Applications of Artificial Intelligence for Metastatic Gastrointestinal Cancer: A Systematic Literature Review
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
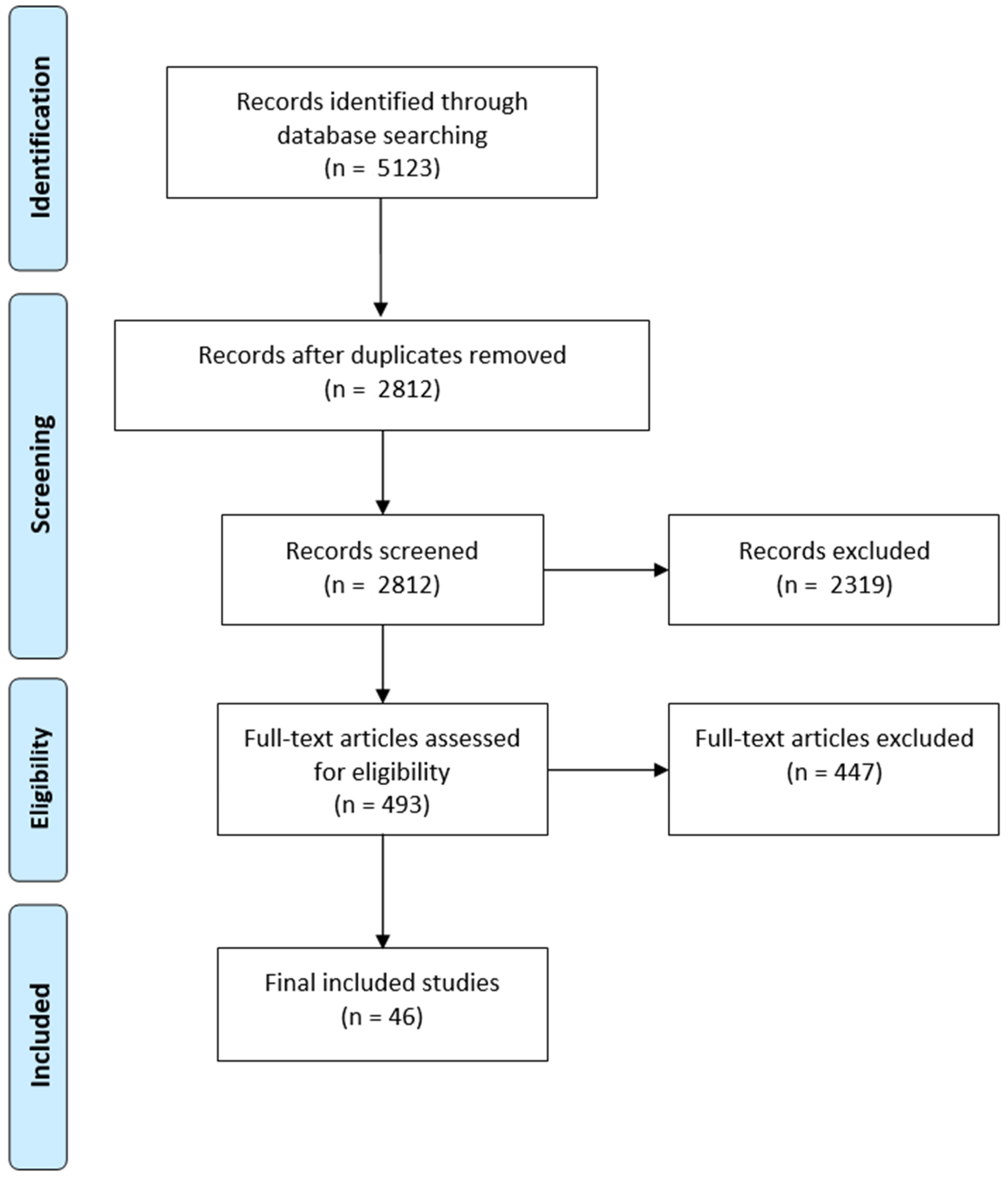
2.1. Search Strategy and Study Selection
2.2. Data Extraction
2.3. Risk of Bias Assessment
3. Results
3.1. Patient Types and Data Collection Period
3.2. Age and Gender Distribution
3.3. Datasets and Features
3.4. Clinical Focus of the Developed Models
3.5. Data Preparation
3.6. AI Models
3.7. Validation and Evaluation
3.8. Risk of Bias Assessment
4. Discussion
4.1. Challenges and Recommendations for Future Research
4.2. Risk of Bias Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, D.; Fang, M.-J.; Tang, L.; Shan, X.-H.; Gao, J.-B.; Giganti, F.; Wang, R.-P.; Chen, X.; Wang, X.-X.; Palumbo, D.; et al. Deep Learning Radiomic Nomogram Can Predict the Number of Lymph Node Metastasis in Locally Advanced Gastric Cancer: An International Multicenter Study. Ann. Oncol. 2020, 31, 912–920. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Li, D.; Li, S.; Li, W.; Tong, T.; Liu, L.; Jiang, D.; Jiang, L.; Ruan, G.; Hai, N.; et al. Deep Learning Radiomics of Dual-Energy Computed Tomography for Predicting Lymph Node Metastases of Pancreatic Ductal Adenocarcinoma. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Eresen, A.; Shangguan, J.; Yang, J.; Lu, Y.; Chen, D.; Wang, J.; Velichko, Y.; Yaghmai, V.; Zhang, Z. Establishment of a New Non-Invasive Imaging Prediction Model for Liver Metastasis in Colon Cancer. Am. J. Cancer Res. 2019, 9, 2482–2492. [Google Scholar] [PubMed]
- Yang, C.; Huang, M.; Li, S.; Chen, J.; Yang, Y.; Qin, N.; Huang, D.; Shu, J. Radiomics Model of Magnetic Resonance Imaging for Predicting Pathological Grading and Lymph Node Metastases of Extrahepatic Cholangiocarcinoma. Cancer Lett. 2020, 470, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Fang, M.; Zhang, L.; Zhong, L.; Dong, D.; Tian, J.; Shan, X. Integrating No. 3 Lymph Nodes and Primary Tumor Radiomics to Predict Lymph Node Metastasis in T1-2 Gastric Cancer. BMC Med. Imaging 2021, 21, 58. [Google Scholar] [CrossRef] [PubMed]
- Starmans, M.P.A.; Buisman, F.E.; Renckens, M.; Willemssen, F.E.J.A.; van der Voort, S.R.; Koerkamp, B.G.; Grünhagen, D.J.; Niessen, W.J.; Vermeulen, P.B.; Verhoef, C.; et al. Distinguishing Pure Histopathological Growth Patterns of Colorectal Liver Metastases on CT Using Deep Learning and Radiomics: A Pilot Study. Clin. Exp. Metastasis 2021, 38, 483–494. [Google Scholar] [CrossRef]
- Cancian, P.; Cortese, N.; Donadon, M.; Di Maio, M.; Soldani, C.; Marchesi, F.; Savevski, V.; Santambrogio, M.D.; Cerina, L.; Laino, M.E.; et al. Development of a Deep-Learning Pipeline to Recognize and Characterize Macrophages in ColoRectal Liver Metastasis. Cancers 2021, 13, 3313. [Google Scholar] [CrossRef]
- Chuang, W.-Y.; Chen, C.-C.; Yu, W.-H.; Yeh, C.-J.; Chang, S.-H.; Ueng, S.-H.; Wang, T.-H.; Hsueh, C.; Kuo, C.-F.; Yeh, C.-Y. Identification of Nodal Micrometastasis in Colorectal Cancer Using Deep Learning on Annotation-Free Whole-Slide Images. Mod. Pathol. 2021, 34, 1901–1911. [Google Scholar] [CrossRef]
- Zhong, Y.-W.; Jiang, Y.; Dong, S.; Wu, W.-J.; Wang, L.-X.; Zhang, J.; Huang, M.-W. Tumor Radiomics Signature for Artificial Neural Network Assisted Detection of Neck Metastasis in Patient with Tongue Cancer. J. Neuroradiol. 2022, 49, 213–218. [Google Scholar] [CrossRef]
- Ariji, Y.; Fukuda, M.; Kise, Y.; Nozawa, M.; Yanashita, Y.; Fujita, H.; Katsumata, A.; Ariji, E. Contrast Enhanced Computed Tomography Image Assessment of Cervical Lymph Node Metastasis in Patients with Oral Cancer by Using a Deep Learning System of Artificial Intelligence. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 127, 458–463. [Google Scholar] [CrossRef]
- Lee, S.; Choe, E.K.; Kim, S.Y.; Kim, H.S.; Park, K.J.; Kim, D. Liver Imaging Features by Convolutional Neural Network to Predict the Metachronous Liver Metastasis in Stage I-III Colorectal Cancer Patients Based on Preoperative Abdominal CT Scan. BMC Bioinform. 2020, 21, 382. [Google Scholar] [CrossRef] [PubMed]
- Azar, A.S.; Ghafari, A.; Najar, M.O.; Rikan, S.B.; Ghafari, R.; Khamene, M.F.; Sheikhzadeh, P. Covidense: Providing a Suitable Solution for Diagnosing COVID-19 Lung Infection Based on Deep Learning from Chest X-Ray Images of Patients. Front. Biomed. Technol. 2021, 8, 131–142. [Google Scholar]
- Moons, K.G.M.; de Groot, J.A.H.; Bouwmeester, W.; Vergouwe, Y.; Mallett, S.; Altman, D.G.; Reitsma, J.B.; Collins, G.S. Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies: The CHARMS Checklist. PLoS Med. 2014, 11, e1001744. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.F.; Moons, K.G.; Riley, R.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S.; for the PROBAST Group. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef]
- Feng, Q.-X.; Liu, C.; Qi, L.; Sun, S.-W.; Song, Y.; Yang, G.; Zhang, Y.-D.; Liu, X.-S. An Intelligent Clinical Decision Support System for Preoperative Prediction of Lymph Node Metastasis in Gastric Cancer. J. Am. Coll. Radiol. 2019, 16, 952–960. [Google Scholar] [CrossRef]
- Zhang, X.-P.; Wang, Z.-L.; Tang, L.; Sun, Y.-S.; Cao, K.; Gao, Y. Support Vector Machine Model for Diagnosis of Lymph Node Metastasis in Gastric Cancer with Multidetector Computed Tomography: A Preliminary Study. BMC Cancer 2011, 11, 10. [Google Scholar] [CrossRef]
