AdhesionScore: A Prognostic Predictor of Breast Cancer Patients Based on a Cell Adhesion-Associated Gene Signature
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. METABRIC Dataset
2.2. TCGA Dataset
2.3. GSE96058 Dataset
2.4. Survival Analysis by Cox Proportional Hazards Model
2.5. Survival Analysis by Kaplan–Meier
2.6. Gene Ontology Overrepresentation Analysis
2.7. Calculation of the AdhesionScore
2.8. Unpaired Samples Non-Parametric Statistical Analyses
2.9. Unsupervised Clustering Analysis
2.10. Gene Expression Analysis
2.11. Receiver Operating Characteristic (ROC) Analysis
3. Results
3.1. Cell Adhesion-Related Phenotypes Associated with the Prognostic Gene Set
3.2. Multivariate Analysis Identifies the Best Combination of Adhesion-Associated Genes for Prognosis Prediction
3.3. Expression and Dispersion of the 61 Adhesion-Related Genes
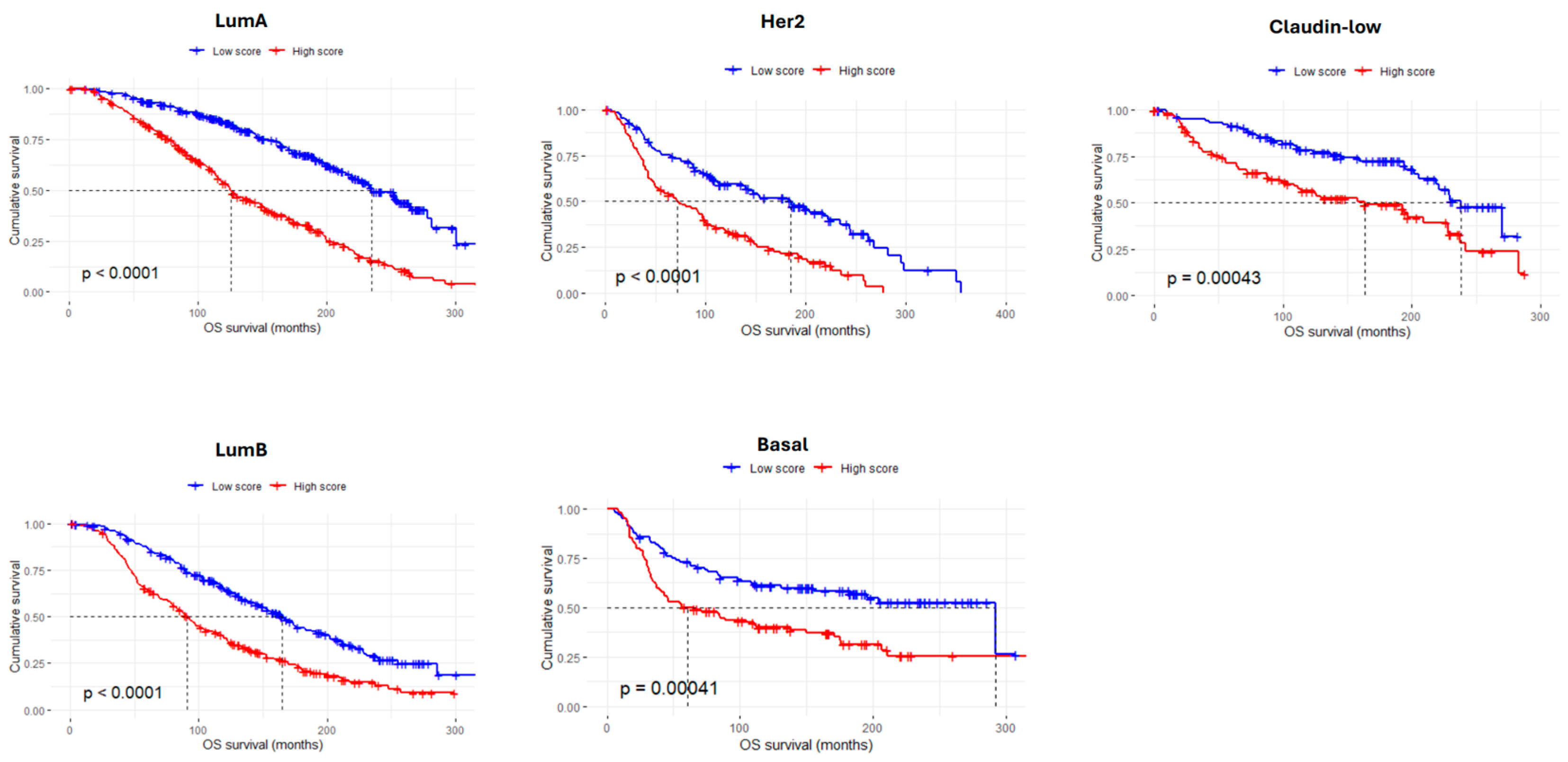
3.4. The AdhesionScore Is a Strong Prognostic Predictor in Breast Cancer
3.5. Validation of the AdhesionScore in Two Independent Breast Cancer Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIC | Akaike Information Criterion |
| AUC | Area Under the Curve |
| BRCA | Breast Invasive Carcinoma |
| CC | Cellular Components |
| C-Index | Concordance-Index |
| CI | CI Confidence Interval |
| Coxph | Cox Proportional Hazards |
| EMT | Epithelial-to-Mesenchymal Transition |
| ER | Estrogen Receptor |
| FDR | False Discovery Rate |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HR | Hazard Ratios |
| HVG | Highly Variant Genes |
| KM | Kaplan–Meier |
| LOH | Loss of Heterozygosity |
| METABRIC | Molecular Taxonomy of Breast Cancer International Consortium |
| OS | Overall Survival |
| PC | Principal Components |
| PCA | Principal Component Analysis |
| ROC | Receiver Operating Characteristic |
| TCGA | The Cancer Genome Atlas |
| TNBC | Triple-Negative Breast Carcinomas |
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Paredes, J.; Figueiredo, J.; Albergaria, A.; Oliveira, P.; Carvalho, J.; Ribeiro, A.S.; Caldeira, J.; Costa, Â.M.; Simões-Correia, J.; Oliveira, M.J.; et al. Epithelial E- and P-cadherins: Role and clinical significance in cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2012, 1826, 297–311. [Google Scholar] [CrossRef]
- Bruner, H.C.; Derksen, P.W. Loss of E-Cadherin-Dependent Cell–Cell Adhesion and the Development and Progression of Cancer. Cold Spring Harb. Perspect. Biol. 2017, 10, a029330. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Blick, T.; Widodo, E.; Hugo, H.; Waltham, M.; Lenburg, M.E.; Neve, R.M.; Thompson, E.W. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 2008, 25, 629–642. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2010, 5, 5–23. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Ribeiro, A.S.; Albergaria, A.; Sousa, B.; Correia, A.L.; Bracke, M.; Seruca, R.; Schmitt, F.C.; Paredes, J. Extracellular cleavage and shedding of P-cadherin: A mechanism underlying the invasive behaviour of breast cancer cells. Oncogene 2009, 29, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.S.; Sousa, B.; Carreto, L.; Mendes, N.; Nobre, A.R.; Ricardo, S.; Albergaria, A.; Cameselle-Teijeiro, J.F.; Gerhard, R.; Söderberg, O.; et al. P-cadherin functional role is dependent on E-cadherin cellular context: A proof of concept using the breast cancer model. J. Pathol. 2012, 229, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.F.; Ricardo, S.; Ablett, M.P.; Dionísio, M.R.; Mendes, N.; Albergaria, A.; Farnie, G.; Gerhard, R.; Cameselle-Teijeiro, J.F.; Seruca, R.; et al. P-Cadherin Is Coexpressed with CD44 and CD49f and Mediates Stem Cell Properties in Basal-like Breast Cancer. STEM CELLS 2012, 30, 854–864. [Google Scholar] [CrossRef]
- Vieira, A.F.; Ribeiro, A.S.; Dionísio, M.R.; Sousa, B.; Nobre, A.R.; Albergaria, A.; Santiago-Gómez, A.; Mendes, N.; Gerhard, R.; Schmitt, F.; et al. P-cadherin signals through the laminin receptor α6β4 integrin to induce stem cell and invasive properties in basal-like breast cancer cells. Oncotarget 2014, 5, 679–692. [Google Scholar] [CrossRef]
- Hwang, P.Y.; Mathur, J.; Cao, Y.; Almeida, J.; Ye, J.; Morikis, V.; Cornish, D.; Clarke, M.; Stewart, S.A.; Pathak, A.; et al. A Cdh3-β-catenin-laminin signaling axis in a subset of breast tumor leader cells control leader cell polarization and directional collective migration. Dev. Cell 2023, 58, 34–50.e9. [Google Scholar] [CrossRef] [PubMed]
- Van’T Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Carney, E.; Zarrella, E.; Steffel, L.; Binns, S.N.; Finkelstein, D.M.; Szymonifka, J.; Bhan, A.K.; Shepherd, L.E.; Zhang, Y.; et al. Prediction of Late Disease Recurrence and Extended Adjuvant Letrozole Benefit by the HOXB13/IL17BR Biomarker. JNCI J. Natl. Cancer Inst. 2013, 105, 1036–1042. [Google Scholar] [CrossRef]
- Ma, X.-J.; Wang, Z.; Ryan, P.D.; Isakoff, S.J.; Barmettler, A.; Fuller, A.; Muir, B.; Mohapatra, G.; Salunga, R.; Tuggle, J.; et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004, 5, 607–616. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Martin, M.; Brase, J.C.; Calvo, L.; Krappmann, K.; Ruiz-Borrego, M.; Fisch, K.; Ruiz, A.; E Weber, K.; Munarriz, B.; Petry, C.; et al. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2− breast cancer patients: Results from the GEICAM 9906 trial. Breast Cancer Res. 2014, 16, R38. [Google Scholar] [CrossRef] [PubMed]
- Ring, B.Z.; Seitz, R.S.; Beck, R.; Shasteen, W.J.; Tarr, S.M.; Cheang, M.C.; Yoder, B.J.; Budd, G.T.; Nielsen, T.O.; Hicks, D.G.; et al. Novel Prognostic Immunohistochemical Biomarker Panel for Estrogen Receptor–Positive Breast Cancer. J. Clin. Oncol. 2006, 24, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Kwa, M.; Makris, A.; Esteva, F.J. Clinical utility of gene-expression signatures in early stage breast cancer. Nat. Rev. Clin. Oncol. 2017, 14, 595–610. [Google Scholar] [CrossRef]
