Characterizing the Clinical and Molecular Profile of SETD2-Mutated Lung Adenocarcinoma
Simple Summary
Abstract
1. Introduction
2. Methods
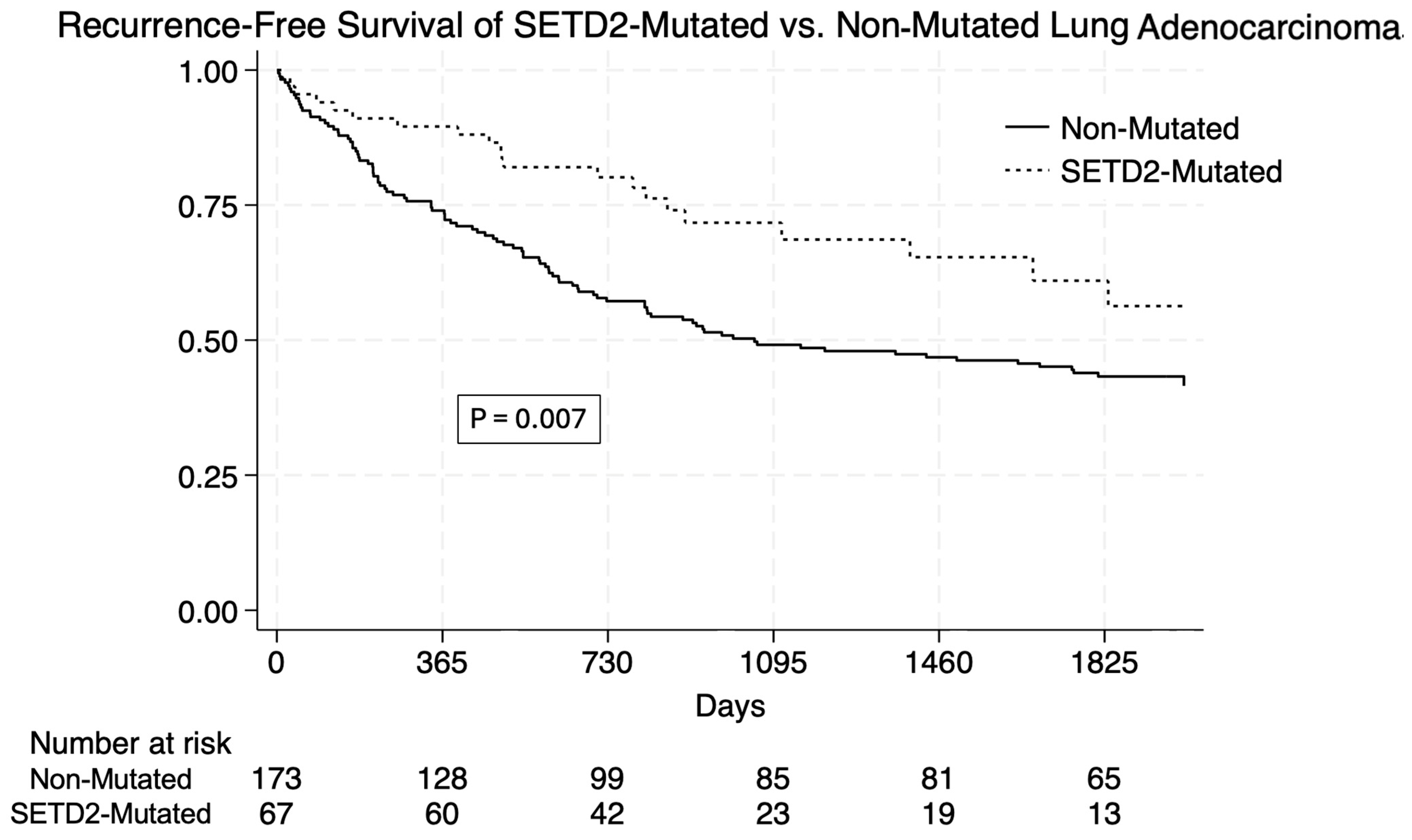
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SETD2 | SET domain containing 2 |
| NSCLC | Non-small cell lung cancer |
| COPD | Chronic obstructive pulmonary disease |
| RFS | Recurrence-free survival |
| IQR | Interquartile range |
| CI | Confidence interval |
| HR | Hazard ratio |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Hendriks, L.E.L.; Remon, J.; Faivre-Finn, C.; Garassino, M.C.; Heymach, J.V.; Kerr, K.M.; Tan, D.S.W.; Veronesi, G.; Reck, M. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 2024, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, N.; Daaboul, N. Lung Cancer: Targeted Therapy in 2025. Curr. Oncol. 2025, 32, 146. [Google Scholar] [CrossRef]
- Cainap, C.; Balacescu, O.; Cainap, S.S.; Pop, L.A. Next Generation Sequencing Technology in Lung Cancer Diagnosis. Biology 2021, 10, 864. [Google Scholar] [CrossRef]
- Carvalho, S.; Raposo, A.C.; Martins, F.B.; Grosso, A.R.; Sridhara, S.C.; Rino, J.; Carmo-Fonseca, M.; de Almeida, S.F. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013, 41, 2881–2893. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, X.; Feng, X.; Bu, J.; Dong, Y.; Lin, P.; Hayashi, Y.; Huang, R.; Olsson, A.; Andreassen, P.R.; et al. Setd2 regulates quiescence and differentiation of adult hematopoietic stem cells by restricting RNA polymerase II elongation. Haematologica 2018, 103, 1110–1123. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zou, Y.; Dong, Y.; Hong, T.; Xu, Q.; Zhang, J. Emerging role of SETD2 in the development and function of immune cells. Genes. Dis. 2025, 12, 101622. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, T.M.; van Leeuwen, F. SETD2: From chromatin modifier to multipronged regulator of the genome and beyond. Cell. Mol. Life Sci. 2022, 79, 346. [Google Scholar] [CrossRef]