- Tomita, H.; Yamashiro, T.; Heianna, J.; Nakasone, T.; Kimura, Y.; Mimura, H.; Murayama, S. Nodal Based Radiomics Analysis for Identifying Cervical Lymph Node Metastasis at Levels I and II in Patients with Oral Squamous Cell Carcinoma Using Contrast Enhanced Computed Tomography. Eur. Radiol. 2021, 31, 7440–7449. [Google Scholar] [CrossRef]
- Taghavi, M.; Trebeschi, S.; Simões, R.; Meek, D.B.; Beckers, R.C.J.; Lambregts, D.M.J.; Verhoef, C.; Houwers, J.B.; van der Heide, U.A.; Beets-Tan, R.G.H.; et al. Machine Learning Based Analysis of CT Radiomics Model for Prediction of Colorectal Metachronous Liver Metastases. Abdom. Radiol. 2021, 46, 249–256. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, D.; Chen, X.; He, D.; Yu, P.; Liu, B.; Wu, B.; Hu, J.; Song, B. Deep Convolu-tional Neural Network Based on Computed Tomography Images for the Preoperative Diagnosis of Occult Peritoneal Metastasis in Advanced Gastric Cancer. Front. Oncol. 2020, 10, 601869. [Google Scholar] [CrossRef]
- Mermod, M.; Jourdan, E.; Gupta, R.; Bongiovanni, M.; Tolstonog, G.; Simon, C.; Clark, J.; Monnier, Y. Development and Validation of a Multivariable Prediction Model for the Identification of Occult Lymph Node Metastasis in Oral Squamous Cell Carcinoma. Head Neck 2020, 42, 1811–1820. [Google Scholar] [CrossRef]
- Schnelldorfer, T.; Ware, M.P.; Liu, L.P.; Sarr, M.G.; Birkett, D.H.; Ruthazer, R. Can We Accurately Identify Peritoneal Metastases Based on Their Appearance? An Assessment of the Current Practice of Intraoperative Gastrointestinal Cancer Staging. Ann. Surg. Oncol. 2019, 26, 1795–1804. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, J.; Xin, B.; Sun, Y.; Feng, D.; Fulham, M.J.; Wang, X.; Song, S. 18F-FDG PET/CT Radiomics for Preoperative Prediction of Lymph Node Metastases and Nodal Staging in Gastric Cancer. Front. Oncol. 2021, 11, 723345. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Kudo, S.-E.; Mori, Y.; Misawa, M.; Kudo, T.; Wakamura, K.; Katagiri, A.; Baba, T.; Hidaka, E.; Ishida, F.; et al. Accuracy of Diagnosing Invasive Colorectal Cancer Using Computer Aided Endocytoscopy. Endoscopy 2017, 49, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Kasai, S.; Shiomi, A.; Kagawa, H.; Hino, H.; Manabe, S.; Yamaoka, Y.; Chen, K.; Nanishi, K.; Kinugasa, Y. The Effectiveness of Machine Learning in Predicting Lateral Lymph Node Metastasis from Lower Rectal Cancer: A Single Center Development and Validation Study. Ann. Gastroenterol. Surg. 2022, 6, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Chen, W.; Yang, B.; Qu, J.; Cheng, Y.; Zhu, Z.; Gao, Y.; Wang, Q.; Liu, Y.; Li, Z.; et al. Prediction of KRAS, NRAS and BRAF Status in Colorectal Cancer Patients with Liver Metastasis Using a Deep Artificial Neural Network Based on Radiomics and Semantic Features. Am. J. Cancer Res. 2020, 10, 4513–4526. [Google Scholar]
- Kang, J.; Choi, Y.J.; Kim, I.-K.; Lee, H.S.; Kim, H.; Baik, S.H.; Kim, N.K.; Lee, K.Y. LASSO-Based Machine Learning Algorithm for Prediction of Lymph Node Metastasis in T1 Colorectal Cancer. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2021, 53, 773–783. [Google Scholar] [CrossRef]
- Mühlberg, A.; Holch, J.W.; Heinemann, V.; Huber, T.; Moltz, J.; Maurus, S.; Jäger, N.; Liu, L.; Froelich, M.F.; Katzmann, A.; et al. The Relevance of CT-Based Geometric and Radiomics Analysis of Whole Liver Tumor Burden to Predict Survival of Patients with Metastatic Colorectal Cancer. Eur. Radiol. 2021, 31, 834–846. [Google Scholar] [CrossRef]
- Mirniaharikandehei, S.; Heidari, M.; Danala, G.; Lakshmivarahan, S.; Zheng, B. Applying a Random Projection Algorithm to Optimize Machine Learning Model for Predicting Peritoneal Metastasis in Gastric Cancer Patients Using CT Images. Comput. Methods Programs Biomed. 2021, 200, 105937. [Google Scholar] [CrossRef]
- Rice, T.W.; Ishwaran, H.; Hofstetter, W.L.; Schipper, P.H.; Kesler, K.A.; Law, S.; Lerut, T.; Denlinger, C.E.; Salo, J.A.; Scott, W.J.; et al. Esophageal Cancer: Associations with (PN+) Lymph Node Metastases. Ann. Surg. 2017, 265, 122–129. [Google Scholar] [CrossRef]
- Chen, W.; Wang, S.; Dong, D.; Gao, X.; Zhou, K.; Li, J.; Lv, B.; Li, H.; Wu, X.; Fang, M.; et al. Evaluation of Lymph Node Metastasis in Advanced Gastric Cancer Using Magnetic Resonance Imaging-Based Radi-omics. Front. Oncol. 2019, 9, 1265. [Google Scholar] [CrossRef]
- Kiritani, S.; Yoshimura, K.; Arita, J.; Kokudo, T.; Hakoda, H.; Tanimoto, M.; Ishizawa, T.; Akamatsu, N.; Kaneko, J.; Takeda, S.; et al. A New Rapid Diagnostic System with Ambient Mass Spectrometry and Machine Learning for Colorectal Liver Metastasis. BMC Cancer 2021, 21, 262. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhou, X.; Tang, X.; Li, S.; Zhang, G. Prediction of Lymph Node Metastasis in Superficial Esophageal Cancer Using a Pattern Recognition Neural Network. Cancer Manag. Res. 2020, 12, 12249–12258. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, Q.; Zhang, C.; Sun, J.; He, K.; Xie, Y.; Zhang, Y.; Fu, Y.; Zhang, H. Multiregional Based Magnetic Resonance Imaging Radiomics Combined with Clinical Data Improves Efficacy in Predicting Lymph Node Metastasis of Rectal Cancer. Front. Oncol. 2021, 10, 585767. [Google Scholar] [CrossRef] [PubMed]
