A Comparative Study Between Copy Number Alterations and PRAME Immunohistochemical Pilot Study in Challenging Melanocytic Lesions
Simple Summary
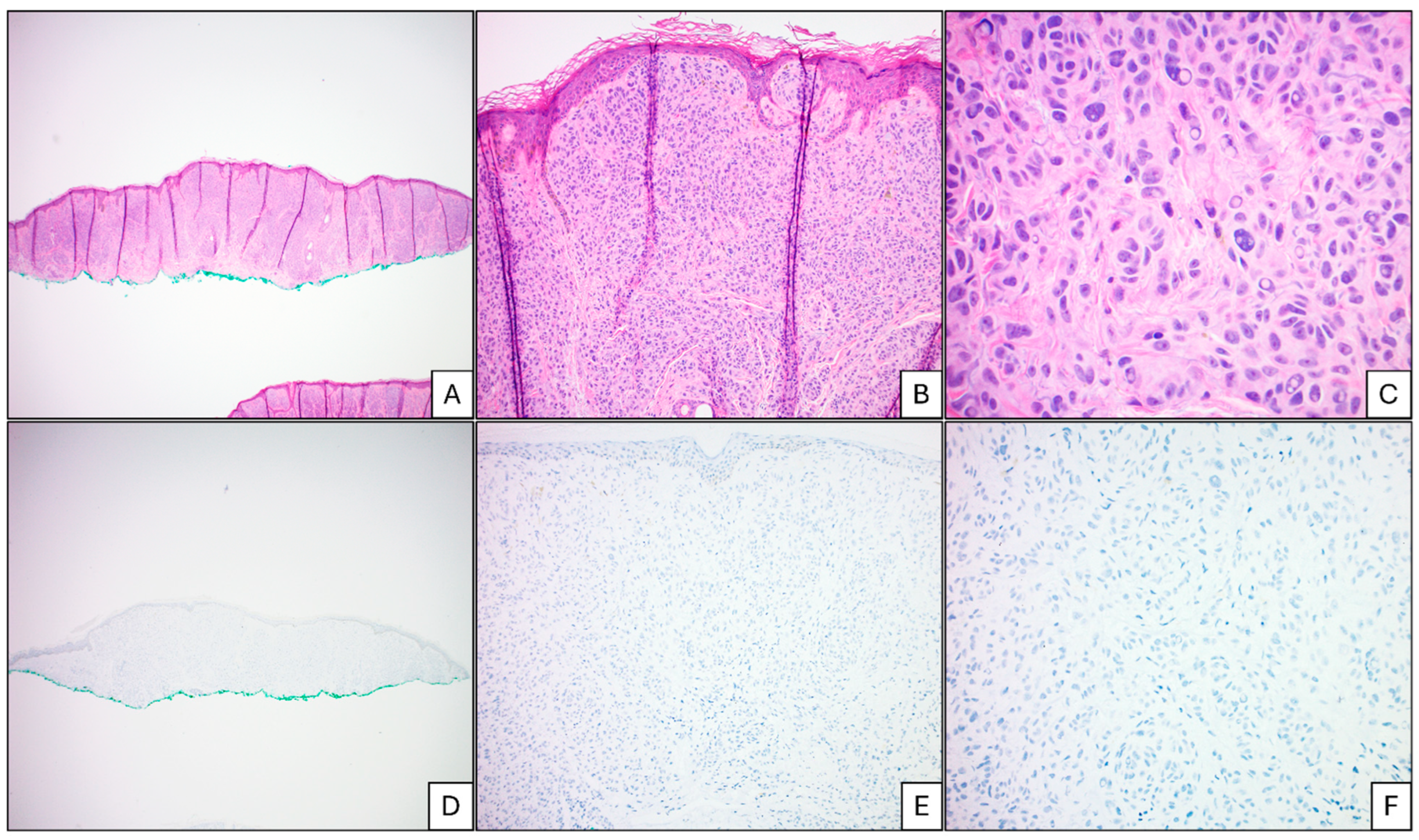
Abstract
1. Introduction
2. Materials/Methods
2.1. Case Selection
2.2. Data Collection
2.3. Data Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Cancer Society. Melanoma Skin Cancer Statistics. Available online: https://www.cancer.org/cancer/types/melanoma-skin-cancer/about/key-statistics.html (accessed on 12 January 2023).
