Recent Studies on Kaposi’s Sarcoma-Associated Herpesvirus Circular RNAs
Simple Summary
Abstract
1. Introduction
2. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV)
3. Host and Microbial circRNAs Found in Nature
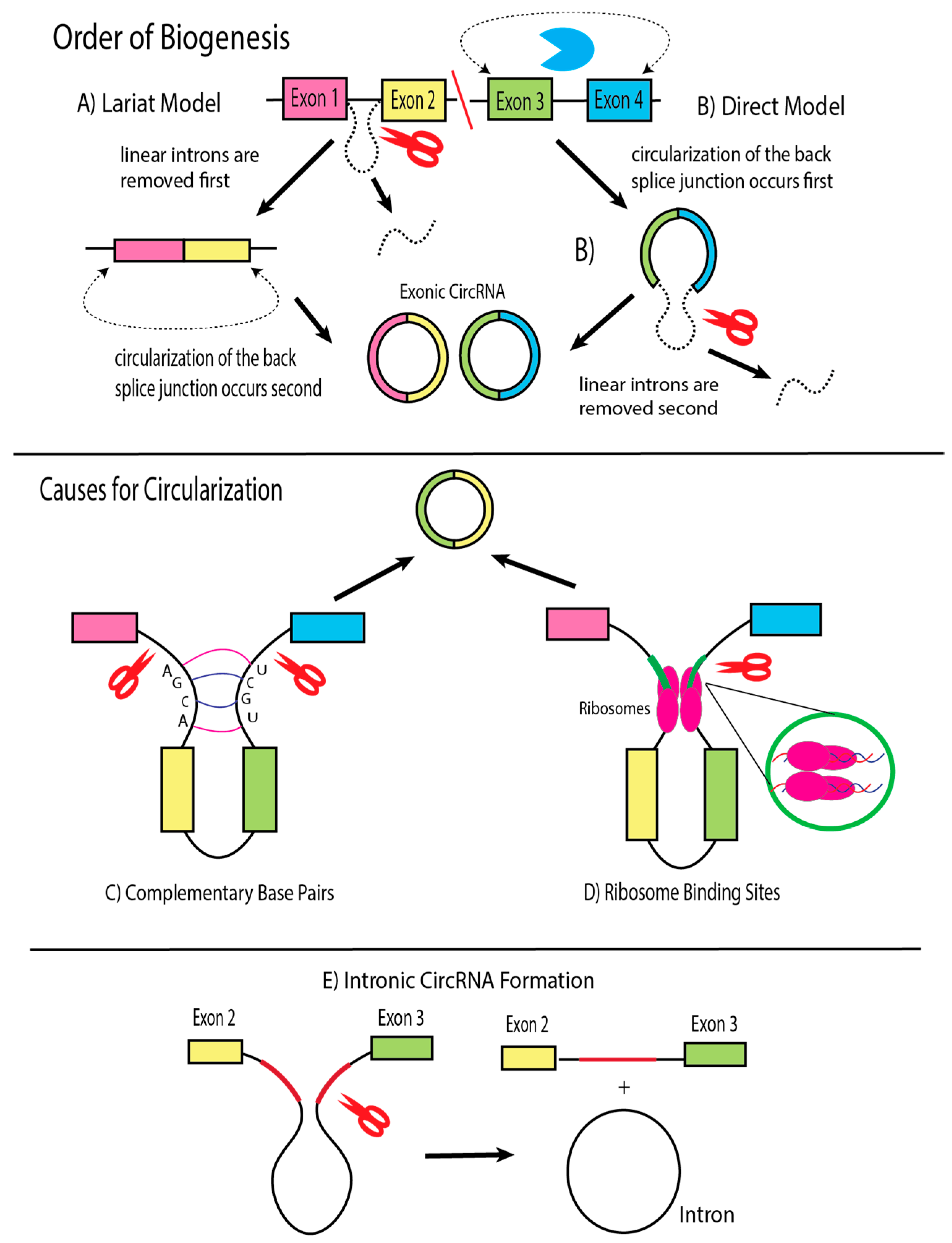
4. Biogenesis of Viral circRNAs and Factors of Influence
5. Identification of KSHV circRNAs and Their Analysis by RNA-Seq Based Strategies
| CircRNA | Location | Localization/Expression Level After Infection | Method of Analysis | KSHV or Human | Potential Functions and Oncogenic Phenotype | Reference |
|---|---|---|---|---|---|---|
| circvIRF4 | K10 locus (vIRF4 gene) | Nucleus, cytoplasm, and detectable in patient sera; high level of abundance | RNA-Seq and ectopic expression | KSHV | Possible relation to oncogenesis | [42] |
| circPANs | PAN/K7.3 locus | High level of abundance | Divergent primers over BSJs BaseScope probing for transcript signals | KSHV | Potential biomarker for initial KSHV lytic activation in cancer cells | [27] |
| circK7.3 | ||||||
| kcirc3 | ORF4, ORF6 | Medium level of abundance | RNA-Seq detection of mapped reads and confirmation of BSJs; Used ectopic forms of circRNA expression from viral plasmids | KSHV | Unknown | [20] |
| kcirc29 | K7/PAN | High level of abundance | KSHV | Unknown | ||
| kcirc38 | ORF21–22 | Medium level of abundance | KSHV | Unknown | ||
| kcirc54 | ORF54 | Low level of abundance | KSHV | Unknown | ||
| kcirc55 | ORF34–36 | KSHV | Potential decoy for defense against antiviral human circRNA | |||
| kcirc57 | ORF34–37 | Medium level of abundance | KSHV | Unknown | ||
| kcirc97 | ORF60–62 | KSHV | Potential decoy for defense against antiviral human circRNA | |||
| hsa_circ_0001400 (circRELL1) | RELL1 | Cytoplasm; upregulated expressioon upon infection | RNA-Seq detection of mapped reads and confirmation of BSJs. | Human | Antiviral circRNA that dysregulates LANA and RTA | [20,46] |
| hsa_circ_0001741 (circTMPO3) | TNPO3 | Unknown localization; upregulated expression upon infection | qRT-PCR analysis on RNA transcripts (Not conducted on KSHV-infected cell lines); RNA fluorescence in situ hybridization (FISH) | Human | miRNA sponge → regulatory roles in ovarian cancer and ESCC | [47] |
| hsa_circ_0005145 | FAM105 | Utilized varying neurological tissues, subsequently transfected and analyzed w/qRT-PCR (Not conducted on KSHV-infected cell lines) | Human | miRNA sponge → regulatory roles in mTLE | [48] | |
| hsa_circ_0001808 (circARFGEF1) | ARFGEF1 | Cytoplasm; upregulated expression upon infection | Comparison of reference in circBase; RT-PCR analysis on amplified regions via divergent primers | Human | miRNA sponge → facilitates pathogenesis of KSHV-related diseases | [41] |
6. KSHV circRNA Transcript Expression Patterns and Potential Functions for Therapeutic Means
6.1. circvIRF4
6.2. circPANs
6.3. K7.3 circRNAs
6.4. ORF-Related circRNAs
6.5. Comparison Between KSHV circRNAs and Existing Biomarkers for KSHV Infections
7. Upregulated Human circRNA Expression upon KSHV Infection
7.1. hsa_circ_0001400
7.2. hsa_circ_0001741
