A Novel Cell-Free DNA Fragmentomic Assay and Its Application for Monitoring Disease Progression in Real Time for Stage IV Cancer Patients
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
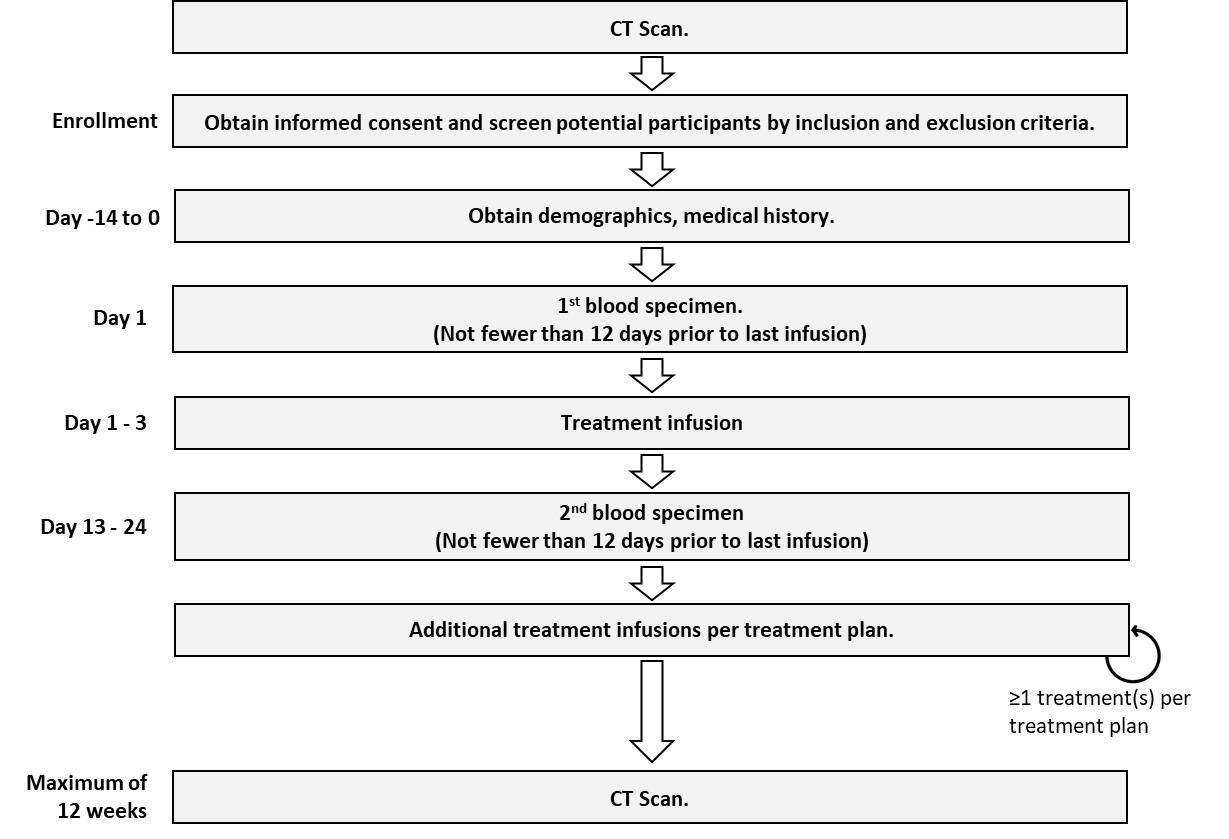
2.1. Study Design
2.2. Participants
2.3. Sample Collection and Transportation
2.4. Plasma Separation
2.5. Cell-Free DNA Extraction
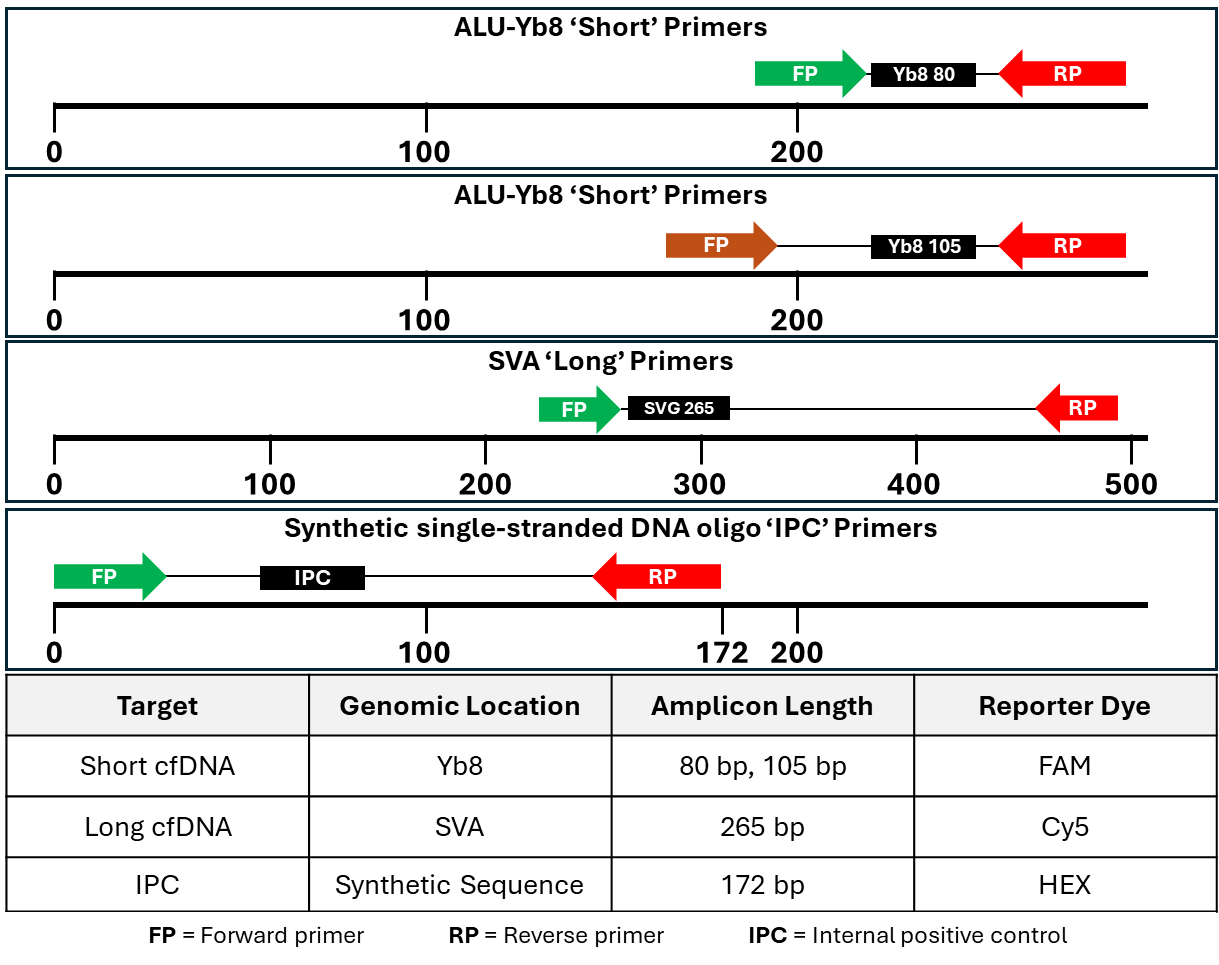
2.6. Analytical Methods
2.7. Statistical Analyses
3. Results
3.1. Model Selection

3.2. Progression Score Cut-Point Selection
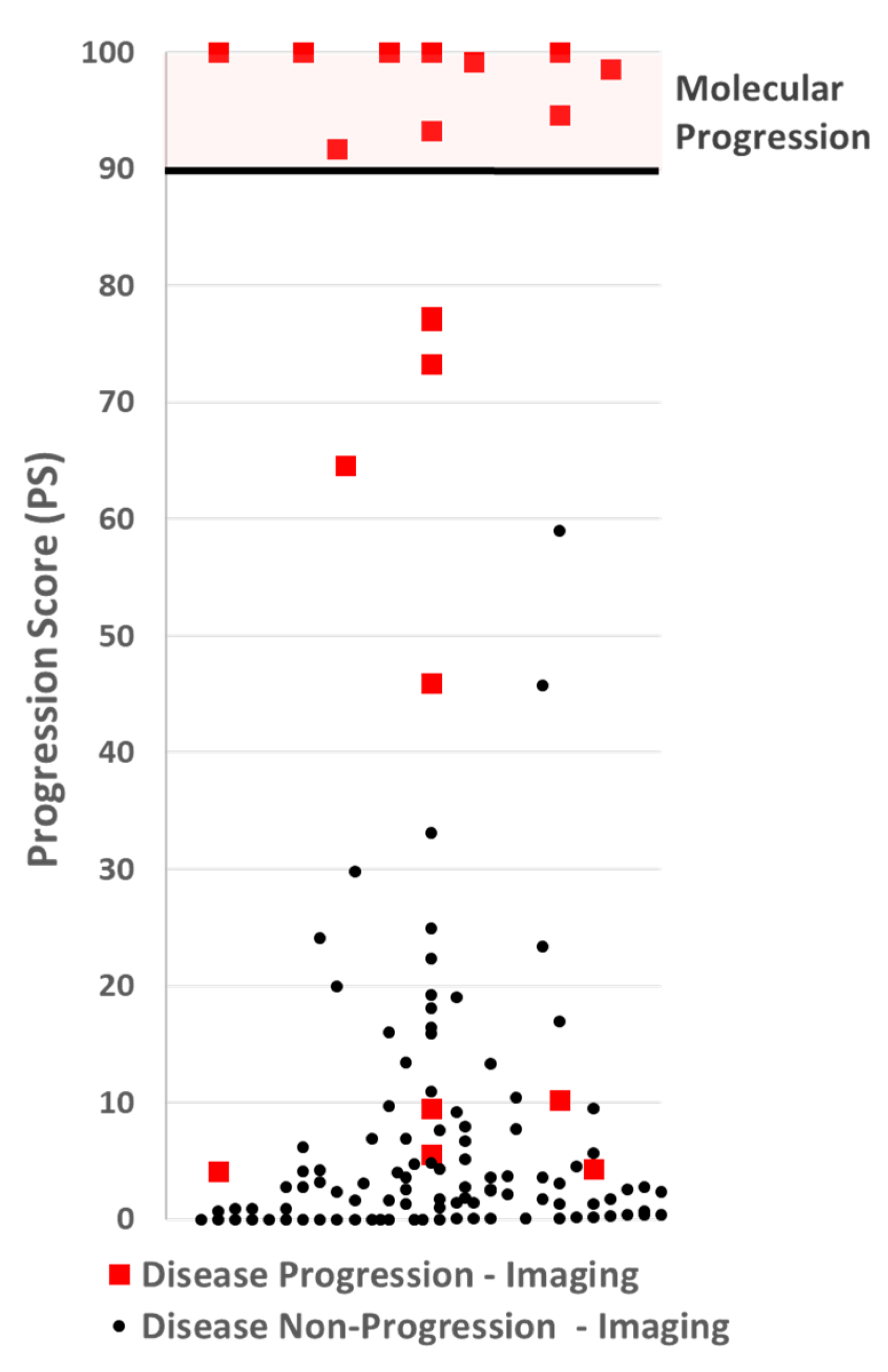
3.3. Assay Performance
4. Discussion
4.1. Role and Limitations of Fragmentomics
4.2. Limitations of This Study
4.3. Opportunities for Improved Clinical Management
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Bp | Base pair |
| ctDNA | Cell-free circulating tumor DNA |
| cfDNA | Cell-free DNA |
| DNA | Deoxyribonucleic acid |
| ICI | Immune checkpoint inhibitors |
| IPC | Internal positive control |
| PCR | Polymerase chain reaction |
| PS | Progression score |
| qPCR | Quantitative polymerase chain reaction |
References
- Akamatsu, S.; Mizuno, K.; Sumiyoshi, T.; Goto, T.; Kobayashi, T. The Current State and Future of Plasma Cell-Free DNA Analysis in Urologic Malignancies. Korean J. Urol. Oncol. 2023, 21, 23–31. [Google Scholar] [CrossRef]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B.; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. CtDNA Applications and Integration in Colorectal Cancer: An NCI Colon and Rectal–Anal Task Forces Whitepaper. Nat. Rev. Clin. Oncol. 2020, 17, 757–770. [Google Scholar] [CrossRef]