- Michail, C.; Rodrigues Lima, F.; Viguier, M.; Deshayes, F. Structure and function of the lysine methyltransferase SETD2 in cancer: From histones to cytoskeleton. Neoplasia 2025, 59, 101090. [Google Scholar] [CrossRef]
- Xiao, C.; Fan, T.; Tian, H.; Zheng, Y.; Zhou, Z.; Li, S.; Li, C.; He, J. H3K36 trimethylation-mediated biological functions in cancer. Clin. Epigenetics 2021, 13, 199. [Google Scholar] [CrossRef]
- Walter, D.M.; Venancio, O.S.; Buza, E.L.; Tobias, J.W.; Deshpande, C.; Gudiel, A.A.; Kim-Kiselak, C.; Cicchini, M.; Yates, T.J.; Feldser, D.M. Systematic In Vivo Inactivation of Chromatin-Regulating Enzymes Identifies Setd2 as a Potent Tumor Suppressor in Lung Adenocarcinoma. Cancer Res. 2017, 77, 1719–1729. [Google Scholar] [CrossRef]
- Gladstein, A.C.; Poltorack, C.D.; Solomon, A.M.C.; Venkatesh, S.; Adler, K.M.; Robertson, M.R.; Stransky, S.; Irizarry-Negron, V.M.; Ruiz, D.A.; Freeburg, N.F.; et al. The H3 (K36M) oncohistone inhibits NSD2 to activate a SETD2-dependent antiviral-like immune response in KRAS-driven lung cancer. bioRxiv 2025. [Google Scholar] [CrossRef]
- Mack, R.J.; Flores, N.M.; Fox, G.C.; Dong, H.; Cebeci, M.; Hausmann, S.; Chasan, T.; Dowen, J.M.; Strahl, B.D.; Mazur, P.K.; et al. SETD2 suppresses tumorigenesis in a KRAS(G12C)-driven lung cancer model, and its catalytic activity is regulated by histone acetylation. Elife 2025, 14, RP107451. [Google Scholar] [CrossRef] [PubMed]
- Walter, D.M.; Gladstein, A.C.; Doerig, K.R.; Natesan, R.; Baskaran, S.G.; Gudiel, A.A.; Adler, K.M.; Acosta, J.O.; Wallace, D.C.; Asangani, I.A.; et al. Setd2 inactivation sensitizes lung adenocarcinoma to inhibitors of oxidative respiration and mTORC1 signaling. Commun. Biol. 2023, 6, 255. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, J.; Li, J.; Li, Y.; Huang, Z.; Han, L.; Xie, C.; Gong, Y. SETD2 regulates gene transcription patterns and is associated with radiosensitivity in lung adenocarcinoma. Front. Genet. 2022, 13, 935601. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Lin, J.; Xiong, J.; Guan, Y.; Lan, B.; Li, Y.; Gao, X.; Fei, Z.; Chen, L.; Chen, L.; et al. SETD2 variation correlates with tumor mutational burden and MSI along with improved response to immunotherapy. BMC Cancer 2023, 23, 686. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.K.; McPherson, J.R.; Tay, S.T.; Das, K.; Tan, I.B.; Ng, C.C.; Chia, N.Y.; Zhang, S.L.; Myint, S.S.; Hu, L.; et al. SETD2 histone modifier loss in aggressive GI stromal tumours. Gut 2016, 65, 1960–1972. [Google Scholar] [CrossRef]
- Niu, N.; Lu, P.; Yang, Y.; He, R.; Zhang, L.; Shi, J.; Wu, J.; Yang, M.; Zhang, Z.-G.; Wang, L.-W.; et al. Loss of Setd2 promotes Kras-induced acinar-to-ductal metaplasia and epithelia–mesenchymal transition during pancreatic carcinogenesis. Gut 2020, 69, 715. [Google Scholar] [CrossRef]
- Kim, Y.; Jee, S.; Kim, H.; Paik, S.S.; Choi, D.; Yoo, S.H.; Shin, S.J. EGFR, HER2, and MET gene amplification and protein expression profiles in biliary tract cancer and their prognostic significance. Oncologist 2024, 29, e1051–e1060. [Google Scholar] [CrossRef]
- Yokouchi, H.; Nishihara, H.; Harada, T.; Ishida, T.; Yamazaki, S.; Kikuchi, H.; Oizumi, S.; Uramoto, H.; Tanaka, F.; Harada, M.; et al. Immunohistochemical profiling of receptor tyrosine kinases, MED12, and TGF-βRII of surgically resected small cell lung cancer, and the potential of c-kit as a prognostic marker. Oncotarget 2017, 8, 39711–39726. [Google Scholar] [CrossRef]
- Bushara, O.; Wester, J.R.; Jacobsen, D.; Sun, L.; Weinberg, S.; Gao, J.; Jennings, L.J.; Wang, L.; Lauberth, S.M.; Yue, F.; et al. Clinical and histopathologic characterization of SETD2-mutated colorectal cancer. Hum. Pathol. 2023, 131, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Howington, J.A.; Blum, M.G.; Chang, A.C.; Balekian, A.A.; Murthy, S.C. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143 (Suppl. 5), e278S–e313S. [Google Scholar] [CrossRef] [PubMed]
- Detterbeck, F.C.; Boffa, D.J.; Kim, A.W.; Tanoue, L.T. The Eighth Edition Lung Cancer Stage Classification. Chest 2017, 151, 193–203. [Google Scholar] [CrossRef] [PubMed]

| Full Cohort | SETD2-Mutated | Control | p Value | ||||
|---|---|---|---|---|---|---|---|
| Patient Characteristics | Median, N | IQR, % | |||||