7.3. hsa_circ_0005145
7.4. hsa_circ_0001808
8. Research Regarding Exogenous Expression and Utilizing Knockouts to Study Functionality of KSHV circRNAs
9. Future Challenges and Direction
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Damania, B.; Cesarman, E. Kaposi’s Sarcoma Herpesvirus. In Fields Virology: DNA Viruses, 7th ed.; Knipe, D.M., Howley, P., Eds.; Wolters Kluwer Health, Lippincott and Williams & Wilkins: Philadelphia, PA, USA, 2021; Volume 1, pp. 513–572. [Google Scholar]
- Minhas, V.; Wood, C. Epidemiology and Transmission of Kaposi’s Sarcoma-Associated Herpesvirus. Viruses 2014, 6, 4178–4194. [Google Scholar] [CrossRef] [PubMed]
- Krug, L.T.; Pellett, P.E. The Family Herpesviridae: A Brief Introduction. In Fields Virology: DNA Viruses, 7th ed.; Knipe, D.M., Howley, P., Eds.; Wolters Kluwer Health, Lippincott and Williams & Wilkins: Philadelphia, PA, USA, 2021; Volume 1, pp. 212–234. [Google Scholar]
- Wen, K.W.; Damania, B. Kaposi sarcoma-associated herpesvirus (KSHV): Molecular biology and oncogenesis. Cancer Lett. 2010, 289, 140–150. [Google Scholar] [CrossRef]
- Fatahzadeh, M. Kaposi sarcoma: Review and medical management update. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2012, 113, 2–16. [Google Scholar] [CrossRef]
- Coen, N.; Duraffour, S.; Snoeck, R.; Andrei, G. KSHV Targeted Therapy: An Update on Inhibitors of Viral Lytic Replication. Viruses 2014, 6, 4731–4759. [Google Scholar] [CrossRef]
- Naimo, E.; Zischke, J.; Schulz, T.F. Recent Advances in Developing Treatments of Kaposi’s Sarcoma Herpesvirus-Related Diseases. Viruses 2021, 13, 1797. [Google Scholar] [CrossRef]
- Yan, L.; Majerciak, V.; Zheng, Z.-M.; Lan, K. Towards Better Understanding of KSHV Life Cycle: From Transcription and Posttranscriptional Regulations to Pathogenesis. Virol. Sin. 2019, 34, 135–161. [Google Scholar] [CrossRef]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.-C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef]
- Jha, H.C.; Lu, J.; Verma, S.C.; Banerjee, S.; Mehta, D.; Robertson, E.S. Kaposi’s Sarcoma-Associated Herpesvirus Genome Programming during the Early Stages of Primary Infection of Peripheral Blood Mononuclear Cells. mBio 2014, 5, e02261-14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.X.; Chong, J.M.; Wu, L.; Yuan, Y. Virion Proteins of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2005, 79, 800–811. [Google Scholar] [CrossRef]
- Shamay, M.; Krithivas, A.; Zhang, J.; Hayward, S.D. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. USA 2006, 103, 14554–14559. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Yang, Z.; Jia, R.; Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 2019, 18, 6. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef]
- Toptan, T.; Abere, B.; Nalesnik, M.A.; Swerdlow, S.H.; Ranganathan, S.; Lee, N.; Shair, K.H.; Moore, P.S.; Chang, Y. Circular DNA tumor viruses make circular RNAs. Proc. Natl. Acad. Sci. USA 2018, 115, E8737–E8745. [Google Scholar] [CrossRef]