- Cisneros-Villanueva, M.; Hidalgo-Pérez, L.; Rios-Romero, M.; Cedro-Tanda, A.; Ruiz-Villavicencio, C.A.; Page, K.; Hastings, R.; Fernandez-Garcia, D.; Allsopp, R.; Fonseca-Montaño, M.A.; et al. Cell-Free DNA Analysis in Current Cancer Clinical Trials: A Review. Br. J. Cancer 2022, 126, 391–400. [Google Scholar] [CrossRef]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef]
- Parikh, A.R.; van Seventer, E.E.; Siravegna, G.; Hartwig, A.V.; Jaimovich, A.; He, Y.; Kanter, K.; Fish, M.G.; Fosbenner, K.D.; Miao, B.; et al. Minimal Residual Disease Detection Using a Plasma-Only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin. Cancer Res. 2021, 27, 5586–5594. [Google Scholar] [CrossRef]
- Fan, W.; Xia, Z.; Chen, R.; Lin, D.; Li, F.; Zheng, Y.; Luo, J.; Xiong, Y.; Yu, P.; Gao, W.; et al. Circulating Tumor DNA Analysis Predicts Recurrence and Avoids Unnecessary Adjuvant Chemotherapy in I–IV Colorectal Cancer. Ther. Adv. Med. Oncol. 2024, 16, 17588359231220607. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wang, Z.; Zhang, J.-T.; Yan, B.; Zhang, C.; Gao, X.; Sun, H.; Li, Y.-S.; Yan, H.-H.; Tu, H.-Y.; et al. Circulating Tumor DNA-Guided De-Escalation Targeted Therapy for Advanced Non-Small Cell Lung Cancer: A Nonrandomized Controlled Trial. JAMA Oncol. 2024, 10, 932–940. [Google Scholar] [CrossRef]
