Transcriptome Analysis of Canine Histiocytic Sarcoma Tumors and Cell Lines Reveals Multiple Targets for Therapy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. HS Tissue and Blood Sample Extraction, Processing, and Sequencing Methods
2.2. HS Cell Line Growth, Maintenance, and RNA Preparation
2.3. RNA-Seq Analysis
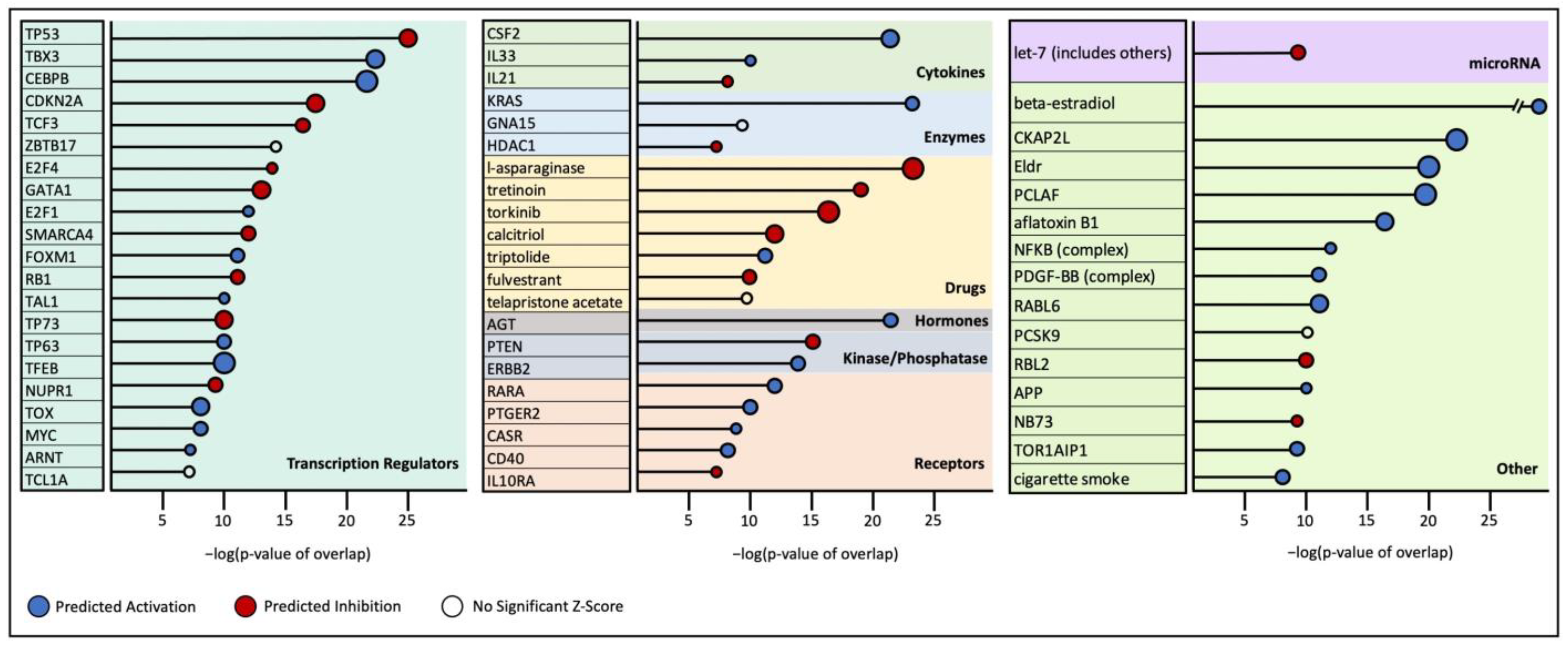
2.4. Upstream Regulator Analysis
2.5. Drug Efficacy Studies
2.6. Cell Viability Assay
2.7. Combination Index (CI)
3. Results
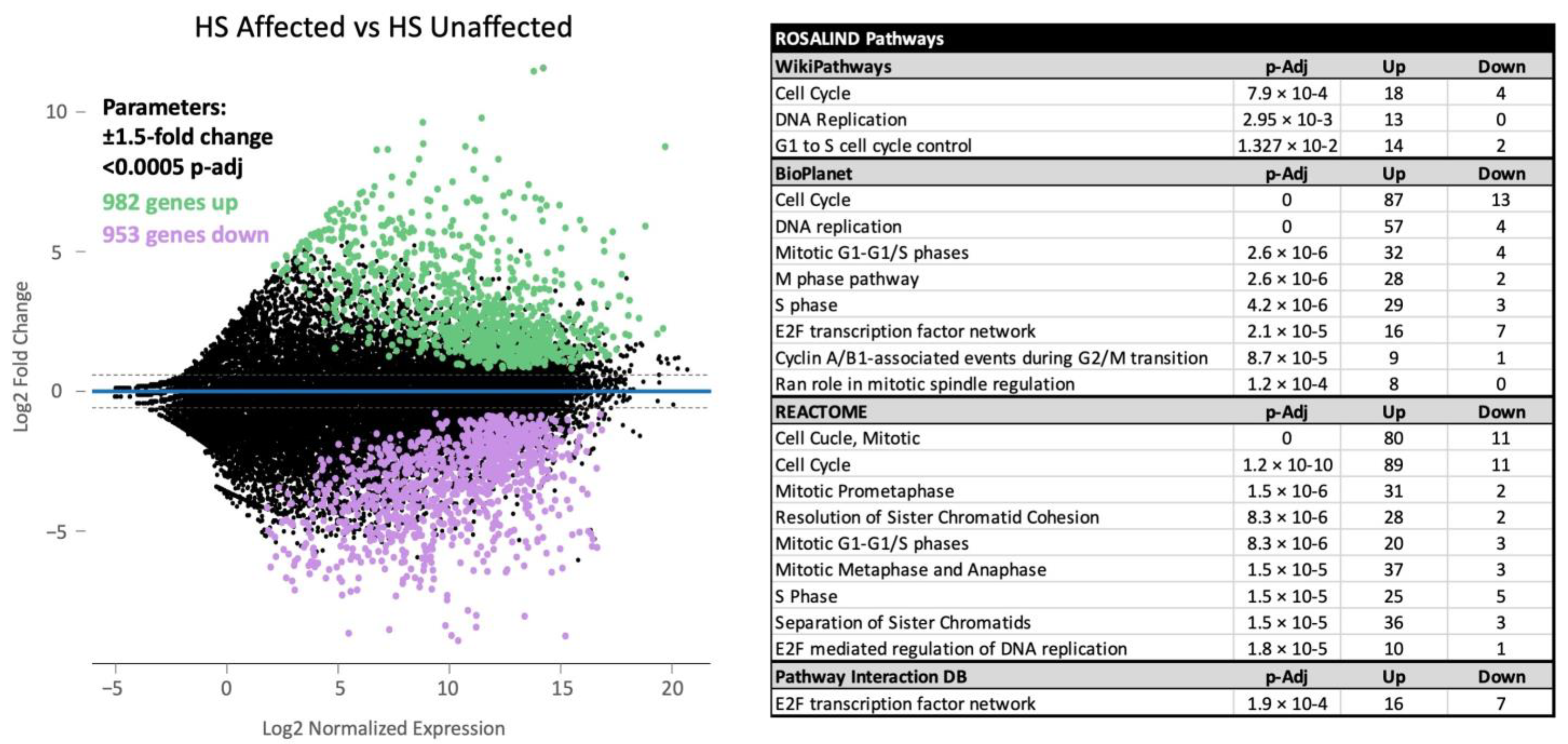
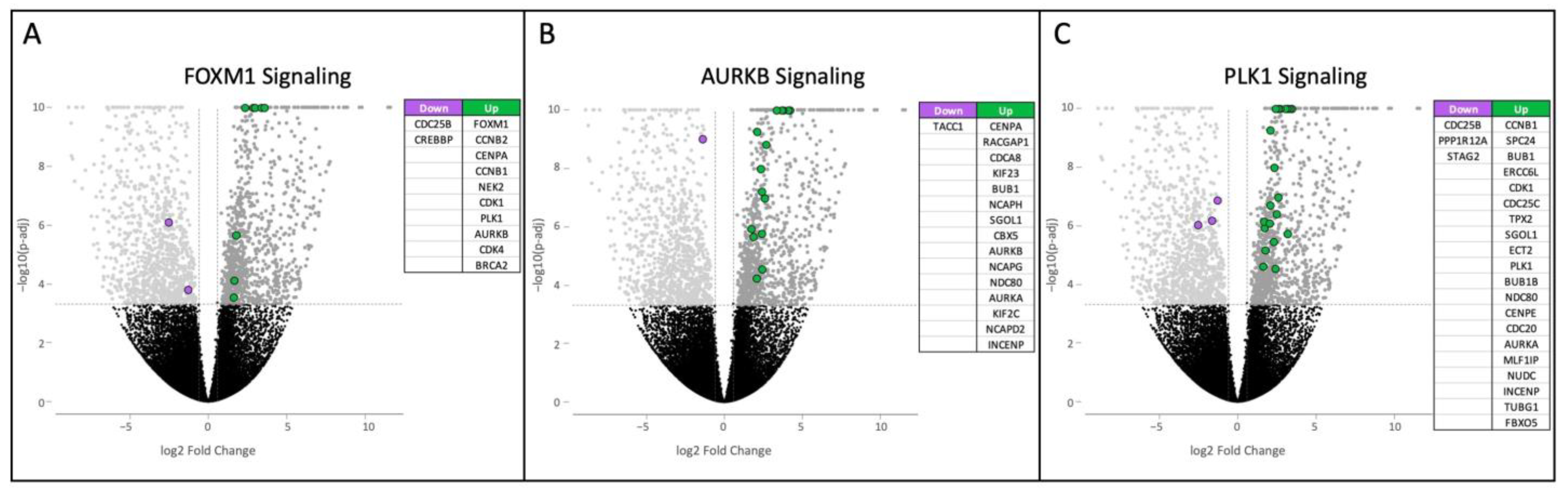
3.1. HS Tumor Gene Expression Results
3.2. ROSALIND HS Cell Line Gene Expression Results
3.3. HS vs. HHS Differential Gene Expression
3.4. Transcriptome Comparison of HS Cases with PTPN11 Variant vs. Without PTPN11 Variant
3.5. Differential Expression of Immune Checkpoint Genes
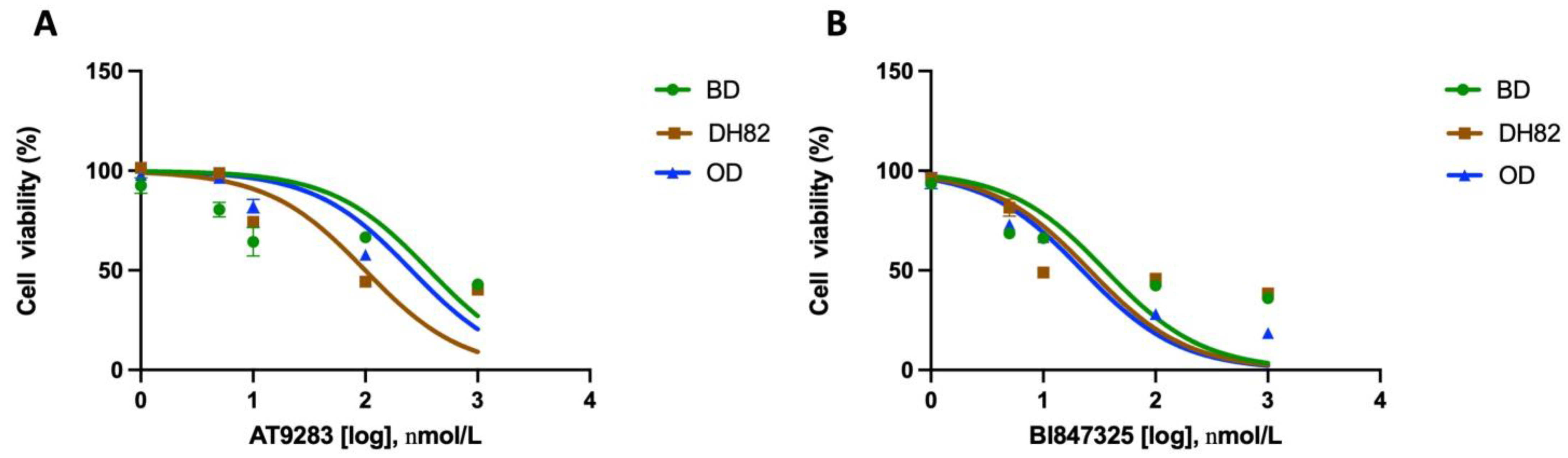
3.6. Effect of AURK Inhibitors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moore, P.F. A review of histiocytic diseases of dogs and cats. Vet. Pathol. 2014, 51, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Abadie, J.; Hedan, B.; Cadieu, E.; De Brito, C.; Devauchelle, P.; Bourgain, C.; Parker, H.G.; Vaysse, A.; Margaritte-Jeannin, P.; Galibert, F.; et al. Epidemiology, pathology, and genetics of histiocytic sarcoma in the Bernese mountain dog breed. J. Hered. 2009, 100 (Suppl. S1), S19–S27. [Google Scholar] [CrossRef]