- Maaref, A.; Romero, F.P.; Montagnon, E.; Cerny, M.; Nguyen, B.; Vandenbroucke, F.; Soucy, G.; Turcotte, S.; Tang, A.; Kadoury, S. Predicting the Response to FOLFOX-Based Chemo-therapy Regimen from Untreated Liver Metastases on Baseline CT: A Deep Neural Network Approach. J. Digit. Imaging 2020, 33, 937–945. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, Y.; Ji, M.-H.; Tong, J.; Yang, J.-J.; Xia, H. Predicting Peritoneal Metasta-sis of Gastric Cancer Patients Based on Machine Learning. Cancer Control 2020, 27, 1073274820968900. [Google Scholar] [CrossRef]
- Zhou, T.; Chen, L.; Guo, J.; Zhang, M.; Zhang, Y.; Cao, S.; Lou, F.; Wang, H. MSIFinder: A Python Package for Detecting MSI Status Using Random Forest Classifier. BMC Bioinform. 2021, 22, 185. [Google Scholar] [CrossRef]
- Bur, A.M.; Holcomb, A.; Goodwin, S.; Woodroof, J.; Karadaghy, O.; Shnayder, Y.; Kakarala, K.; Brant, J.; Shew, M. Machine Learning to Predict Occult Nodal Metastasis in Early Oral Squamous Cell Carci-noma. Oral Oncol. 2019, 92, 20–25. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kwak, M.S.; Lee, H.H.; Cha, J.M.; Shin, H.P.; Jeon, J.W.; Yoon, J.Y. Development of a Novel Prognostic Model for Predicting Lymph Node Metastasis in Early Colorectal Cancer: Analysis Based on the Surveillance, Epidemiology, and End Results Database. Front. Oncol. 2021, 11, 614398. [Google Scholar] [CrossRef]
- Ariji, Y.; Sugita, Y.; Nagao, T.; Nakayama, A.; Fukuda, M.; Kise, Y.; Nozawa, M.; Nishiyama, M.; Katumata, A.; Ariji, E. CT Evaluation of Extranodal Extension of Cervical Lymph Node Metastases in Patients with Oral Squamous Cell Carcinoma Using Deep Learning Classification. Oral Radiol. 2020, 36, 148–155. [Google Scholar] [CrossRef]
- Dercle, L.; Lu, L.; Schwartz, L.H.; Qian, M.; Tejpar, S.; Eggleton, P.; Zhao, B.; Piessevaux, H. Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway. JNCI J. Natl. Cancer Inst. 2020, 112, 902–912. [Google Scholar] [CrossRef]
- Kwak, M.S.; Eun, Y.; Lee, J.; Lee, Y.C. Development of a Machine Learning Model for the Prediction of Nodal Metastasis in Early T Classification Oral Squamous Cell Carcinoma: SEER-based Population Study. Head Neck 2021, 43, 2316–2324. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, M.; Staal, F.; Munoz, F.G.; Imani, F.; Meek, D.B.; Simões, R.; Klompenhouwer, L.G.; van der Heide, U.A.; Beets-Tan, R.G.H.; Maas, M. CT-Based Radiomics Analysis before Thermal Ablation to Predict Local Tumor Progression for Colorectal Liver Metastases. Cardiovasc. Interv. Radiol. 2021, 44, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Chiang, S.-F.; Sahoo, P.K.; Mohapatra, S.K.; You, J.-F.; Onthoni, D.D.; Hung, H.-Y.; Chiang, J.-M.; Huang, Y.; Tsai, W.-S.; et al. Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers 2019, 11, 2007. [Google Scholar] [CrossRef] [PubMed]
- Sitnik, D.; Aralica, G.; Hadžija, M.; Hadžija, M.P.; Pačić, A.; Periša, M.M.; Manojlović, L.; Krstanac, K.; Plavetić, A.; Kopriva, I. A Dataset and a Methodology for Intraoperative Computer-Aided Diagnosis of a Metastatic Colon Cancer in a Liver. Biomed. Signal Process. Control 2021, 66, 102402. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Y.; Wang, P.; Zhao, H.; Wang, X.; Tang, N.; Luan, K. Deep Transfer Learning Based on Magnetic Resonance Imaging Can Improve the Diagnosis of Lymph Node Metastasis in Patients with Rectal Cancer. Quant. Imaging Med. Surg. 2021, 11, 2477–2485. [Google Scholar] [CrossRef]
- Li, J.; Wang, P.; Zhou, Y.; Liang, H.; Luan, K. Different Machine Learning and Deep Learning Methods for the Classification of Colorectal Cancer Lymph Node Metastasis Images. Front. Bioeng. Biotechnol. 2021, 8, 620257. [Google Scholar] [CrossRef]
- Shuwen, H.; Xi, Y.; Qing, Z.; Jing, Z.; Wei, W. Predicting Biomarkers from Classifier for Liver Metastasis of Colorectal Adenocarcinomas Using Machine Learning Models. Cancer Med. 2020, 9, 6667–6678. [Google Scholar] [CrossRef]
- Takamatsu, M.; Yamamoto, N.; Kawachi, H.; Chino, A.; Saito, S.; Ueno, M.; Ishikawa, Y.; Takazawa, Y.; Takeuchi, K. Prediction of Early Colorectal Cancer Metastasis by Machine Learning Using Digi-tal Slide Images. Comput. Methods Programs Biomed. 2019, 178, 155–161. [Google Scholar] [CrossRef]
- Mao, B.; Ma, J.; Duan, S.; Xia, Y.; Tao, Y.; Zhang, L. Preoperative Classification of Primary and Metastatic Liver Cancer via Machine Learning Based Ultrasound Radiomics. Eur. Radiol. 2021, 31, 4576–4586. [Google Scholar] [CrossRef]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Song, Y.-Y.; Lu, Y. Decision Tree Methods: Applications for Classification and Prediction. Shanghai Arch. Psychiatry 2015, 27, 130–135. [Google Scholar]
- Breiman, L. Classification and Regression Trees; Routledge: New York, NY, USA, 2017. [Google Scholar]
- Kramer, O. Dimensionality Reduction with Unsupervised Nearest Neighbors; Springer: Berlin/Heidelberg, Germany, 2013; Volume 51. [Google Scholar]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Natekin, A.; Knoll, A. Gradient Boosting Machines, a Tutorial. Front. Neurorobot. 2013, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-Art in Artificial Neural Network Applications: A Survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Zhao, P.; Yu, B. On Model Selection Consistency of Lasso. J. Mach. Learn. Res. 2006, 7, 2541–2563. [Google Scholar]