- Bredno, J.; Lipson, J.; Venn, O.; Aravanis, A.M.; Jamshidi, A. Clinical Correlates of Circulating Cell-Free DNA Tumor Fraction. PLoS ONE 2021, 16, e0256436. [Google Scholar] [CrossRef]
- Liu, X.; Liu, L.; Ji, Y.; Li, C.; Wei, T.; Yang, X.; Zhang, Y.; Cai, X.; Gao, Y.; Xu, W.; et al. Enrichment of Short Mutant Cell-Free DNA Fragments Enhanced Detection of Pancreatic Cancer. EBioMedicine 2019, 41, 345–356. [Google Scholar] [CrossRef]
- Han, D.S.C.; Ni, M.; Chan, R.W.Y.; Chan, V.W.H.; Lui, K.O.; Chiu, R.W.K.; Lo, Y.M.D. The Biology of Cell-Free DNA Fragmentation and the Roles of DNASE1, DNASE1L3, and DFFB. Am. J. Hum. Genet. 2020, 106, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Higazi, A.M.; El Hini, S.H.; El-Sharkawy, E.A.; Gayyed, M.F.; Aziz, N.A.; Matta, R.A. Diagnostic Role of Cell-Free DNA Integrity in Thyroid Cancer Particularly for Bethesda IV Cytology. Endocr. Pract. 2021, 27, 673–681. [Google Scholar] [CrossRef]
- Leal, A.I.C.; Mathios, D.; Jakubowski, D.; Johansen, J.S.; Lau, A.; Wu, T.; Cristiano, S.; Medina, J.E.; Phallen, J.; Bruhm, D.C.; et al. Cell-Free DNA Fragmentomes in the Diagnostic Evaluation of Patients With Symptoms Suggestive of Lung Cancer. Chest 2023, 164, 1019–1027. [Google Scholar] [CrossRef]
- Ding, S.C.; Lo, Y.M.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978. [Google Scholar] [CrossRef]