- Venet, D.; Dumont, J.E.; Detours, V. Most Random Gene Expression Signatures Are Significantly Associated with Breast Cancer Outcome. PLOS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Saal, L.H.; Vallon-Christersson, J.; Häkkinen, J.; Hegardt, C.; Grabau, D.; Winter, C.; Brueffer, C.; Tang, M.-H.E.; Reuterswärd, C.; Schulz, R.; et al. The Sweden Cancerome Analysis Network—Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015, 7, 20. [Google Scholar] [CrossRef]
- Brueffer, C.; Vallon-Christersson, J.; Grabau, D.; Ehinger, A.; Häkkinen, J.; Hegardt, C.; Malina, J.; Chen, Y.; Bendahl, P.-O.; Manjer, J.; et al. Clinical Value of RNA Sequencing–Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report From the Population-Based Multicenter Sweden Cancerome Analysis Network—Breast Initiative. JCO Precis. Oncol. 2018, 2, 1–18. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 1 October 2024).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.M.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e411. [Google Scholar] [CrossRef] [PubMed]
- Makrilia, N.; Kollias, A.; Manolopoulos, L.; Syrigos, K. Cell Adhesion Molecules: Role and Clinical Significance in Cancer. Cancer Investig. 2009, 27, 1023–1037. [Google Scholar] [CrossRef]
- Xiong, L.; Edwards, C.K.; Zhou, L. The Biological Function and Clinical Utilization of CD147 in Human Diseases: A Review of the Current Scientific Literature. Int. J. Mol. Sci. 2014, 15, 17411–17441. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Partridge, M.A.; Li, B.; Hong, M.; Liao, W.; Cheng, S.K.; Zhao, Y.; Calaf, G.M.; Liu, T.; Zhou, J.; et al. TGFBI expression reduces in vitro and in vivo metastatic potential of lung and breast tumor cells. Cancer Lett. 2011, 308, 23–32. [Google Scholar] [CrossRef]
- Tumbarello, D.A.; Temple, J.; Brenton, J.D. β3 integrin modulates transforming growth factor beta induced (TGFBI) function and paclitaxel response in ovarian cancer cells. Mol. Cancer 2012, 11, 36. [Google Scholar] [CrossRef]
- Gnosa, S.P.; Blasco, L.P.; Piotrowski, K.B.; Freiberg, M.L.; Savickas, S.; Madsen, D.H.; Keller, U.A.D.; Kronqvist, P.; Kveiborg, M. ADAM17-mediated EGFR ligand shedding directs macrophage-promoted cancer cell invasion. J. Clin. Investig. 2022, 7, e155296. [Google Scholar] [CrossRef]
- Huang, Y.; Benaich, N.; Tape, C.; Kwok, H.F.; Murphy, G. Targeting the Sheddase Activity of ADAM17 by an Anti-ADAM17 Antibody D1(A12) Inhibits Head and Neck Squamous Cell Carcinoma Cell Proliferation and Motility via Blockage of Bradykinin Induced HERs Transactivation. Int. J. Biol. Sci. 2014, 10, 702–714. [Google Scholar] [CrossRef]