- Tagawa, T.; Gao, S.; Koparde, V.N.; Gonzalez, M.; Spouge, J.L.; Serquina, A.P.; Lurain, K.; Ramaswami, R.; Uldrick, T.S.; Yarchoan, R.; et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. USA 2018, 115, 12805–12810. [Google Scholar] [CrossRef]
- Yang, L.; Wilusz, J.E.; Chen, L.L. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022, 38, 263–289. [Google Scholar] [CrossRef]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going circular: History, present, and future of circRNAs in cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ RNA s. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Abere, B.; Zhou, H.; Li, J.; Cao, S.; Toptan, T.; Grundhoff, A.; Fischer, N.; Moore, P.S.; Chang, Y. Merkel Cell Polyomavirus Encodes Circular RNAs (circRNAs) Enabling a Dynamic circRNA/microRNA/mRNA Regulatory Network. mBio 2020, 11, e03059-20. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, Y. Biological functions and applications of circRNAs-next generation of RNA-based therapy. J. Mol. Cell Biol. 2023, 15, mjad031. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hetuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Abere, B.; Li, J.; Zhou, H.; Toptan, T.; Moore, P.S.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded circRNAs Are Expressed in Infected Tumor Tissues and Are Incorporated into Virions. mBio 2020, 11, e03027-19. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, B.; Li, F.; Yu, K.; Bai, Y. The mechanism and detection of alternative splicing events in circular RNAs. PeerJ 2020, 8, e10032. [Google Scholar] [CrossRef]
- Karimi, R.; Javandoost, E.; Asadmasjedi, N.; Atashi, A.; Soleimani, A.; Behzadifard, M. Circular RNAs: History, metabolism, mechanisms of function, and regulatory roles at a glance. Ann. Med. Surg. 2025, 87, 141–150. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, Q. Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer. Cancers 2021, 13, 4185. [Google Scholar] [CrossRef]
- Dremel, S.E.; Koparde, V.N.; Arbuckle, J.H.; Hogan, C.H.; Kristie, T.M.; Krug, L.T.; Conrad, N.K.; Ziegelbauer, J.M. Noncanonical circRNA biogenesis driven by alpha and gamma herpesviruses. EMBO J. 2025, 44, 2323–2352. [Google Scholar] [CrossRef]
- Majerciak, V.; Alvarado-Hernandez, B.; Lobanov, A.; Cam, M.; Zheng, Z.-M. Genome-wide regulation of KSHV RNA splicing by viral RNA-binding protein ORF57. PLOS Pathog. 2022, 18, e1010311. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Cai, Z.; Fan, Y.; Zhang, Z.; Lu, C.; Zhu, Z.; Jiang, T.; Shan, T.; Peng, Y. VirusCircBase: A database of virus circular RNAs. Brief. Bioinform. 2021, 22, 2182–2190. [Google Scholar] [CrossRef]
- Majerciak, V.; Ni, T.; Yang, W.; Meng, B.; Zhu, J.; Zheng, Z.-M. A Viral Genome Landscape of RNA Polyadenylation from KSHV Latent to Lytic Infection. PLoS Pathog. 2013, 9, e1003749. [Google Scholar] [CrossRef]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e821. [Google Scholar] [CrossRef]
- Tagawa, T.; Koparde, V.N.; Ziegelbauer, J.M. Identifying and characterizing virus-encoded circular RNAs. Methods 2021, 196, 129–137. [Google Scholar] [CrossRef]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef]