- Qi, T.; Pan, M.; Shi, H.; Wang, L.; Bai, Y.; Ge, Q. Cell-Free DNA Fragmentomics: The Novel Promising Biomarker. Int. J. Mol. Sci. 2023, 24, 1503. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-Wide Cell-Free DNA Fragmentation in Patients with Cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, N.; Wu, X.; Tang, W.; Bao, H.; Si, C.; Shao, P.; Li, D.; Zhou, X.; Zhu, D.; et al. Multidimensional Fragmentomics Enables Early and Accurate Detection of Colorectal Cancer. Cancer Res. 2024, 84, 3286–3295. [Google Scholar] [CrossRef]
- Bruhm, D.C.; Vulpescu, N.A.; Foda, Z.H.; Phallen, J.; Scharpf, R.B.; Velculescu, V.E. Genomic and Fragmentomic Landscapes of Cell-Free DNA for Early Cancer Detection. Nat. Rev. Cancer 2025, 25, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Yang, S.; Chen, X.; Dong, G.; Mao, Y.; Wu, S.; Cheng, X.; Wu, X.; Tang, W.; Wu, M.; et al. Early Detection of Multiple Cancer Types Using Multidimensional Cell-Free DNA Fragmentomics. Nat. Med. 2025, 31, 2737–2745. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meng, F.; Li, M.; Bao, H.; Chen, X.; Zhu, M.; Liu, R.; Xu, X.; Yang, S.; Wu, X.; et al. Multidimensional Cell-Free DNA Fragmentomic Assay for Detection of Early-Stage Lung Cancer. Am. J. Respir. Crit. Care Med. 2023, 207, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R. Circulating DNA Fragmentomics and Cancer Screening. Cell Genom. 