo, G., Rega, D., Gili, S., Maiello, F., Barberis, A., ... Azzolina, D. (2025). Dynamic Prediction of Rectal Cancer Relapse and Mortality Using a Landmarking-Based Machine Learning Model: A Multicenter Retrospective Study from the Italian Society of Surgical Oncology—Colorectal Cancer Network Collaborative Group. Cancers, 17(8), 1294. https://doi.org/10.3390/cancers17081294