- American Cancer Society. Risk Factors for Melanoma Skin Cancer American Cancer Society. 2000. Available online: https://www.cancer.org/cancer/melanoma-skin-cancer/causes-risks-prevention/risk-factors.html (accessed on 12 January 2023).
- Bataille, V. Early detection of melanoma improves survival. Practitioner 2009, 253, 29–32. [Google Scholar]
- Elmore, J.G.; Barnhill, R.L.; Elder, D.E.; Longton, G.M.; Pepe, M.S.; Reisch, L.M.; Carney, P.A.; Titus, L.J.; Nelson, H.D.; Onega, T.; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357, j2813. [Google Scholar] [CrossRef]
- Auer, B.; Oris, B. Distinguishing melanocytic nevi from melanoma by DNA copy number changes: Comparative genomic hybridization as a research and diagnostic tool. Dermatol. Ther. 2006, 19, 40–49. [Google Scholar]
- Gerami, P.; Jewell, S.S.; Morrison, L.E.; Blondin, B.B.; Schulz, J.B.; Ruffalo, T.B.; Matushek, P.I.; Legator, M.B.; Jacobson, K.M.; Dalton, S.R.; et al. Fluorescence In Situ Hybridization (FISH) as an Ancillary Diagnostic Tool in the Diagnosis of Melanoma. Am. J. Surg. Pathol. 2009, 33, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Gerami, P.; Li, G.; Pouryazdanparast, P.; Blondin, B.; Beilfuss, B.; Slenk, C.; Du, J.; Guitart, J.; Jewell, S.; Pestova, K. A Highly Specific and Discriminatory FISH Assay for Distinguishing Between Benign and Malignant Melanocytic Neoplasms. Am. J. Surg. Pathol. 2012, 36, 808–817. [Google Scholar] [CrossRef]
- Gerami, P.; Zembowicz, A. Update on Fluorescence In Situ Hybridization in Melanoma: State of the Art. Arch. Pathol. Lab. Med. 2011, 135, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Lezcano, C.; Jungbluth, A.A.; Busam, K.J. PRAME Immunohistochemistry as an Ancillary Test for the Assessment of Melanocytic Lesions. Surg. Pathol. Clin. 2021, 14, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Harvey, N.T.F.; Peverall, J.B.; Acott, N.B.; Ardakani, N.F.M.; Leecy, T.N.F.; Iacobelli, J.F.; McCallum, D.F.; Van Vliet, C.F.; Wood, B.A.F. Correlation of FISH and PRAME Immunohistochemistry in Ambiguous Superficial Cutaneous Melanocytic Proliferations. Am. J. Dermatopathol. 2021, 43, 913–920. [Google Scholar] [CrossRef]
- Epping, M.T.; Wang, L.; Edel, M.J.; Carlée, L.; Hernandez, M.; Bernards, R. The Human Tumor Antigen PRAME Is a Dominant Repressor of Retinoic Acid Receptor Signaling. Cell 2005, 122, 835–847. [Google Scholar] [CrossRef]
- Schenk, T.; Stengel, S.; Goellner, S.; Steinbach, D.; Saluz, H.P. Hypomethylation of PRAME is responsible for its aberrant overexpression in human malignancies. Genes Chromosom. Cancer 2007, 46, 796–804. [Google Scholar] [CrossRef]
- Epping, M.T.; Bernards, R. A Causal Role for the Human Tumor Antigen Preferentially Expressed Antigen of Melanoma in Cancer. Cancer Res. 2006, 66, 10639–10642. [Google Scholar] [CrossRef] [PubMed]
- Alomari, A.K.; Miedema, J.R.; Carter, M.D.; Harms, P.W.; Lowe, L.; Durham, A.B.; Fullen, D.R.; Patel, R.M.; Hristov, A.C.; Chan, M.P.; et al. DNA copy number changes correlate with clinical behavior in melanocytic neoplasms: Proposal of an algorithmic approach. Mod. Pathol. 2020, 33, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, A. Discordance among expert pathologists in diagnosis of melanocytic neoplasms. Hum. Pathol. 1996, 27, 1115–1116. [Google Scholar] [CrossRef]
- Farmer, E.R.; Gonin, R.; Hanna, M.P. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum. Pathol. 1996, 27, 528–531. [Google Scholar] [CrossRef]