- Rikan, S.B.; Azar, A.S.; Naemi, A.; Mohasefi, J.B.; Pirnejad, H.; Wiil, U.K. Survival Prediction of Glioblastoma Patients Using Modern Deep Learning and Machine Learning Techniques. Sci. Rep. 2024, 14, 2371. [Google Scholar]
- Rainio, O.; Teuho, J.; Klén, R. Evaluation Metrics and Statistical Tests for Machine Learning. Sci. Rep. 2024, 14, 6086. [Google Scholar] [CrossRef]
- Naemi, A.; Schmidt, T.; Mansourvar, M.; Ebrahimi, A.; Wiil, U.K. Quantifying the Impact of Ad-dressing Data Challenges in Prediction of Length of Stay. BMC Med. Inform. Decis. Mak. 2021, 21, 298. [Google Scholar] [CrossRef]
- Naemi, A.; Schmidt, T.; Mansourvar, M.; Naghavi-Behzad, M.; Ebrahimi, A.; Wiil, U.K. Machine Learning Techniques for Mortality Prediction in Emergency Departments: A Systematic Review. BMJ Open 2021, 11, e052663. [Google Scholar] [CrossRef]
- Xie, H.; Jia, Y.; Liu, S. Integration of Artificial Intelligence in Clinical Laboratory Medicine: Advance-ments and Challenges. Interdiscip. Med. 2024, 2, e20230056. [Google Scholar] [CrossRef]
- Ireland, P.A. Resource Review. J. Am. Assoc. Med. Transcr. 2007, 26, 37–38. [Google Scholar] [CrossRef]
| Q1 | What AI techniques have been used in different applications of metastatic gastrointestinal cancers? |
| Q2 | What are the common clinical features used in studies? |
| Q3 | What are the common preprocessing steps for AI models’ development? |
| Q4 | What are the methodology settings? |
| Q5 | What are the challenges and research gaps in this domain? |
| G1—AI keywords | Artificial Intelligence, machine learning, learning algorithms, deep learning, unsupervised machine learning, supervised learning |
| G2—Medical keywords | Gastrointestinal Neoplasms, Digestive System Neoplasms, Esophageal Neoplasms, Stomach Neoplasms, Colorectal Neoplasms, Liver Neoplasms, Rectal Neoplasms, Biliary Tract Neoplasms, Pancreatic Neoplasms, Peritoneal Neoplasms, Cancer, Metastasis, Neoplasm Metastasis |
| G3—Document type | Journal |
| G4—Publication year | 1 January 2010–1 January 2022 |
| G5—Final result | G1 AND G2 AND G3 AND G4 |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Cohort should be metastasis gastrointestinal cancer patients. | Studies with traditional statistical models. |
| Developing AI techniques for metastatic gastrointestinal cancer should be the main aim. | Not journal articles. |
| Articles should be journal publications in English. | Not English language publications. |
| Studies with metastatic gastrointestinal cancer patients as a subgroup of the cohort. | |
| The main focus of the study is not an explicit application for metastatic gastrointestinal cancer patients. |
| Number of Patients | Number of Studies |
|---|---|
| <100 | 9 |
| 101–200 | 13 |
| 201–300 | 3 |
| 301–400 | 5 |
| 401–500 | 1 |
| 501–600 | 1 |
| 601–700 | 1 |
| 701–800 | 2 |
| 1001–2000 | 4 |
| 2001–4000 | 3 |
| >4001 | 4 |
| ID | Authors | Year | Country | Study Type | Outcome | Age (Year) Mean ± Std or Median | Male (%) | |
|---|---|---|---|---|---|---|---|---|
| Population | Patient Type | |||||||
| A1 | An et al. [2] | 2022 | China | Retrospective | 148 | PDA | 59.20 ± 10.60 | 49.32 |
| A2 | Tomita et al. [17] | 2021 | Japan | Retrospective | 201 lymph nodes in 23 OSCC cases | OSCC | 52.00 ± 8.00 | 56.52 |
| A3 | Feng et al. [15] | 2019 | China | Retrospective | 490 | GC | 61.80 ± 10.40 | 74.08 |
| A4 | Taghavi et al. [18] | 2020 | The Netherlands | Retrospective | 91 | CC | 64.00 ± 11.00 | 60.43 |
| A5 | Huang et al. [19] | 2020 | China | Retrospective | 544 | AGC | 60.0 (median) | 65.99 |
| A6 | Dong et al. [1] | 2020 | China, Italy | Retrospective | 730 | LAGC | 74.22 ± 13.95 | 64.52 |
| A7 | Mermod et al. [20] | 2020 | Switzerland, Australia | Retrospective | 168 | Early-stage OSCC | 62.09 | 62.50 |
| A8 | Schnelldorfe et al. [21] | 2019 | USA | Prospective | 35 | GC | 67.00 | 65.71 |
| A9 | Liu et al. [22] | 2021 | China | Retrospective | 185 | GC | 62.00 | 68.60 |
| A10 | Takeda et al. [23] | 2017 | Japan | Retrospective | 242 | Non-neoplasms, adenomas, and invasive cancers | 64.75 ± 11.45 | 61.98 |
| A11 | Kasai et al. [24] | 2021 | Japan | Prospective | 323 | Primary RC surgery with LLND | 65.00 | NA |
| A12 | Wang et al. [5] | 2021 | China | Retrospective | 159 | T1-2 GC | 61.78 ± 10.47 | 71.06 |
| A13 | Zhang et al. [16] | 2011 | China | Retrospective | 175 | GC | 59.80 | 71.42 |
| A14 | Shi et al. [25] | 2020 | China | Retrospective | 159 | CRLM | NA | 61.00 |
| A15 | Chuang et al. [8] | 2021 | Taiwan | Retrospective | 1051 | CC | NA | NA |
| A16 | Kang et al. [26] | 2021 | Republic of Korea | Retrospective | 316 | AGC and T1 CRC | NA | 57.59 |
| A17 | Mühlberg et al. [27] | 2021 | Germany | Retrospective | 103 | CRLM | 61.00 ± 11.20 | 53.40 |
| A18 | Mirniaharikandehei et al. [28] | 2021 | USA | Retrospective | 159 | With and without PM | Cases with PM = 59.49 ± 11.97 Cases without PM = 59.11 ± 8.75 | Cases with PM = 59.10 Cases without PM = 18.80 |
| A19 | Rice et al. [29] | 2017 | USA, China, Finland, Canada, Spain | Retrospective | 5806 | Esophagectomy alone | 63.00 ± 11.00 | 77.00 |
| A20 | Chen et al. [30] | 2019 | China | Retrospective | 146 | AGC | 64.94 ± 11.11 | Center 1: 76.05 Center: 80.85 |
| A21 | Cancian et al. [7] | 2021 | Italy, UK | Retrospective | 303 | CLRM | NA | NA |
| A22 | Yang et al. [4] | 2019 | China | Retrospective | 100 | ECC | 57.10 ± 10.00 | 54.00 |