- Evans, J.M.; Parker, H.G.; Rutteman, G.R.; Plassais, J.; Grinwis, G.C.M.; Harris, A.C.; Lana, S.E.; Ostrander, E.A. Multi-omics approach identifies germline regulatory variants associated with hematopoietic malignancies in retriever dog breeds. PLoS Genet. 2021, 17, e1009543. [Google Scholar] [CrossRef]
- Hedan, B.; Cadieu, E.; Rimbault, M.; Vaysse, A.; Dufaure de Citres, C.; Devauchelle, P.; Botherel, N.; Abadie, J.; Quignon, P.; Derrien, T.; et al. Identification of common predisposing loci to hematopoietic cancers in four dog breeds. PLoS Genet. 2021, 17, e1009395. [Google Scholar] [CrossRef] [PubMed]
- Shearin, A.L.; Hedan, B.; Cadieu, E.; Erich, S.A.; Schmidt, E.V.; Faden, D.L.; Cullen, J.; Abadie, J.; Kwon, E.M.; Grone, A.; et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Boerkamp, K.M.; van der Kooij, M.; van Steenbeek, F.G.; van Wolferen, M.E.; Groot Koerkamp, M.J.; van Leenen, D.; Grinwis, G.C.; Penning, L.C.; Wiemer, E.A.; Rutteman, G.R. Gene expression profiling of histiocytic sarcomas in a canine model: The predisposed flatcoated retriever dog. PLoS ONE 2013, 8, e71094. [Google Scholar] [CrossRef]
- Takada, M.; Smyth, L.A.; Thaiwong, T.; Richter, M.; Corner, S.M.; Schall, P.Z.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas. Genes 2019, 10, 505. [Google Scholar] [CrossRef]
- Yang, Y.T.; Engleberg, A.I.; Kapoor, I.; Kitagawa, K.; Hilburger, S.A.; Thaiwong-Nebelung, T.; Yuzbasiyan-Gurkan, V. Canine Histiocytic and Hemophagocytic Histiocytic Sarcomas Display KRAS and Extensive PTPN11/SHP2 Mutations and Respond In Vitro to MEK Inhibition by Cobimetinib. Genes 2024, 15, 1050. [Google Scholar] [CrossRef]