2023, 3, 100242. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Li, M.; Liu, W.; Lu, L.; Li, Y.; Chen, X.; Yang, S.; Liu, T.; Cheng, W.; et al. Ultra-Short Cell-Free DNA Fragments Enhance Cancer Early Detection in a Multi-Analyte Blood Test Combining Mutation, Protein and Fragmentomics. Clin. Chem. Lab. Med. 2024, 62, 168–177. [Google Scholar] [CrossRef]
- Jiang, P.; Sun, K.; Peng, W.; Cheng, S.H.; Ni, M.; Yeung, P.C.; Heung, M.M.S.; Xie, T.; Shang, H.; Zhou, Z.; et al. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov. 2020, 10, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Foda, Z.H.; Annapragada, A.V.; Boyapati, K.; Bruhm, D.C.; Vulpescu, N.A.; Medina, J.E.; Mathios, D.; Cristiano, S.; Niknafs, N.; Luu, H.T.; et al. Detecting Liver Cancer Using Cell-Free DNA Fragmentomes. Cancer Discov. 2023, 13, 616–631. [Google Scholar] [CrossRef]
- Gianni, C.; Palleschi, M.; Merloni, F.; Di Menna, G.; Sirico, M.; Sarti, S.; Virga, A.; Ulivi, P.; Cecconetto, L.; Mariotti, M.; et al. Cell-Free DNA Fragmentomics: A Promising Biomarker for Diagnosis, Prognosis and Prediction of Response in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 14197. [Google Scholar] [CrossRef]
- Wang, P.I. Genome-Wide Cell-Free DNA Fragmentation: A Potential Low-Cost Cancer Screening Method. 2019. Available online: https://www.cancer.gov/ccg/blog/2019/interview-dna-frag (accessed on 3 November 2025).