| A23 | Kiritani et al. [31] | 2021 | Japan | Retrospective and prospective | 183 | With and without CRLM | 68.00 | 59.20 |
| A24 | Chen et al. [32] | 2020 | China | Retrospective | 733 | Superficial esophageal squamous cell carcinoma (SESCC) | 62.80 | 69.98 |
| A25 | Liu et al. [33] | 2021 | China | Retrospective | 186 | Rectal adenocarcinoma | 59.22 ± 5.72 | 68.81 |
| A26 | Maaref et al. [34] | 2020 | Canada | Retrospective | 202 | CRLM | NA | NA |
| A27 | Zhou et al. [35] | 2020 | China | Retrospective | 1080 | GC with CT | 63.7 ± 11.65 | 77.68 |
| A28 | Starmans et al. [6] | 2021 | The Netherlands, Belgium | Retrospective | 76 | Pure HGPs | 68.00 | 57.89 |
| A29 | Zhou et al. [36] | 2021 | China | Retrospective | 30 | Metastatic solid tumors | NA | NA |
| A30 | Bur et al. [37] | 2019 | USA | Retrospective | 2032 | Clinically node negative OCSCC | NCDB: 61.90, single institution: 58.10 | NA |
| A31 | Ahn et al. [38] | 2021 | Republic of Korea | Retrospective | 26,733 | Early CRC (T1) | NA | 52.81 |
| A32 | Ariji et al. [39] | 2020 | Japan | Retrospective | 51 | CLNM from OCSCC | 64.00 | 52.94 |
| A33 | Ariji et al. [10] | 2018 | Japan | Retrospective | 45 | OCSCC | 63.00 | 53.33 |
| A34 | Dercle et al. [40] | 2020 | USA, France, Belgium, Germany, | Retrospective | 667 | Liver metastatic CRC | NA | 64.72 |
| A35 | Kwak et al. [41] | 2021 | Republic of Korea | Retrospective | 16,878 | Gastric metastasis | metastasis: 64.60 ± 14.40, non-metastasis: 62.60 ± 13.70 | metastasis: 58.60, non-metastasis: 61.40 |
| A36 | Taghavi et al. [42] | 2021 | The Netherlands, Spain | Retrospective | 90 | Colorectal liver metastases | 62.00 ± 11.00 | 57.77 |
| A37 | Gupta et al. [43] | 2019 | Taiwan | Retrospective | 4021 | CRC | NA | 56.93 |
| A38 | Zhong et al. [9] | 2021 | China, Germany | Retrospective | 313 | SCC | 55.07 ± 12.46 | 60.38 |
| A39 | Sitnik et al. [44] | 2021 | Croatia | Retrospective | 19 | Metastatic colon cancer | NA | NA |
| A40 | Li et al. [45] | 2021 | China | Retrospective | 129 | Rectal cancer | 58.40 ± 10.27 | 64.34 |
| A41 | Li et al. [3] | 2019 | China, USA | Retrospective | 48 | Liver metastasis (LM) in colon cancer (CC) | 61.52 ± 12.53 | 62.50 |
| A42 | Li et al. [46] | 2020 | China | Retrospective | 3364 | CRC | NA | NA |
| A43 | Shuwen et al. [47] | 2020 | China | Retrospective | 1186 | CAD | NA | NA |
| A44 | Takamatsu et al. [48] | 2019 | Japan | Retrospective | 397 | CRC | Training: 61.30 ± 11.45 Test: NA | Training: 51.51 |
| A45 | Mao et al. [49] | 2021 | China | Retrospective | 114 | Metastatic liver cancer | 59.10 | 61.40% |
| A46 | Lee et al. [11] | 2020 | Republic of Korea, USA | Retrospective | 2019 | CRC | 62.70 ± 9.35 | 62.85 |
| Id | AI Algorithm | Evaluation Metrics | Handling Missing Values | Hyperparameter Optimization | Approach | Validation |
|---|---|---|---|---|---|---|
| A1 | LR, SVM, Resnet 18 | AUC = 0.92, Confidence, Accuracy = 0.86, Sensitivity = 0.92, Specificity = 0.78, PPV = 0.8, NPV = 0.93 | No | NA | Classification | Internal |
| A2 | SVM | p-value = 0.05, AUC = (0.820 at level I/II, 0.820 at level I, and 0.930 at level II), Cutoff, Accuracy, Sensitivity, Specificity | No | SVM-RBF | Classification | Internal |
| A3 | SVM | AUC = (0.699–0.833), Sensitivity, Specificity, PPV, NPV, Accuracy = 71.3%, Cutoff Value | No | 10-fold cross-validation, Monte Carlo cross-validation (200 repeats), RBF | Classification | Internal |
| A4 | RF | AUC = 71% and 86%, F1-score, CI = (69–72%, 85–87%) | No | Bayesian hyperparameter optimization | Classification | Internal |
| A5 | DCNN | Sensitivity = 81%, AUC = 0.670, 95% CI: 0.615–0.739; p < 0.001, Specificity = 87.5% | No | NA | Forecasting | Internal |
| A6 | DLRN | C-index = 0.821, confusion matrix | No | NA | Forecasting | Internal and external |
| A7 | RF, Lasso LR, SVM, C5.0 | Sensitivity, Specificity, NPV, PPV, Accuracy = 0.88, AUC = 0.89 | No | NA | Classification | External |
| A8 | DNN | Sensitivity, Specificity, PPV, NPV, Accuracy, AUC = 0.47 | No | NA | Classification | Internal |
| A9 | Balanced Bagging Ensemble Classifier | Accuracy = 0.852, AUC = 0.822, Sensitivity = 0.733, Specificity = 0.891, PPV = 0.688, NPV = 0.911 | No | NA | Classification | Internal |
| A10 | SVM | Sensitivity = 89.4%, Specificity = 98.9%, Accuracy = 94.1%, PPV = 98.8%, NPV = 90.1% | No | NA | Classification | Internal |
| A11 | Prediction One (Sony Network Communications) Software | AUC = 0.754, U-test = 0.022, Accuracy = 80.4%, Sensitivity = 90.0%, Specificity = 79.4%, PPV, NPV, p-value = 0.022 | No | NA | Classification | Internal |
| A12 | LR | Confusion matrix, Accuracy = 0.899, Sensitivity = 0.882, Specificity = 0.903, PPV = 0.714, NPV = 0.966, AUC = 0.908, p-value | No | NA | Forecasting | Internal |
| A13 | SVM | T-test, U-test, AUC, AUC = 0.876, Sensitivity = 88.5%, Specificity = 78.5%, p-value = 0.002, p-value < 0.001 | No | 5-fold cross-validation | Classification | Internal |
| A14 | ANN, KNN, SVM, Bayes, LR, AdaBoost, GB | AUC = 0.95, Accuracy = 87.10%, Sensitivity = 89.19%, Specificity = 84.00%, PPV = 89.19%, NPV = 84.00%, p-value | No | NA | Classification | Internal |
| A15 | ResNet-50 | AUC = 0.9724, Accuracy = 98.50% | No | NA | Classification | Internal |