- Hedan, B.; Rault, M.; Abadie, J.; Ulve, R.; Botherel, N.; Devauchelle, P.; Copie-Bergman, C.; Cadieu, E.; Parrens, M.; Alten, J.; et al. PTPN11 mutations in canine and human disseminated histiocytic sarcoma. Int. J. Cancer 2020, 147, 1657–1665. [Google Scholar] [CrossRef]
- Takada, M.; Hix, J.M.L.; Corner, S.; Schall, P.Z.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Targeting MEK in a Translational Model of Histiocytic Sarcoma. Mol. Cancer Ther. 2018, 17, 2439–2450. [Google Scholar] [CrossRef]
- Takada, M.; Smyth, L.A.; Hix, J.M.; Corner, S.M.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Development of an Orthotopic Intrasplenic Xenograft Mouse Model of Canine Histiocytic Sarcoma and Its Use in Evaluating the Efficacy of Treatment with Dasatinib. Comp. Med. 2019, 69, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Takada, M.; Kitagawa, K.; Zhang, Y.; Bulitta, J.B.; Moirano, S.; Jones, A.; Borgen, J.; Onsager, A.; Thaiwong, T.; Vail, D.M. Population Pharmacokinetics, Pharmacodynamics and Safety Properties of Trametinib in Dogs With Cancer: A Phase I Dose Escalating Clinical Trial. Vet. Comp. Oncol. 2024, 22, 410–421. [Google Scholar] [CrossRef]
- Rassnick, K.M.; Moore, A.S.; Russell, D.S.; Northrup, N.C.; Kristal, O.; Bailey, D.B.; Flory, A.B.; Kiselow, M.A.; Intile, J.L. Phase II, open-label trial of single-agent CCNU in dogs with previously untreated histiocytic sarcoma. J. Vet. Intern. Med. 2010, 24, 1528–1531. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Skorupski, K.A.; Clifford, C.A.; Paoloni, M.C.; Lara-Garcia, A.; Barber, L.; Kent, M.S.; LeBlanc, A.K.; Sabhlok, A.; Mauldin, E.A.; Shofer, F.S.; et al. CCNU for the treatment of dogs with histiocytic sarcoma. J. Vet. Intern. Med. 2007, 21, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.; Thomas, R.; Durrant, J.; Jiang, T.; Motsinger-Reif, A.; Breen, M. Genome-wide DNA copy number analysis and targeted transcriptional analysis of canine histiocytic malignancies identifies diagnostic signatures and highlights disruption of spindle assembly complex. Chromosome Res. 2019, 27, 179–202. [Google Scholar] [CrossRef]
- Asada, H.; Tani, A.; Sakuma, H.; Hirabayashi, M.; Matsumoto, Y.; Watanabe, K.; Tsuboi, M.; Yoshida, S.; Harada, K.; Uchikai, T.; et al. Whole exome and transcriptome analysis revealed the activation of ERK and Akt signaling pathway in canine histiocytic sarcoma. Sci. Rep. 2023, 13, 8512. [Google Scholar] [CrossRef]
- Takada, M.; Parys, M.; Gregory-Bryson, E.; Vilar Saavedra, P.; Kiupel, M.; Yuzbasiyan-Gurkan, V. A novel canine histiocytic sarcoma cell line: Initial characterization and utilization for drug screening studies. BMC Cancer 2018, 18, 237. [Google Scholar] [CrossRef]
- Clinton, J.S.B. In Vitro Differentiation of Macrophages and Dendritic Cells from Primary Human CD14+ Monocytes; ATCC: Manassas, VA, USA, 2015. [Google Scholar]
- Christian, H. Cran-Package fpc. Available online: https://cran.r-project.org/web/packages/fpc/index.html (accessed on 15 August 2024).