- Pantano, F.; Croset, M.; Driouch, K.; Bednarz-Knoll, N.; Iuliani, M.; Ribelli, G.; Bonnelye, E.; Wikman, H.; Geraci, S.; Bonin, F.; et al. Integrin alpha5 in human breast cancer is a mediator of bone metastasis and a therapeutic target for the treatment of osteolytic lesions. Oncogene 2021, 40, 1284–1299. [Google Scholar] [CrossRef]
- Leong, A.; Kim, M. The Angiopoietin-2 and TIE Pathway as a Therapeutic Target for Enhancing Antiangiogenic Therapy and Immunotherapy in Patients with Advanced Cancer. Int. J. Mol. Sci. 2020, 21, 8689. [Google Scholar] [CrossRef]
- Xu, H.; Niu, M.; Yuan, X.; Wu, K.; Liu, A. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, E.; Lecomte, N.; Durand, L.; de Pinieux, G.; Decaudin, D.; Chomienne, C.; Smadja-Joffe, F.; Poupon, M.-F. CD44 targeting reduces tumour growth and prevents post-chemotherapy relapse of human breast cancers xenografts. Br. J. Cancer 2009, 100, 918–922. [Google Scholar] [CrossRef]
- Hersey, P.; Sosman, J.; O’Day, S.; Richards, J.; Bedikian, A.; Gonzalez, R.; Sharfman, W.; Weber, R.; Logan, T.; Buzoianu, M.; et al. A randomized phase 2 study of etaracizumab, a monoclonal antibody against integrin αvβ3, ± dacarbazine in patients with stage IV metastatic melanoma. Cancer 2010, 116, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Wu, F.T.; Cruz-Munoz, W.; Kerbel, R.S. Ang2 inhibitors and Tie2 activators: Potential therapeutics in perioperative treatment of early stage cancer. EMBO Mol. Med. 2021, 13, e08253. [Google Scholar] [CrossRef]
- Hyman, D.M.; Rizvi, N.A.; Natale, R.B.; Armstrong, D.K.; Birrer, M.J.; Recht, L.; Dotan, E.; Makker, V.; Kaley, T.J.; Kuruvilla, D.; et al. Phase I Study of MEDI3617, a Selective Angiopoietin-2 Inhibitor Alone and Combined with Carboplatin/Paclitaxel, Paclitaxel, or Bevacizumab for Advanced Solid Tumors. Clin. Cancer Res. 2018, 24, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Girnius, N.; Henstridge, A.Z.; Marks, B.; Yu, J.K.; Gray, G.K.; Sander, C.; Zervantonakis, I.K.; Luna, A. Cilengitide sensitivity is predicted by overall integrin expression in breast cancer. Breast Cancer Res. 2024, 26, 187. [Google Scholar] [CrossRef]
- Sung, J.; Wang, Y.; Chandrasekaran, S.; Witten, D.M.; Price, N.D. Molecular signatures from omics data: From chaos to consensus. Biotechnol. J. 2012, 7, 946–957. [Google Scholar] [CrossRef]
- Paredes, J.; Albergaria, A.; Oliveira, J.T.; JerόnImo, C.; Milanezi, F.; Schmitt, F.C. P-Cadherin Overexpression Is an Indicator of Clinical Outcome in Invasive Breast Carcinomas and Is Associated with CDH3 Promoter Hypomethylation. Clin. Cancer Res. 2005, 11, 5869–5877. [Google Scholar] [CrossRef]