- Ungerleider, N.A.; Jain, V.; Wang, Y.; Maness, N.J.; Blair, R.V.; Alvarez, X.; Midkiff, C.; Kolson, D.; Bai, S.; Roberts, C.; et al. Comparative Analysis of Gammaherpesvirus Circular RNA Repertoires: Conserved and Unique Viral Circular RNAs. J. Virol. 2019, 93, e01952-18. [Google Scholar] [CrossRef]
- Niu, M.; Ju, Y.; Lin, C.; Zou, Q. Characterizing viral circRNAs and their application in identifying circRNAs in viruses. Brief. Bioinform. 2022, 23, bbab404. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Jia, X.; Wang, F.; Sheng, L.; Song, P.; Cao, Y.; Shi, H.; Fan, W.; Ding, X.; Gao, S.-J.; et al. CircRNA ARFGEF1 functions as a ceRNA to promote oncogenic KSHV-encoded viral interferon regulatory factor induction of cell invasion and angiogenesis by upregulating glutaredoxin 3. PLOS Pathog. 2021, 17, e1009294. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.; Jain, V.; Stribling, D.; Gay, L.A.; Naeem, M.; Baddoo, M.; Flemington, E.K.; Tibbetts, S.A.; Renne, R. A viral circular RNA in Kaposi’s sarcoma-associated herpesvirus modulates viral and host gene expression during latent and lytic replication. Explor. Target. Anti-Tumor Ther. 2025, 6, 1002320. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, X.; Wang, M.; Cao, D.; Jaijyan, D.K.; Enescu, N.; Liu, J.; Wu, S.; Wang, S.; Sun, W.; et al. Circular RNAs Represent a Novel Class of Human Cytomegalovirus Transcripts. Microbiol. Spectr. 2022, 10, e01106-22. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.K.; Wang, M.R.; Liu, C.X.; Dong, R.; Carmichael, G.G.; Chen, L.L.; Yang, L. CIRCexplorer3: A CLEAR Pipeline for Direct Comparison of Circular and Linear RNA Expression. Genom. Proteom. Bioinform. 2019, 17, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Dremel, S.E.; Tagawa, T.; Koparde, V.N.; Hernandez-Perez, C.; Arbuckle, J.H.; Kristie, T.M.; Krug, L.T.; Ziegelbauer, J.M. Interferon induced circRNAs escape herpesvirus host shutoff and suppress lytic infection. EMBO Rep. 2024, 25, 1541–1569. [Google Scholar] [CrossRef]
- Tagawa, T.; Oh, D.; Dremel, S.; Mahesh, G.; Koparde, V.N.; Duncan, G.; Andresson, T.; Ziegelbauer, J.M. A virus-induced circular RNA maintains latent infection of Kaposi’s sarcoma herpesvirus. Proc. Natl. Acad. Sci. USA 2023, 120, e2212864120. [Google Scholar] [CrossRef]
- Li, L.; Lei, K.; Lyu, Y.; Tan, B.; Liang, R.; Wu, D.; Wang, K.; Wang, W.; Lin, H.; Wang, M. hsa_circ_0001741 promotes esophageal squamous cell carcinoma stemness, invasion and migration by sponging miR-491-5p to upregulate NOTCH3 expression. Am. J. Cancer Res. 2022, 12, 2012–2031. [Google Scholar]
- Gong, B.; Li, M.; Wang, Z.; Hao, G.; Sun, L.; Zhang, J.; Yuan, L. Integrated analysis of circRNA- related ceRNA network targeting neuroinflammation in medial temporal lobe epilepsy. Brain Res. Bull. 2024, 209, 110908. [Google Scholar] [CrossRef] [PubMed]