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Goswami, S.; Raychaudhuri, D.; Siddiqui, B.A.; Singh, P.; Nagarajan, A.; Liu, J.; Subudhi, S.K.; Poon, C.; Gant, K.L.; et al. Immune checkpoint therapy-current perspectives and future directions. Cell 2023, 186, 1652–1669. [Google Scholar] [CrossRef]
- Foran, J.; Ravandi, F.; Wierda, W.; Garcia-Manero, G.; Verstovsek, S.; Kadia, T.; Burger, J.; Yule, M.; Langford, G.; Lyons, J.; et al. A phase I and pharmacodynamic study of AT9283, a small-molecule inhibitor of aurora kinases in patients with relapsed/refractory leukemia or myelofibrosis. Clin. Lymphoma Myeloma Leuk. 2014, 14, 223–230. [Google Scholar] [CrossRef]
- Sini, P.; Gurtler, U.; Zahn, S.K.; Baumann, C.; Rudolph, D.; Baumgartinger, R.; Strauss, E.; Haslinger, C.; Tontsch-Grunt, U.; Waizenegger, I.C.; et al. Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases. Mol. Cancer Ther. 2016, 15, 2388–2398. [Google Scholar] [CrossRef]
- Egan, C.; Nicolae, A.; Lack, J.; Chung, H.J.; Skarshaug, S.; Pham, T.A.; Navarro, W.; Abdullaev, Z.; Aguilera, N.S.; Xi, L.; et al. Genomic profiling of primary histiocytic sarcoma reveals two molecular subgroups. Haematologica 2020, 105, 951–960. [Google Scholar] [CrossRef]
- Diamond, E.L.; Durham, B.H.; Ulaner, G.A.; Drill, E.; Buthorn, J.; Ki, M.; Bitner, L.; Cho, H.; Young, R.J.; Francis, J.H.; et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 2019, 567, 521–524. [Google Scholar] [CrossRef]
- Shanmugam, V.; Griffin, G.K.; Jacobsen, E.D.; Fletcher, C.D.M.; Sholl, L.M.; Hornick, J.L. Identification of diverse activating mutations of the RAS-MAPK pathway in histiocytic sarcoma. Mod. Pathol. 2019, 32, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Stoecker, M.M.; Wang, E. Histiocytic/dendritic cell transformation of B-cell neoplasms: Pathologic evidence of lineage conversion in differentiated hematolymphoid malignancies. Arch. Pathol. Lab. Med. 2013, 137, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Fernandez-Hazoury, D.; Yoshikawa, G.; Minja, A.; Huang, H.; Hwang, A.; Qing, X. Transformation of Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma to Histiocytic/Dendritic Cell Sarcoma. J. Hematol. 2024, 13, 216–223. [Google Scholar] [CrossRef]
- Reiner, A.S.; Durham, B.H.; Yabe, M.; Petrova-Drus, K.; Francis, J.H.; Rampal, R.K.; Lacouture, M.E.; Rotemberg, V.; Abdel-Wahab, O.; Panageas, K.S.; et al. Outcomes after interruption of targeted therapy in patients with histiocytic neoplasms. Br. J. Haematol. 2023, 203, 389–394. [Google Scholar] [CrossRef]
- Koh, K.N.; Yoon, S.H.; Kang, S.H.; Kim, H.; Im, H.J. Advancements in the understanding and management of histiocytic neoplasms. Blood Res. 2024, 59, 22. [Google Scholar] [CrossRef]
- Friedman, J.S.; Durham, B.H.; Reiner, A.S.; Yabe, M.; Petrova-Drus, K.; Dogan, A.; Pulitzer, M.; Busam, K.J.; Francis, J.H.; Rampal, R.K.; et al. Mixed histiocytic neoplasms: A multicentre series revealing diverse somatic mutations and responses to targeted therapy. Br. J. Haematol. 2024, 205, 127–137. [Google Scholar] [CrossRef]
- Costa, R.H. FoxM1 dances with mitosis. Nat. Cell Biol. 2005, 7, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Laoukili, J.; Kooistra, M.R.; Bras, A.; Kauw, J.; Kerkhoven, R.M.; Morrison, A.; Clevers, H.; Medema, R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005, 7, 126–136. [Google Scholar] [CrossRef]