- Kempf, W.M.; Haeffner, A.C.M.; Mueller, B.B.; Panizzon, R.G.M.; Burg, G.M. Experts and Gold Standards in Dermatopathology. Am. J. Dermatopathol. 1998, 20, 478–482. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Nehal, K.S.; Hollmann, T.J.; Busam, K.J. PRAME Expression in Melanocytic Tumors. Am. J. Surg. Pathol. 2018, 42, 1456–1465. [Google Scholar] [CrossRef]
- Koh, S.S.; Opel, M.L.; Wei, J.-P.J.; Yau, K.; Shah, R.; Gorre, M.E.; Whitman, E.; Shitabata, P.K.; Tao, Y.; Cochran, A.J.; et al. Molecular classification of melanomas and nevi using gene expression microarray signatures and formalin-fixed and paraffin-embedded tissue. Mod. Pathol. 2009, 22, 538–546. [Google Scholar] [CrossRef]
- Bastian, B.C.; Olshen, A.B.; LeBoit, P.E.; Pinkel, D. Classifying Melanocytic Tumors Based on DNA Copy Number Changes. Am. J. Pathol. 2003, 163, 1765–1770. [Google Scholar] [CrossRef]
- Moore, M.W.; Gasparini, R. FISH as an effective diagnostic tool for the management of challenging melanocytic lesions. Diagn. Pathol. 2011, 6, 76. [Google Scholar] [CrossRef]
- Casillas, A.C.B.; Muhlbauer, A.; Barragan, V.A.B.; Jefferson, I.B.; Speiser, J.J. A Comparison of Preferentially Expressed Antigen in Melanoma Immunohistochemistry and Diagnostic Gene Expression-Profiling Assay in Challenging Melanocytic Proliferations. Am. J. Dermatopathol. 2023, 46, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Fattori, A.; de la Fouchardière, A.; Cribier, B.; Mitcov, M. Preferentially expressed Antigen in MElanoma immunohistochemistry as an adjunct for evaluating ambiguous melanocytic proliferation. Hum. Pathol. 2022, 121, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Alomari, A.K.; Tharp, A.W.; Umphress, B.; Kowal, R.P. The utility of PRAME immunohistochemistry in the evaluation of challenging melanocytic tumors. J. Cutan. Pathol. 2021, 48, 1115–1123. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Busam, K.J. Comparison of Immunohistochemistry for PRAME With Cytogenetic Test Results in the Evaluation of Challenging Melanocytic Tumors. Am. J. Surg. Pathol. 2020, 44, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.S.; Lau, S.K.; Scapa, J.V.; Cassarino, D.S. PRAME immunohistochemistry of spitzoid neoplasms. J. Cutan. Pathol. 2022, 49, 709–716. [Google Scholar] [CrossRef]
- Warbasse, E.; Mehregan, D.; Utz, S.; Stansfield, R.B.; Abrams, J. PRAME immunohistochemistry compared to traditional FISH testing in spitzoid neoplasms and other difficult to diagnose melanocytic neoplasms. Front. Med. 2023, 10, 1265827. [Google Scholar] [CrossRef]

| Demographics | |
|---|---|
| Total Number of Cases | 43 |
| Age (years) | |
| Mean | 45.1 |
| Range | 2–92 |
| Gender | |
| Male | 18 |
| Female | 25 |
| Location | |
| Trunk | 23 |
| Extremities | 11 |
| Head/Neck | 9 |
| Non-Diagnostically Challenging Melanomas | 9 |
| Diagnostically Challenging Melanocytic Lesions | 34 |
| Treated as Severely Atypical Nevi | 26 |
| Treated as Melanoma | 8 |
| Non- Diagnostically Challenging Melanoma | Diagnostically Challenging Melanocytic Lesions Treated as Melanoma | Diagnostically Challenging Melanocytic Lesions Treated as Severely Atypical Nevus | |
|---|---|---|---|
| Total cases (n) | 9 | 8 | 26 |
| Age (years) | |||
| Range | 66–92 | 2–69 | 15–61 |
| Mean | 77.89 | 38.1 | 36.7 |
| Median | 78 | 37.5 | 37 |
| Gender | |||
| Male | 6 | 4 | 8 |
| Female | 3 | 4 | 18 |
| Location | |||
| Trunk | 3 | 4 | 14 |
| Extremities | 4 | 4 | 6 |
| Head/Neck | 2 | 0 | 6 |
| Breslow thickness (mm) | |||