| A16 | Lasso regression | AUROC (0.765 vs. 0.518, p = 0.003), NRI (0.447, p = 0.039)/IDI (0.121, p = 0.034), DCA, p-value | No | log (λ), where λ is a tuning hyperparameter, cross-validation | Classification | Internal |
| A17 | LR, RF | AUC = 0.70, Z-value, p-value, Odds ratio, C-index = [0.56, 0.90] | No | 10-fold cross-validation | Classification | Internal |
| A18 | DT, RF, SVM, LR, GBM | Precision = 65.78%, Sensitivity = 43.10%, Specificity = 87.12%, Accuracy = 71.2%, AUC = 0.69 ± 0.019 | SMOTE | Cross-validation | Classification | Internal |
| A19 | RF | Minimal depth, variable importance, probability | No | NA | Classification | Internal |
| A20 | LASSO, LR, and Learning Vector Quantization (LVQ) | U-test, p-value, AUC = 0.657, Accuracy = 0.745, Sensitivity = 0.853, Specificity = 0.462, confusion matrix | No | Cross-validation | Classification and Regression | Internal and external |
| A21 | UNet, SegNet, DeepLab-v3 | IoU mean = 89.13, IoU StDev = 3.85, SBD mean = 79.00, SBD StDev = 3.72 | No | NA | Classification | Internal |
| A22 | RF | AUC = 0.80 and 0.90 | SMOTE | NA | Classification | Internal |
| A23 | LR | Specificity = 100%, Sensitivity = 99%, Accuracy = 99.5%, p-value | No | 10-fold cross-validation | Classification | Internal |
| A24 | LR, ANN | Specificity = 91.20%, Sensitivity = 87.06%, Accuracy = 90.72%, p-value, AUC = 0.915, PPV = 56.49%, NPV = 98.17%, NRI = −1.1%, IDI = 23.3% | No | Cross-validation | Classification | Internal |
| A25 | SVM | AUC = 0.827, p-value, Sensitivity = 0.815, Specificity = 0.694, PPV = 0.667, NPV = 0.833 | No | NA | Classification | Internal |
| A26 | DCNN | AUC, Sensitivity, Specificity, Accuracy = 91%,78% | No | NA | Classification | Internal |
| A27 | Light Gradient Boosting Machine, GradientBoosting, RF, Logistic, and DT | AUC = 0.745, Accuracy = 0.907, MSE = 0.093 | No | Tuning parameter for each model | Classification | Internal |
| A28 | CNN, LR, SVM, RF, Quadratic Discriminant Analysis, AdaBoost, Extreme Gradient Boosting | AUC = 0.72, Accuracy = 0.65, Sensitivity = 0.62, Specificity = 0.68 | No | Cross-validation | Classification | Internal |
| A29 | RFC and SVM | AUC = 0.999, Sensitivity = 1, Specificity = 0.997, Accuracy = 0.998, PPV = 0.954, F1 = 0.977 | No | Cross-validation | Classification | Internal |
| A30 | LR, RF, SVM, GB | Specificity, Sensitivity, AUC = 0.840, p-value | No | Cross-validation | Classification | Internal and external |
| A31 | LR, XGB, KNN, CARTs, SVM, NN, RF | AUC = 0.991, Accuracy = 0.960, Sensitivity = 0.997, Specificity = 0.929, Precision (PPV) = 0.919, NPV = 0.998, FDR = 0.081, AP = 0.995, F1-score = 0.956, MCC = 0.922 | Random oversampling | Cross-validation | Classification | Internal |
| A32 | AlexNet | AUC, Accuracy = 84.0%, Sensitivity, Specificity, PPV, NPV | No | NA | Classification | Internal |
| A33 | CNN | Accuracy = 78.2%, Sensitivity = 75.4%, Specificity = 81.0%, PPV = 79.9%, NPV = 77.1%, AUC = 0.80 | No | Cross-validation | Classification | Internal and external |
| A34 | Deep learning, RF | AUC = 0.80 | No | NA | Classification | Internal |
| A35 | CART, KNN, LR, RF, SVM, XGB | AUC = 0.956, Sensitivity, Specificity, AP, F1-score, MCC | No | Cross-validation | Classification | Internal |
| A36 | Three machine learning survival models | C-index = 0.77–0.79, p-value | No | Bayesian hyperparameter optimization, cross-validation | Classification | Internal and external |
| A37 | RF, SVM, LR, MLP, KNN, AdaBoost | Accuracy = 0.89, Precision = 0.89, Recall = 0.88, F-measure = 0.89, AUC = 0.94 | No | Scikit-Optimize, Cross-validation | Classification | Internal |
| A38 | ANN | Accuracy = 84.1%, Sensitivity = 93.1%, Specificity = 76.5%, AUC = 0.943, net reclassification index (NRI) = 40% | No | NA | Classification | Internal |
| A39 | SVM, KNN, U-Net, U-Net++, DeepLabv3 | F1-score = 83.67%, Accuracy = 89.34%, TPR, TNR, BACC, PPV = 81.11% | No | Cross-validation | Classification | Internal |
| A40 | Inception-v3 | Accuracy = 95.7%, PPV = 95.2%, NPV = 95.3%, Sensitivity = 95.3%, Specificity = 95.2%, AUC = 0.994, confusion matrix, p-value > 0.05 | No | NA | Classification | Internal |
| A41 | SVM | Accuracy = 69.50%, Specificity = 83.14%, Sensitivity = 62.00%, area under the curve (AUC) = 0.69 | No | Cross-validation | Classification | Internal |
| A42 | AB, MLP, LeNet, DT, NB, AlexNet, KNN, SGD, AlexNet Pre-trained, LR, SVM | Accuracy = 0.7583, AUC = 0.7941, Sensitivity = 0.8004, Specificity = 0.7997, PPV = 0.7992, NPV = 0.8009 | No | freezing and fine-tuning parameters of CNN models | Classification | Internal |
| A43 | LR, NN, SVM, RF, GBDT, Catboost | Accuracy = 1, AUC = 1 | No | Cross-validation | Classification | Internal and external |
| A44 | RF, LR | AUC = 0.94 | No | Cross-validation | Classification | Internal |
| A45 | KNN, SVM, RF, LR, MLP | AUC = 0.816 ± 0.088, Accuracy = 0.843 ± 0.078, F1-score, Specificity = 0.880 ± 0.117, Sensitivity = 0.768 ± 0.232, Precision | No | Cross-validation | Classification | Internal |
| A46 | VGG16, Logistic Regression, Random Forest | AUC = 0.747 ± 0.036 | SMOTE | Cross-validation | Classification | Internal |
| Algorithm | Description | Pros | Cons |
|---|---|---|---|
| LR [50] | LR is a supervised ML algorithm adopted from linear regression. It can be used for classification problems and finding the probability of an event happening. | Fast training, good for small datasets, and easy to understand. | Not very accurate, not proper for non-linear problems, high chance of overfitting, and not flexible enough to adopt to complex datasets. |