| Age | 68 | [62–74] | 68 | [62–74] | 68 | [62–74] | 0.92 |
| BMI | 26.5 | [23.1–31.1] | 27.1 | [23.6–31.6] | 26.2 | [22.6–30.8] | 0.39 |
| Gender (M) | 96 | 39.8% | 22 | 32.8% | 74 | 42.5% | 0.19 |
| Diabetes | 52 | 21.6% | 11 | 16.4% | 41 | 19.9% | 0.29 |
| History of Tobacco Smoking | 200 | 83.0% | 53 | 79.1% | 147 | 71.4% | 0.34 |
| COPD | 95 | 39.4% | 25 | 37.3% | 70 | 34.0% | 0.77 |
| Asthma | 49 | 20.3% | 15 | 22.4% | 34 | 16.5% | 0.72 |
| Asbestos | 19 | 7.9% | 5 | 7.5% | 14 | 6.8% | 0.61 |
| Tumor Characteristics | Median, N | IQR, % | |||||
| T Stage | <0.001 | ||||||
| T1 | 85 | 35.3% | 37 | 55.2% | 48 | 27.6% | |
| T2 | 44 | 18.3% | 11 | 16.4% | 33 | 19.0% | |
| T3 | 29 | 12.0% | 8 | 11.9% | 21 | 12.1% | |
| T4 | 40 | 16.6% | 7 | 10.4% | 33 | 19.0% | |
| TX | 43 | 17.8% | 4 | 6.0% | 39 | 22.4% | |
| N Stage | <0.001 | ||||||
| N0 | 108 | 44.8% | 46 | 68.7% | 62 | 35.6% | |
| N1 | 13 | 5.4% | 4 | 6.0% | 9 | 5.2% | |
| N2 | 50 | 20.7% | 11 | 16.4% | 39 | 22.4% | |
| N3 | 27 | 11.2% | 2 | 3.0% | 25 | 14.4% | |
| NX | 43 | 17.8% | 4 | 6.0% | 39 | 22.4% | |
| M Stage | <0.001 | ||||||
| M0 | 125 | 51.9% | 20 | 29.9% | 83 | 47.7% | |
| M1 | 85 | 35.3% | 8 | 11.9% | 67 | 38.5% | |
| MX | 63 | 26.1% | 39 | 58.2% | 24 | 13.8% | |
| Clinical Stage | <0.001 | ||||||
| I | 68 | 28.2% | 37 | 55.2% | 31 | 17.8% | |
| II | 30 | 12.4% | 9 | 13.4% | 21 | 12.1% | |
| III | 51 | 21.2% | 13 | 19.4% | 38 | 21.8% | |
| IV | 92 | 38.2% | 8 | 11.9% | 84 | 48.3% | |
| PD-L1 | |||||||
| Median Expression | 2 | IQR = [0–50] | 1.5 | [0–40] | 2 | [0–60] | 0.45 |
| Mean Expression | 24.6 | SD = 34.7 | 20.5 | 30.8 | 26.2 | 35.9 | 0.45 |
| PD-L1-Positive (>10%) | 99 | 41.1% | 24 | 35.8% | 75 | 43.1% | 0.33 |
| Gene Mutations | 8 | [6–12] | 11 | [9–16] | 7 | [5–11] | <0.001 |
| SETD2-Mutated | Non-SETD2-Mutated | ||||
|---|---|---|---|---|---|
| Gene | N | % | N | % | p Value |
| TMB | 48 | 71.6% | 46 | 26.4% | <0.001 |
| BRAF | 37 | 55.2% | 113 | 64.9% | 0.1 |
| KRAS | 32 | 47.8% | 59 | 33.9% | 0.03 |
| TP53 | 31 | 46.3% | 111 | 63.8% | 0.01 |