| Range | 0.3–5.2 | 0.6–4.2 | Na |
| Mean | 1.63 | 1.74 | Na |
| Median | 0.95 | 1.49 | Na |
| Pathologic stage | |||
| T | |||
| Range | 1a–3b | 1–4 | Na |
| N | |||
| Range | 0–2c | 0–1a | Na |
| M | |||
| Range | 0–1d | 0 | Na |
| Follow-up duration (months) | |||
| Range | 7–64 | 3–61 | 21–64 |
| Mean | 43.33 | 31.1 | 44.2 |
| Median | 50 | 28 | 51 |
| Follow-up information | |||
| Recurrent local disease | 1 | 0 | 1 |
| Metastatic melanoma | 3 | 1 | 0 |
| Alive with disease | 3 | 8 | 26 |
| Died of disease | 2 | 0 | 0 |
| Died of other causes | 3 | 0 | Na |
| Lost to follow up | 0 | 0 | 0 |
| Non-Challenging Melanomas (n = 9) | |
|---|---|
| Copy Number Alterations (%) | 100% |
| Positive FISH (n) | 9 |
| Positive SNP array (n) | 0 |
| PRAME Positivity (%) | 77.8% |
| Positive PRAME (n) | 9 |
| Diagnostically Challenging Melanocytic Lesions (n = 34) | |
| Copy Number Alterations (%) | 17.6% |
| Positive FISH (n) | 1 |
| Positive SNP array (n) | 5 |
| PRAME Positivity (%) | 5.9% |
| Positive PRAME (n) | 2 |
| Non-challenging melanomas (n = 9) | |
| Diagnosis: PRAME Concordance (%) | 77.8% |
| Diagnosis: Molecular Concordance (%) | 100% |
| Diagnostically challenging melanocytic lesions (n = 34) | |
| Diagnosis: PRAME Concordance (%) | 76.5% |
| Diagnosis: Molecular Concordance (%) | 94.1% |
| Diagnostically challenging severely atypical nevi (n = 26) | |
| Diagnosis: PRAME Concordance (%) | 96.2% |
| Diagnosis: Molecular Concordance (%) | 100% |
| Diagnostically challenging melanomas (n = 8) | |
| Diagnosis: PRAME Concordance (%) | 12.5% |
| Diagnosis: Molecular concordance (%) | 75% |
| Diagnostically Challenging Melanocytic Lesions: Prame Staining Results | Diagnostically Challenging Melanocytic Lesions: Copy Number Alteration Results | |||||
| PRAME + | PRAME − | Total | FISH/SNP + | FISH/SNP − | Total | |
| Treated as Severely Atypical Nevus | 1 | 25 | 26 | 0 | 26 | 26 |
| Treated as Melanoma | 1 | 7 | 8 | 6 | 2 | 8 |
| PRAME performance metrics: Diagnostically challenging melanocytic lesions | Molecular studies performance metrics: Diagnostically challenging melanocytic lesions | |||||
| Sensitivity | 12.50% | 75% | ||||
| Specificity | 96.20% | 100% | ||||
| PPV | 50% | 100% | ||||
| NPV | 78.10% | 92.90% | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chun, J.; Scholl, A.R.; Crimmins, J.; Schneider, M.M.; Selim, M.A.; Al-Rohil, R.N. A Comparative Study Between Copy Number Alterations and PRAME Immunohistochemical Pilot Study in Challenging Melanocytic Lesions. Cancers 2025, 17, 1218. https://doi.org/10.3390/cancers17071218
Chun J, Scholl AR, Crimmins J, Schneider MM, Selim MA, Al-Rohil RN. A Comparative Study Between Copy Number Alterations and PRAME Immunohistochemical Pilot Study in Challenging Melanocytic Lesions. Cancers. 2025; 17(7):1218. https://doi.org/10.3390/cancers17071218
Chicago/Turabian StyleChun, Jeana, Ashley R. Scholl, Jennifer Crimmins, Michelle M. Schneider, M. Angelica Selim, and Rami N. Al-Rohil. 2025. "A Comparative Study Between Copy Number Alterations and PRAME Immunohistochemical Pilot Study in Challenging Melanocytic Lesions" Cancers 17, no. 7: 1218. https://doi.org/10.3390/cancers17071218
APA StyleChun, J., Scholl, A. R., Crimmins, J., Schneider, M. M., Selim, M. A., & Al-Rohil, R. N. (2025). A Comparative Study Between Copy Number Alterations and PRAME Immunohistochemical Pilot Study in Challenging Melanocytic Lesions. Cancers, 17(7), 1218. https://doi.org/10.3390/cancers17071218