| DT [51] | DT is a supervised ML algorithm that solves a problem by transforming the data into a tree representation where each internal node represents an attribute and each leaf denotes a class label. CART is also a decision tree algorithm used for both classification and regression tasks. It is a supervised learning algorithm that learns from labeled data to predict unseen data. | Easy to understand and interpret, robust to outliers, no standardization or normalization required, and useful for regression and classification. | High risk of overfitting, not suitable for large datasets, and adding new samples leads to the regeneration of the whole tree. |
| CART [52] | CART is used for classification and regression by splitting the data into subsets to achieve the highest information gain or lowest variance. | Easy to understand and interpret, robust to outliers, no standardization or normalization required, and useful for regression and classification. | High risk of overfitting, not suitable for small datasets; adding new samples leads to the regeneration of the whole model. |
| KNN [53] | KNN is a supervised and instance-based ML algorithm. It can be used when we want to forecast a label of a new sample based on similar samples with known labels. Different similarity or distance measures such as Euclidean can be used. | Simple and easy to understand, easy to implement, no need for training, and useful for regression and classification. | Memory-intensive, costly, slow performance, and all training data might be involved in decision-making. |
| SVM [54] | SVM is an instance-based and supervised ML technique that generates a boundary between classes known as a hyperplane. Maximizing the margin between classes is the main goal of this technique. | Efficient in high-dimensional spaces. Effective when the number of dimensions exceeds the number of samples, useful for regression and classification, regularization capabilities that prevent overfitting, and handling non-linear data. | Not suitable for large datasets, not suitable for noisy datasets, regularization capabilities that prevent overfitting, handling non-linear data, and long training time. |
| GB [55] | GB is a supervised ML algorithm, which produces a model in the form of an ensemble of weak prediction models, usually DT. GB is an iterative gradient technique that minimizes a loss function by iteratively selecting a function that points toward the negative gradient. | High accuracy, high flexibility, fast execution, and useful for regression and classification, robust to missing values and overfitting. | Sensitive to outliers, not suitable for small datasets, and many parameters to optimize. |
| RF [56] | RF is an ensemble and supervised ML algorithm that is based on the bagging technique, which means that many subsets of data are randomly selected with replacements and each model such as DT is trained using one subset. The output is the average of all predictions of various single models. | High accuracy, fast execution, useful for regression and classification, and robust to missing values and overfitting. | Not suitable for limited datasets; may change considerably by a small change in the data. |
| ANN [57] | ANN is a family of supervised ML algorithms. It is inspired by the biological neural network of the human brain. ANN consists of input, hidden, and output layers and multiple neurons (nodes) carry data from the input layer to the output layer. | Accurate; suitable for complex non-linear classification and regression problems. | Very slow to train and test, large amounts of essential data, computationally expensive, and prone to overfitting. |
| DNN [58] | DNN is a family of supervised ML algorithms. DNN is based on NNs where the adjective ’deep’ comes from the use of multiple layers in the network. Usually having two or more hidden layers counts as a DNN. Some specific training algorithms and architectures exist, such as LSTM, GAN, and CNN for DNNs. DNNs provide the opportunity to solve complex problems when the data are very diverse, unstructured, and interconnected. | High accuracy, features are automatically deduced and optimally tuned, robust to noise, and architecture is flexible. | Needs a very large amount of data, computationally expensive, not easy to understand, no standard theory in selecting the right settings, and difficult for less skilled researchers. |
| Lasso [59] | LASSO is a regularization technique used in statistical modeling and machine learning for estimating the relationships between variables and making predictions. | Simplicity, feature selection, and robustness. | Introduces bias into the estimates; low performance when the number of observations is less than the number of features or there is high multicollinearity among the features. |
| Metric | Description |
|---|---|
| Accuracy [61] | Accuracy is a general metric that quantifies the proportion of correctly classified instances (both positive and negative) out of the total instances in the dataset. While Accuracy provides a simple overall performance measure, it may be misleading for imbalanced datasets, as it does not differentiate between the types of errors (false positives vs. false negatives). |