- Helzer, K.T.; Sharifi, M.N.; Sperger, J.M.; Shi, Y.; Annala, M.; Bootsma, M.L.; Reese, S.R.; Taylor, A.; Kaufmann, K.R.; Krause, H.K.; et al. Fragmentomic Analysis of Circulating Tumor DNA-Targeted Cancer Panels. Ann. Oncol. 2023, 34, 813–825. [Google Scholar] [CrossRef]
- Sangphukieo, A.; Noisagul, P.; Thongkumkoon, P.; Chaiyawat, P. Ultra-Low Coverage Fragmentomic Model of Cell-Free DNA for Cancer Detection Based on Whole-Exome Regions. medRxiv 2024. [Google Scholar] [CrossRef]
- Zhou, Q.; Kang, G.; Jiang, P.; Qiao, R.; Lam, W.K.J.; Yu, S.C.Y.; Ma, M.-J.L.; Ji, L.; Cheng, S.H.; Gai, W.; et al. Epigenetic Analysis of Cell-Free DNA by Fragmentomic Profiling. Proc. Natl. Acad. Sci. USA 2022, 119, e2209852119. [Google Scholar] [CrossRef] [PubMed]
- van ’t Erve, I.; Alipanahi, B.; Lumbard, K.; Skidmore, Z.L.; Rinaldi, L.; Millberg, L.K.; Carey, J.; Chesnick, B.; Cristiano, S.; Portwood, C.; et al. Cancer Treatment Monitoring Using Cell-Free DNA Fragmentomes. Nat. Commun. 2024, 15, 8801. [Google Scholar] [CrossRef] [PubMed]
- Stutheit-Zhao, E.Y.; Sanz-Garcia, E.; Liu, Z.; Wong, D.; Marsh, K.; Abdul Razak, A.R.; Spreafico, A.; Bedard, P.L.; Hansen, A.R.; Lheureux, S.; et al. Early Changes in Tumor-Naive Cell-Free Methylomes and Fragmentomes Predict Outcomes in Pembrolizumab-Treated Solid Tumors. Cancer Discov. 2024, 14, 1048–1063. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, S.; Rao, W.; Mei, S.; Hu, G.; Liu, L.; Wang, Z.; Tang, J. Comprehensive Fragmentation of Cell-Free Repetitive DNA for Enhanced Cancer Detection in Plasma. Front. Cell Dev. Biol. 2025, 13, 1630231. [Google Scholar] [CrossRef]
- Pereira, A.A.L.; Lam, M.; Kanikarla Marie, P.; Raghav, K.P.S.; Morris, V.K.; Brown, H.; Windham, J.; Duose, D.Y.; Overman, M.J.; Vilar Sanchez, E.; et al. Circulating Tumor DNA (CtDNA) as an Early Marker to Monitor Clinical Benefit of Regorafenib and TAS-102 in Patients with Metastatic Colorectal Cancer (MCRC). J. Clin. Oncol. 2018, 36 (Suppl. S15), 3533. [Google Scholar] [CrossRef]
- Choi, H.; Charnsangavej, C.; Faria, S.C.; Macapinlac, H.A.; Burgess, M.A.; Patel, S.R.; Chen, L.L.; Podoloff, D.A.; Benjamin, R.S. Correlation of Computed Tomography and Positron Emission Tomography in Patients With Metastatic Gastrointestinal Stromal Tumor Treated at a Single Institution With Imatinib Mesylate: Proposal of New Computed Tomography Response Criteria. J. Clin. Oncol. 2007, 25, 1753–1759. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Villaruz, L.C.; Socinski, M.A. The Clinical Viewpoint: Definitions, Limitations of RECIST, Practical Considerations of Measurement. Clin. Cancer Res. 2013, 19, 2629–2636. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating Tumor DNA to Monitor Treatment Response in Solid Tumors and Advance Precision Oncology. npj Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef]
- Frank, M.S.; Andersen, C.S.A.; Ahlborn, L.B.; Pallisgaard, N.; Bodtger, U.; Gehl, J. Circulating Tumor DNA Monitoring Reveals Molecular Progression before Radiologic Progression in a Real-Life Cohort of Patients with Advanced Non–Small Cell Lung Cancer. Cancer Res. Commun. 2022, 2, 1174–1187. [Google Scholar] [CrossRef]