- Rebolledo, C.; Silva, J.P.; Saavedra, N.; Maracaja-Coutinho, V. Computational approaches for circRNAs prediction and in silico characterization. Brief. Bioinform. 2023, 24, bbad154. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Teng, P.; Ku, L.; Chen, L.; Feng, Y.; Yao, B. Accurate identification of circRNA landscape and complexity reveals their pivotal roles in human oligodendroglia differentiation. Genome Biol. 2022, 23, 48. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Yang, J.; Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 2020, 11, 90. [Google Scholar] [CrossRef]
- Xin, R.; Gao, Y.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Zhang, J.; Zhao, F. Full-length circular RNA profiling by nanopore sequencing with CIRI-long. Nat. Protoc. 2023, 18, 1795–1813. [Google Scholar] [CrossRef]
- Cui, J.; Chen, M.; Zhang, L.; Huang, S.; Xiao, F.; Zou, L. Circular RNAs: Biomarkers of cancer. Cancer Innov. 2022, 1, 197–206. [Google Scholar] [CrossRef]
- Avilala, J.; Becnel, D.; Abdelghani, R.; Nanbo, A.; Kahn, J.; Li, L.; Lin, Z. Role of Virally Encoded Circular RNAs in the Pathogenicity of Human Oncogenic Viruses. Front. Microbiol. 2021, 12, 657036. [Google Scholar] [CrossRef] [PubMed]
- Kouhsar, M.; Azimzadeh Jamalkandi, S.; Moeini, A.; Masoudi-Nejad, A. Detection of novel biomarkers for early detection of Non-Muscle-Invasive Bladder Cancer using Competing Endogenous RNA network analysis. Sci. Rep. 2019, 9, 8434. [Google Scholar] [CrossRef]
- Tagawa, T.; Oh, D.; Santos, J.; Dremel, S.; Mahesh, G.; Uldrick, T.S.; Yarchoan, R.; Kopardé, V.N.; Ziegelbauer, J.M. Characterizing Expression and Regulation of Gamma-Herpesviral Circular RNAs. Front. Microbiol. 2021, 12, 670542. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Steitz, J.A. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005, 24, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.; Pari, G. PAN’s Labyrinth: Molecular Biology of Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) PAN RNA, a Multifunctional Long Noncoding RNA. Viruses 2014, 6, 4212–4226. [Google Scholar] [CrossRef]
- Grundhoff, A.; Ganem, D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 2004, 113, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Fan, J.; He, J.; Fan, W.; Che, X.; Wang, X.; Han, C. Circular RNA as Diagnostic and Prognostic Biomarkers in Hematological Malignancies:Systematic Review. Technol. Cancer Res. Treat. 2024, 23, 15330338241285149. [Google Scholar] [CrossRef]
- Lan, K.; Kuppers, D.A.; Verma, S.C.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: A potential mechanism for virus-mediated control of latency. J. Virol. 2004, 78, 6585–6594. [Google Scholar] [CrossRef]
- Bu, W.; Palmeri, D.; Krishnan, R.; Marin, R.; Aris, V.M.; Soteropoulos, P.; Lukac, D.M. Identification of direct transcriptional targets of the Kaposi’s sarcoma-associated herpesvirus Rta lytic switch protein by conditional nuclear localization. J. Virol. 2008, 82, 10709–10723. [Google Scholar] [CrossRef]