| Precision [61] | Precision evaluates the proportion of correctly predicted positive cases out of all cases predicted as positive by the model. High Precision indicates that the model is accurate in its positive predictions, minimizing false positives. Precision is especially relevant in scenarios where the cost of false positives is high. |
| Sensitivity [61] | Sensitivity, also known as the true positive rate, measures the model’s ability to correctly identify positive cases out of all actual positive cases in the dataset. A high Sensitivity value indicates that the model effectively identifies most of the true positive cases, making it particularly important in applications where minimizing false negatives is critical, such as in disease diagnosis. |
| Specificity [61] | Specificity measures the model’s ability to correctly identify negative cases out of all actual negative cases. A high Specificity value signifies that the model can accurately exclude non-relevant cases, reducing the occurrence of false positives. Specificity is critical in contexts where false positives may lead to unnecessary interventions. |
| F1-Score [61] | The F1-score is the harmonic mean of Precision and Sensitivity, providing a balanced metric that considers both false positives and false negatives. |
| AUC (ROC) [61] | The AUC (ROC) is a widely used metric to evaluate the performance of binary classification models by measuring their ability to distinguish between two classes. The ROC curve plots the true positive rate (Sensitivity) against the false positive rate (1-specificity) at various threshold levels. The AUC quantifies the area under this curve, providing a single scalar value ranging from 0 to 1. A higher AUC indicates better model performance, with 1 representing a perfect classifier and 0.5 reflecting no discriminatory power (equivalent to random guessing). The AUC is particularly useful for imbalanced datasets, as it evaluates the model’s performance across different classification thresholds. |
| Id | Domain 1: Participants | Domain 2: Predictors | Domain 3: Outcome | Domain 4: Analysis |
|---|---|---|---|---|
| A1 | High | Low | Low | High |
| A2 | High | Low | Low | High |
| A3 | High | Low | Low | High |
| A4 | High | Low | Low | High |
| A5 | Low | Low | Low | Unclear |
| A6 | High | Low | Low | Low |
| A7 | High | Low | Low | High |
| A8 | High | Low | Low | High |
| A9 | High | Low | Low | High |
| A10 | High | Low | Low | High |
| A11 | High | Low | Low | High |
| A12 | High | Low | Low | High |
| A13 | High | Low | Low | High |
| A14 | High | Low | Low | High |
| A15 | Low | Low | Low | High |
| A16 | Unclear | Low | Low | High |
| A17 | High | Low | Low | High |
| A18 | High | Low | Low | High |
| A19 | Low | Low | Low | Unclear |
| A20 | High | Low | Low | Unclear |
| A21 | Low | Low | Low | High |
| A22 | High | Low | Low | High |
| A23 | High | Low | Low | High |
| A24 | Low | Low | Low | High |
| A25 | High | Low | Low | High |
| A26 | High | Low | Low | High |
| A27 | Low | Low | Low | Unclear |
| A28 | Low | Low | Low | High |
| A29 | High | Low | Low | High |
| A30 | Low | Low | Low | Low |
| A31 | Low | Low | Low | Unclear |
| A32 | High | Low | Low | High |
| A33 | Low | Low | Low | Low |
| A34 | Low | Low | Low | High |
| A35 | High | Low | Low | High |
| A36 | High | Low | Low | High |
| A37 | Low | Low | Low | Unclear |
| A38 | Unclear | Low | Low | High |
| A39 | Low | Low | Low | High |
| A40 | Unclear | Low | Low | High |
| A41 | High | Low | Low | High |
| A42 | Low | Low | Low | High |
| A43 | High | Low | Low | High |
| A44 | High | Low | Low | High |
| A45 | High | Low | Low | High |
| A46 | Low | Low | Low | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naemi, A.; Tashk, A.; Sorayaie Azar, A.; Samimi, T.; Tavassoli, G.; Bagherzadeh Mohasefi, A.; Nasiri Khanshan, E.; Heshmat Najafabad, M.; Tarighi, V.; Wiil, U.K.; et al. Applications of Artificial Intelligence for Metastatic Gastrointestinal Cancer: A Systematic Literature Review. Cancers 2025, 17, 558. https://doi.org/10.3390/cancers17030558
Naemi A, Tashk A, Sorayaie Azar A, Samimi T, Tavassoli G, Bagherzadeh Mohasefi A, Nasiri Khanshan E, Heshmat Najafabad M, Tarighi V, Wiil UK, et al. Applications of Artificial Intelligence for Metastatic Gastrointestinal Cancer: A Systematic Literature Review. Cancers. 2025; 17(3):558. https://doi.org/10.3390/cancers17030558
Chicago/Turabian StyleNaemi, Amin, Ashkan Tashk, Amir Sorayaie Azar, Tahereh Samimi, Ghanbar Tavassoli, Anita Bagherzadeh Mohasefi, Elaheh Nasiri Khanshan, Mehrdad Heshmat Najafabad, Vafa Tarighi, Uffe Kock Wiil, and et al. 2025. "Applications of Artificial Intelligence for Metastatic Gastrointestinal Cancer: A Systematic Literature Review" Cancers 17, no. 3: 558. https://doi.org/10.3390/cancers17030558
APA StyleNaemi, A., Tashk, A., Sorayaie Azar, A., Samimi, T., Tavassoli, G., Bagherzadeh Mohasefi, A., Nasiri Khanshan, E., Heshmat Najafabad, M., Tarighi, V., Wiil, U. K., Bagherzadeh Mohasefi, J., Pirnejad, H., & Niazkhani, Z. (2025). Applications of Artificial Intelligence for Metastatic Gastrointestinal Cancer: A Systematic Literature Review. Cancers, 17(3), 558. https://doi.org/10.3390/cancers17030558