- Anagnostou, V.; Ho, C.; Nicholas, G.; Juergens, R.A.; Sacher, A.; Fung, A.S.; Wheatley-Price, P.; Laurie, S.A.; Levy, B.; Brahmer, J.R.; et al. CtDNA Response after Pembrolizumab in Non-Small Cell Lung Cancer: Phase 2 Adaptive Trial Results. Nat. Med. 2023, 29, 2559–2569. [Google Scholar] [CrossRef]
- Ozdogan, M.; Okan Cakir, M.; Kirca, O.; Gunduz, S. Hyperprogression after Immunotherapy: A Comprehensive Review. Off. J. Balk. Union Oncol. 2019, 24, 2232–2241. [Google Scholar]
- Fritsche, H.A.; Bast, R.C. CA 125 in Ovarian Cancer: Advances and Controversy. Clin. Chem. 1998, 44, 1379–1380. [Google Scholar] [CrossRef]
- Fakih, M.G.; Padmanabhan, A. CEA Monitoring in Colorectal Cancer. Oncology 2006, 20, 579–587. [Google Scholar]
- Sinha, S.; Brown, H.; Tabak, J.; Fang, Z.; Tertre, M.C.d.; McNamara, S.; Gambaro, K.; Batist, G.; Buell, J.F. Multiplexed Real-Time Polymerase Chain Reaction Cell-Free DNA Assay as a Potential Method to Monitor Stage IV Colorectal Cancer. Surgery 2019, 166, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Brown, H.; Knopf, K.B.; Hall, P.; Shannon, W.G.; Haack, W. Development of a Novel Cell-Free DNA Fragmentomics Assay for Monitoring Disease Progression in Real-Time for Stage IV Cancer Patients. J. Clin. Oncol. 2024, 42 (Suppl. S16), e14544. [Google Scholar] [CrossRef]
- Gerber, T.; Taschner-Mandl, S.; Saloberger-Sindhöringer, L.; Popitsch, N.; Heitzer, E.; Witt, V.; Geyeregger, R.; Hutter, C.; Schwentner, R.; Ambros, I.M.; et al. Assessment of Pre-Analytical Sample Handling Conditions for Comprehensive Liquid Biopsy Analysis. J. Mol. Diagn. 2020, 22, 1070–1086. [Google Scholar] [CrossRef] [PubMed]
- Pineda, G.M.; Montgomery, A.H.; Thompson, R.; Indest, B.; Carroll, M.; Sinha, S.K. Development and Validation of InnoQuantTM, a Sensitive Human DNA Quantitation and Degradation Assessment Method for Forensic Samples Using High Copy Number Mobile Elements Alu and SVA. Forensic Sci. Int. Genet. 2014, 13, 224–235. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. Alu Repeats and Human Genomic Diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Deininger, P.L.; Batzer, M.A. Mammalian Retroelements. Genome Res. 2002, 12, 1455–1465. [Google Scholar] [CrossRef]
- Shewale, J.G.; Schneida, E.; Wilson, J.; Walker, J.A.; Batzer, M.A.; Sinha, S.K. Human Genomic DNA Quantitation System, H-Quant: Development and Validation for Use in Forensic Casework. J. Forensic Sci. 2007, 52, 364–370. [Google Scholar] [CrossRef]
- Efron, B. The Bootstrap and Modern Statistics. J. Am. Stat. Assoc. 2000, 95, 1293–1296. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Watson, D.; Bryant, J.; Costantino, J.; Wolmark, N. Expression of the 21 genes in the Recurrence Score assay and tamoxifen clinical benefit in the NSABP study B-14 of node negative, estrogen receptor positive breast cancer. J. Clin. Oncol. 2005, 23 (Suppl. S16), 510. [Google Scholar] [CrossRef]