- Wei, J.; Li, M.; Xue, C.; Chen, S.; Zheng, L.; Deng, H.; Tang, F.; Li, G.; Xiong, W.; Zeng, Z.; et al. Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression. J. Exp. Clin. Cancer Res. 2023, 42. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, S.; Wang, Q.; Li, P.; Liu, Y. Circ_0001741 exerts as a tumor promoter in ovarian cancer through the regulation of miR-491-5p/PRSS8 axis. Discov. Oncol. 2024, 15, 643. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, C.; Lin, J.; Dong, Y.; Wang, Y.; Xia, L. Hsa_circ_0001741 Suppresses Ovarian Cancer Cell Proliferations Through Adsorption of miR-188-5p and Promotion of FOXN2 Expression. Mol. Biotechnol. 2024, 66, 1477–1483. [Google Scholar] [CrossRef]
- Cao, D.; Wu, S.; Wang, X.; Li, Y.; Xu, H.; Pan, Z.; Wu, Z.; Yang, L.; Tan, X.; Li, D. Kaposi’s sarcoma-associated herpesvirus infection promotes proliferation of SH-SY5Y cells by the Notch signaling pathway. Cancer Cell Int. 2021, 21, 577. [Google Scholar] [CrossRef]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.-O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, Y.; Wang, C.; Zhang, X.; He, D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death Amp; Dis. 2020, 11, 284. [Google Scholar] [CrossRef]
- Zhou, J.; Yuan, M.; Zhao, Y.; Quan, Q.; Yu, D.; Yang, H.; Tang, X.; Xin, X.; Cai, G.; Qian, Q.; et al. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. J. 2021, 19, 1240–1252. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ma, X.-K.; Li, X.; Li, G.-W.; Liu, C.-X.; Zhang, J.; Wang, Y.; Wei, J.; Chen, J.; Chen, L.-L.; et al. Knockout of circRNAs by base editing back-splice sites of circularized exons. Genome Biol. 2022, 23, 16. [Google Scholar] [CrossRef]
- Karstentischer, B.; Von Einem, J.; Kaufer, B.; Osterrieder, N. Two-Step Red-Mediated Recombination for versatile High-Efficiency Markerless DNA Manipulation in Escherichia Coli. BioTechniques 2006, 40, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z.; Lee, S.H.; Lee, H.R.; Myoung, J.; Ganem, D.; et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C.; De, S.; Grammatikakis, I.; Munk, R.; Yang, X.; Piao, Y.; Dudekula, D.B.; Abdelmohsen, K.; Gorospe, M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017, 45, e116. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.F.; Bindereif, A.; Bozzoni, I.; Hanan, M.; Hansen, T.B.; Irimia, M.; Kadener, S.; Kristensen, L.S.; Legnini, I.; Morlando, M.; et al. Best practice standards for circular RNA research. Nat. Methods 2022, 19, 1208–1220. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagtalunan, C.J.; Zhang, I.; Turley, A.; Liu, F. Recent Studies on Kaposi’s Sarcoma-Associated Herpesvirus Circular RNAs. Cancers 2025, 17, 3743. https://doi.org/10.3390/cancers17233743
Pagtalunan CJ, Zhang I, Turley A, Liu F. Recent Studies on Kaposi’s Sarcoma-Associated Herpesvirus Circular RNAs. Cancers. 2025; 17(23):3743. https://doi.org/10.3390/cancers17233743
Chicago/Turabian StylePagtalunan, Cristian J., Isadora Zhang, Ariella Turley, and Fenyong Liu. 2025. "Recent Studies on Kaposi’s Sarcoma-Associated Herpesvirus Circular RNAs" Cancers 17, no. 23: 3743. https://doi.org/10.3390/cancers17233743
APA StylePagtalunan, C. J., Zhang, I., Turley, A., & Liu, F. (2025). Recent Studies on Kaposi’s Sarcoma-Associated Herpesvirus Circular RNAs. Cancers, 17(23), 3743. https://doi.org/10.3390/cancers17233743