- Fu, Z.; Malureanu, L.; Huang, J.; Wang, W.; Li, H.; van Deursen, J.M.; Tindall, D.J.; Chen, J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 2008, 10, 1076–1082. [Google Scholar] [CrossRef]
- Ma, R.Y.; Tong, T.H.; Leung, W.Y.; Yao, K.M. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1. Methods Mol. Biol. 2010, 647, 113–123. [Google Scholar] [CrossRef]
- Puig-Butille, J.A.; Vinyals, A.; Ferreres, J.R.; Aguilera, P.; Cabre, E.; Tell-Marti, G.; Marcoval, J.; Mateo, F.; Palomero, L.; Badenas, C.; et al. AURKA Overexpression Is Driven by FOXM1 and MAPK/ERK Activation in Melanoma Cells Harboring BRAF or NRAS Mutations: Impact on Melanoma Prognosis and Therapy. J. Investig. Dermatol. 2017, 137, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Teh, M.T.; Gemenetzidis, E.; Patel, D.; Tariq, R.; Nadir, A.; Bahta, A.W.; Waseem, A.; Hutchison, I.L. FOXM1 induces a global methylation signature that mimics the cancer epigenome in head and neck squamous cell carcinoma. PLoS ONE 2012, 7, e34329. [Google Scholar] [CrossRef] [PubMed]
- Pfister, K.; Pipka, J.L.; Chiang, C.; Liu, Y.; Clark, R.A.; Keller, R.; Skoglund, P.; Guertin, M.J.; Hall, I.M.; Stukenberg, P.T. Identification of Drivers of Aneuploidy in Breast Tumors. Cell Rep. 2018, 23, 2758–2769. [Google Scholar] [CrossRef]
- Barger, C.J.; Zhang, W.; Hillman, J.; Stablewski, A.B.; Higgins, M.J.; Vanderhyden, B.C.; Odunsi, K.; Karpf, A.R. Genetic determinants of FOXM1 overexpression in epithelial ovarian cancer and functional contribution to cell cycle progression. Oncotarget 2015, 6, 27613–27627. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Vorderbruggen, M.; Munoz-Trujillo, C.; Kim, S.H.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S.; Karpf, A.R. NB compounds are potent and efficacious FOXM1 inhibitors in high-grade serous ovarian cancer cells. J. Ovarian Res. 2024, 17, 94. [Google Scholar] [CrossRef]
- Carmena, M.; Ruchaud, S.; Earnshaw, W.C. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009, 21, 796–805. [Google Scholar] [CrossRef]
- Cannon, C.M.; Pozniak, J.; Scott, M.C.; Ito, D.; Gorden, B.H.; Graef, A.J.; Modiano, J.F. Canine osteosarcoma cells exhibit resistance to aurora kinase inhibitors. Vet. Comp. Oncol. 2015, 13, 48–59. [Google Scholar] [CrossRef]
- Katayama, H.; Brinkley, W.R.; Sen, S. The Aurora kinases: Role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003, 22, 451–464. [Google Scholar] [CrossRef]
- Bellelli, R.; Castellone, M.D.; Garcia-Rostan, G.; Ugolini, C.; Nucera, C.; Sadow, P.M.; Nappi, T.C.; Salerno, P.; Cantisani, M.C.; Basolo, F.; et al. FOXM1 is a molecular determinant of the mitogenic and invasive phenotype of anaplastic thyroid carcinoma. Endocr. Relat. Cancer 2012, 19, 695–710. [Google Scholar] [CrossRef]
- Fu, J.; Bian, M.; Jiang, Q.; Zhang, C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 2007, 5, 1–10. [Google Scholar] [CrossRef]
- Hindriksen, S.; Lens, S.M.A.; Hadders, M.A. The Ins and Outs of Aurora B Inner Centromere Localization. Front. Cell Dev. Biol. 2017, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Chen, Q.; Yan, H.; Zhang, M.; Liang, C.; Xiang, X.; Pan, X.; Wang, F. Aurora B kinase activity-dependent and -independent functions of the chromosomal passenger complex in regulating sister chromatid cohesion. J. Biol. Chem. 2019, 294, 2021–2035. [Google Scholar] [CrossRef]