- Fujihara, J.; Takinami, Y.; Kimura-Kataoka, K.; Kawai, Y.; Takeshita, H. Cell-Free DNA Release in the Plasma of Patients with Cardiac Disease Is Associated with Cell Death Processes. Indian J. Clin. Biochem. 2023, 38, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Zhang, Y.; Gong, Y.; Sun, R.; Su, L.; Lin, X.; Shen, A.; Zhou, J.; Caiji, Z.; Wang, X.; et al. Diagnosis of Sepsis with Cell-Free DNA by Next-Generation Sequencing Technology in ICU Patients. Arch. Med. Res. 2016, 47, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, Q.; Dong, Q.; Zhan, L.; Zhang, J. How to Differentiate Pseudoprogression from True Progression in Cancer Patients Treated with Immunotherapy. Am. J. Cancer. Res. 2019, 9, 1546–1553. Available online: https://pubmed.ncbi.nlm.nih.gov/31497342/ (accessed on 3 November 2025). [PubMed]
| Model | Hypothesis |
|---|---|
| FragDiff | Change in tumor burden indicates changes in tumor size. |
| Frag1 + FragDiff | Initial tumor burden as measured by Frag1 adds predictive power. |
| SM1 + FragDiff | Initial tumor burden as measured by SM1 adds predictive power. |
| MMDiff + FragDiff | Add MMDiff to the above models. MMDiff is expected to refine FragDiff by helping to clean up noise (i.e., negative coefficient) from non-cancer sources of cfDNA. |
| Frag1 + MMDiff + FragDiff | |
| SM1 + MMDiff + FragDiff |
| Model | AUC | Note |
|---|---|---|
| FragDiff | 0.802 | |
| Frag1 + FragDiff | 0.847 | |
| SM1 + FragDiff | 0.910 | |
| MMDiff + FragDiff | 0.934 | Best model |
| Frag1 + MMDiff + FragDiff | 0.936 | Frag1 is not statistically significant. |
| SM1 + MMDiff + FragDiff | 0.934 | SM1 is not statistically significant. |
| Progression Score (PS) | n | Interpretation | Performance |
|---|---|---|---|
| ≥90 | 11 | Progression | PPV = 100% |
| <90 | 117 | Likely non-progression | NPV = 92% |
| Participant | PD by Imaging | Δ > 80 bp cfDNA | PS |
|---|---|---|---|
| 2022 | No | 10.1-fold increase | 8.6 |
| 4019 | No | 8.8-fold increase | 0.0 |
| 8003 | No | 3.8-fold increase | 0.1 |
| 2021 | Yes | 2.2% decrease | 100.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinha, S.K.; Brown, H.; Knopf, K.; Hall, P.; Shannon, W.D.; Haack, W. A Novel Cell-Free DNA Fragmentomic Assay and Its Application for Monitoring Disease Progression in Real Time for Stage IV Cancer Patients. Cancers 2025, 17, 3583. https://doi.org/10.3390/cancers17213583
Sinha SK, Brown H, Knopf K, Hall P, Shannon WD, Haack W. A Novel Cell-Free DNA Fragmentomic Assay and Its Application for Monitoring Disease Progression in Real Time for Stage IV Cancer Patients. Cancers. 2025; 17(21):3583. https://doi.org/10.3390/cancers17213583
Chicago/Turabian StyleSinha, Sudhir K., Hiromi Brown, Kevin Knopf, Patrick Hall, William D. Shannon, and William Haack. 2025. "A Novel Cell-Free DNA Fragmentomic Assay and Its Application for Monitoring Disease Progression in Real Time for Stage IV Cancer Patients" Cancers 17, no. 21: 3583. https://doi.org/10.3390/cancers17213583
APA StyleSinha, S. K., Brown, H., Knopf, K., Hall, P., Shannon, W. D., & Haack, W. (2025). A Novel Cell-Free DNA Fragmentomic Assay and Its Application for Monitoring Disease Progression in Real Time for Stage IV Cancer Patients. Cancers, 17(21), 3583. https://doi.org/10.3390/cancers17213583