- Popescu, C.I.; Giuşcă, S.E.; Liliac, L.; Avadanei, R.; Ceauşu, R.; Cîmpean, A.M.; Balan, R.; Amălinei, C.; Apostol, D.C.; Căruntu, I.D. E-cadherin expression in molecular types of breast carcinoma. Rom. J. Morphol. Embryol. 2013, 54, 267–273. [Google Scholar]
- Vieira, A.F.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests—Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Schaafsma, E.; Cheng, C. Gene signature-based prediction of triple-negative breast cancer patient response to Neoadjuvant chemotherapy. Cancer Med. 2020, 9, 6281–6295. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wu, D.; Li, F.; Zhang, P.; Feng, Y.; He, A. Identification of key biomarkers associated with cell adhesion in multiple myeloma by integrated bioinformatics analysis. Cancer Cell Int. 2020, 20, 262. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Z.; Hua, Z.; Ji, Y.; Ye, Q.; Zhang, H.; Yan, W. Prognostic signature and immunotherapeutic relevance of Focal adhesion signaling pathway-related genes in osteosarcoma. Heliyon 2024, 10, e38523. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Miao, D.; Xu, Q.; Wang, X.; Yu, F. A novel focal adhesion related gene signature for prognostic prediction in hepatocellular carcinoma. Aging 2021, 13, 10724–10748. [Google Scholar] [CrossRef]
- Chang, P.-Y.; Liao, Y.-P.; Wang, H.-C.; Chen, Y.-C.; Huang, R.-L.; Wang, Y.-C.; Yuan, C.-C.; Lai, H.-C. An epigenetic signature of adhesion molecules predicts poor prognosis of ovarian cancer patients. Oncotarget 2017, 8, 53432–53449. [Google Scholar] [CrossRef]
- Mao, D.; Xu, R.; Chen, H.; Chen, X.; Li, D.; Song, S.; He, Y.; Wei, Z.; Zhang, C. Cross-Talk of Focal Adhesion-Related Gene Defines Prognosis and the Immune Microenvironment in Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 716461. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esquível, C.; Ribeiro, R.; Ribeiro, A.S.; Ferreira, P.G.; Paredes, J. AdhesionScore: A Prognostic Predictor of Breast Cancer Patients Based on a Cell Adhesion-Associated Gene Signature. Cancers 2025, 17, 3731. https://doi.org/10.3390/cancers17233731
Esquível C, Ribeiro R, Ribeiro AS, Ferreira PG, Paredes J. AdhesionScore: A Prognostic Predictor of Breast Cancer Patients Based on a Cell Adhesion-Associated Gene Signature. Cancers. 2025; 17(23):3731. https://doi.org/10.3390/cancers17233731
Chicago/Turabian StyleEsquível, Catarina, Rogério Ribeiro, Ana Sofia Ribeiro, Pedro G. Ferreira, and Joana Paredes. 2025. "AdhesionScore: A Prognostic Predictor of Breast Cancer Patients Based on a Cell Adhesion-Associated Gene Signature" Cancers 17, no. 23: 3731. https://doi.org/10.3390/cancers17233731
APA StyleEsquível, C., Ribeiro, R., Ribeiro, A. S., Ferreira, P. G., & Paredes, J. (2025). AdhesionScore: A Prognostic Predictor of Breast Cancer Patients Based on a Cell Adhesion-Associated Gene Signature. Cancers, 17(23), 3731. https://doi.org/10.3390/cancers17233731