- Klein, U.R.; Nigg, E.A.; Gruneberg, U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell 2006, 17, 2547–2558. [Google Scholar] [CrossRef]
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285. [Google Scholar] [CrossRef]
- Munoz-Barrera, M.; Monje-Casas, F. Increased Aurora B activity causes continuous disruption of kinetochore-microtubule attachments and spindle instability. Proc. Natl. Acad. Sci. USA 2014, 111, E3996–E4005. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Loyola, A.; Fernandez-Miranda, G.; Trakala, M.; Partida, D.; Samejima, K.; Ogawa, H.; Canamero, M.; de Martino, A.; Martinez-Ramirez, A.; de Carcer, G.; et al. Aurora B Overexpression Causes Aneuploidy and p21Cip1 Repression during Tumor Development. Mol. Cell Biol. 2015, 35, 3566–3578. [Google Scholar] [CrossRef]
- Garlapati, C.; Joshi, S.; Bhattarai, S.; Krishnamurthy, J.; Turaga, R.C.; Nguyen, T.; Li, X.; Aneja, R. PLK1 and AURKB phosphorylate survivin differentially to affect proliferation in racially distinct triple-negative breast cancer. Cell Death Dis. 2023, 14, 12. [Google Scholar] [CrossRef]
- Wang, Q.; Bode, A.M.; Zhang, T. Targeting CDK1 in cancer: Mechanisms and implications. NPJ Precis. Oncol. 2023, 7, 58. [Google Scholar] [CrossRef]
- Georgieva, I.; Koychev, D.; Wang, Y.; Holstein, J.; Hopfenmuller, W.; Zeitz, M.; Grabowski, P. ZM447439, a novel promising aurora kinase inhibitor, provokes antiproliferative and proapoptotic effects alone and in combination with bio- and chemotherapeutic agents in gastroenteropancreatic neuroendocrine tumor cell lines. Neuroendocrinology 2010, 91, 121–130. [Google Scholar] [CrossRef]
- Hauf, S.; Cole, R.W.; LaTerra, S.; Zimmer, C.; Schnapp, G.; Walter, R.; Heckel, A.; van Meel, J.; Rieder, C.L.; Peters, J.M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003, 161, 281–294. [Google Scholar] [CrossRef]
- Harrington, E.A.; Bebbington, D.; Moore, J.; Rasmussen, R.K.; Ajose-Adeogun, A.O.; Nakayama, T.; Graham, J.A.; Demur, C.; Hercend, T.; Diu-Hercend, A.; et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004, 10, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Borah, N.A.; Reddy, M.M. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021, 26, 1981. [Google Scholar] [CrossRef] [PubMed]
- Chiappa, M.; Petrella, S.; Damia, G.; Broggini, M.; Guffanti, F.; Ricci, F. Present and Future Perspective on PLK1 Inhibition in Cancer Treatment. Front. Oncol. 2022, 12, 903016. [Google Scholar] [CrossRef] [PubMed]
- Vormoor, B.; Veal, G.J.; Griffin, M.J.; Boddy, A.V.; Irving, J.; Minto, L.; Case, M.; Banerji, U.; Swales, K.E.; Tall, J.R.; et al. A phase I/II trial of AT9283, a selective inhibitor of aurora kinase in children with relapsed or refractory acute leukemia: Challenges to run early phase clinical trials for children with leukemia. Pediatr. Blood Cancer 2017, 64, e26351. [Google Scholar] [CrossRef]
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183. [Google Scholar] [CrossRef]
- Waksal, J.A.; Bruedigam, C.; Komrokji, R.S.; Jamieson, C.H.M.; Mascarenhas, J.O. Telomerase-targeted therapies in myeloid malignancies. Blood Adv. 2023, 7, 4302–4314. [Google Scholar] [CrossRef]
- Lenz, J.A.; Peng, B.; Assenmacher, C.A.; King, A.; Zhang, P.J.; Maki, R.G.; Blanco, M.A.; Radaelli, E.; Atherton, M.J. Identification of immune suppressor candidates utilizing comparative transcriptional profiling in histiocytic sarcoma. Cancer Immunol. Immunother. 2025, 74, 61. [Google Scholar] [CrossRef]
- Merck Animal Health. Announces Availability of Novel Canine Oncology Therapy to Veterinary Specialists Practicing Oncology Nationwide. Gilvetmab is Available to Veterinary Specialists Practicing Oncology to Treat Dogs with Mast Cell Tumors and Melanoma; Merck Animal Health: Rahway, NJ, USA, 2023. [Google Scholar]
- Mason, N.J.; Chester, N.; Xiong, A.; Rotolo, A.; Wu, Y.; Yoshimoto, S.; Glassman, P.; Gulendran, G.; Siegel, D.L. Development of a fully canine anti-canine CTLA4 monoclonal antibody for comparative translational research in dogs with spontaneous tumors. MAbs 2021, 13, 2004638. [Google Scholar] [CrossRef]
- Dow, S. A Role for Dogs in Advancing Cancer Immunotherapy Research. Front. Immunol. 2019, 10, 2935. [Google Scholar] [CrossRef]
| Inhibitory Molecule | |||||
|---|---|---|---|---|---|
| Alias | Gene Name | Average Log2 Expression | Average Log2 Fold Change | Significance (p-adj) | Compared to Control |
| A2AR | ADORA2A | 10.87 | −1.84 | 1.78 × 10−3 | Down |
| A2BR | ADORA2B | 9.25 | 0.711 | N.S. | |
| PD-L1 | CD274 | 13.64 | 2.36 | 1.84 × 10−4 | Up |
| PD-L2 | PDCD1LG2 | 12.61 | 2.39 | 2.62 × 10−4 | Up |
| PD-1 | PDCD1 | 8.50 | 0.128 | N.S. | |
| TIM3 | HAVCR2 | 8.73 | 0.145 | N.S. | |
| VISTA | VSIR | 14.27 | −3.14 | 7.31 × 10−7 | Down |
| LAG3 | LAG3 | 10.71 | 0.497 | N.S. | |
| IDO | IDO1 | 14.08 | 0.646 | N.S. | |
| CTLA-4 | CTLA4 | 10.55 | 0.264 | N.S. | |
| BTLA | BTLA | 11.10 | −3.46 | 1.7 × 10−4 | Down |
| B7-H4 | VTCN1 | 7.17 | −3.33 | 1.08 × 10−3 | Down |
| B7-H3 | CD276 | 12.98 | 2.45 | 9.01 × 10−5 | Up |
| AT9283 | BI847325 | |
|---|---|---|
| BD (IC50) | 285.7 nM | 31.4 nM |
| DH82 (IC50) | 117.5 nM | 19.7 nM |
| OD (IC50) | 269.1 nM | 23.8 nM |
| Cmax | 488.2 nM | 500 nM (in mice) |
| Molecular Targets | Aurora A/B JAK2/3 ABL FLT3 | Aurora C MEK 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engleberg, A.I.; Yang, Y.-T.; Schall, P.Z.; Takada, M.; Thaiwong-Nebelung, T.; Evans, J.M.; Ostrander, E.A.; Yuzbasiyan-Gurkan, V. Transcriptome Analysis of Canine Histiocytic Sarcoma Tumors and Cell Lines Reveals Multiple Targets for Therapy. Cancers 2025, 17, 954. https://doi.org/10.3390/cancers17060954
Engleberg AI, Yang Y-T, Schall PZ, Takada M, Thaiwong-Nebelung T, Evans JM, Ostrander EA, Yuzbasiyan-Gurkan V. Transcriptome Analysis of Canine Histiocytic Sarcoma Tumors and Cell Lines Reveals Multiple Targets for Therapy. Cancers. 2025; 17(6):954. https://doi.org/10.3390/cancers17060954
Chicago/Turabian StyleEngleberg, Alexander I., Ya-Ting Yang, Peter Z. Schall, Marilia Takada, Tuddow Thaiwong-Nebelung, Jacquelyn M. Evans, Elaine A. Ostrander, and Vilma Yuzbasiyan-Gurkan. 2025. "Transcriptome Analysis of Canine Histiocytic Sarcoma Tumors and Cell Lines Reveals Multiple Targets for Therapy" Cancers 17, no. 6: 954. https://doi.org/10.3390/cancers17060954
APA StyleEngleberg, A. I., Yang, Y.-T., Schall, P. Z., Takada, M., Thaiwong-Nebelung, T., Evans, J. M., Ostrander, E. A., & Yuzbasiyan-Gurkan, V. (2025). Transcriptome Analysis of Canine Histiocytic Sarcoma Tumors and Cell Lines Reveals Multiple Targets for Therapy. Cancers, 17(6), 954. https://doi.org/10.3390/cancers17060954