| MET | 28 | 41.8% | 119 | 68.4% | <0.001 |
| EGFR | 27 | 40.3% | 128 | 73.6% | <0.001 |
| ERBB2 | 25 | 37.3% | 116 | 66.7% | <0.001 |
| RET | 15 | 22.4% | 24 | 13.8% | 0.08 |
| ATM | 13 | 19.4% | 21 | 12.1% | 0.1 |
| KMT2C | 13 | 19.4% | 15 | 8.6% | 0.02 |
| SLIT2 | 12 | 17.9% | 7 | 4.0% | 0.001 |
| ARID1A | 10 | 14.9% | 15 | 8.6% | 0.11 |
| DNMT3A | 10 | 14.9% | 12 | 6.9% | 0.05 |
| NF1 | 10 | 14.9% | 13 | 7.5% | 0.07 |
| NOTCH2 | 10 | 14.9% | 8 | 4.6% | 0.01 |
| MED12 | 9 | 13.4% | 2 | 1.1% | <0.001 |
| NOTCH3 | 9 | 13.4% | 11 | 6.3% | 0.07 |
| NTRK3 | 9 | 13.4% | 13 | 7.5% | 0.12 |
| TSC2 | 9 | 13.4% | 6 | 3.4% | 0.007 |
| ALK | 8 | 11.9% | 12 | 6.9% | 0.16 |
| BRCA2 | 8 | 11.9% | 9 | 5.2% | 0.06 |
| NTRK1 | 8 | 11.9% | 3 | 1.7% | 0.002 |
| STK11 | 8 | 11.9% | 21 | 12.1% | 0.059 |
| BRCA1 | 7 | 10.4% | 7 | 4.0% | 0.06 |
| ERBB4 | 7 | 10.4% | 10 | 5.7% | 0.16 |
| KMT2D | 7 | 10.4% | 0 | 0.0% | <0.001 |
| PIK3CA | 7 | 10.4% | 12 | 6.9% | 0.25 |
| SMAD4 | 7 | 10.4% | 11 | 6.3% | 0.2 |
| TET2 | 7 | 10.4% | 7 | 4.0% | 0.06 |
| TSHR | 7 | 10.4% | 3 | 1.7% | 0.006 |
| Unadjusted Survival | N (%) | SETD2-Mutated | Non-Mutated | p = 0.007 |
|---|---|---|---|---|
| One-Year | 189 (78.33%) | 60 (89.5%) | 129 (73.9%) | |
| Three-Year | 132 (54.9%) | 48 (71.7%) | 85 (49.1%) | |
| Five-Year | 115 (48%) | 41 (61%) | 75 (43.2%) | |
| Unadjusted Univariate Hazard Ratios | HR | 95% CI | p Value | |
| SETD2 Mutation | 0.53 | 0.33 | 0.85 | 0.008 |
| Age | 1.16 | 0.98 | 1.38 | 0.08 |
| Male | 1.03 | 0.72 | 1.48 | 0.86 |
| BMI | 1.10 | 0.85 | 1.44 | 0.46 |
| Diabetes | 1.37 | 0.91 | 2.06 | 0.13 |
| Smoking | 1.27 | 0.78 | 2.11 | 0.33 |
| COPD | 1.40 | 0.98 | 2.00 | 0.06 |
| Asthma | 1.18 | 0.78 | 1.82 | 0.43 |
| Asbestos | 1.00 | 1.00 | 1.00 | 0.65 |
| Advanced Stage | 1.59 | 1.11 | 2.33 | 0.01 |
| PD-L1 | 0.99 | 1.00 | 1.00 | 0.33 |
| Total | 0.99 | 0.96 | 1.03 | 0.76 |
| Adjusted Multivariate Cox Hazard Ratios | ||||
| SETD2 Mutation | 0.71 | 0.43 | 1.19 | 0.10 |
| Age | 1.20 | 0.99 | 1.45 | 0.06 |
| Diabetes | 1.29 | 0.85 | 1.95 | 0.29 |
| COPD | 1.23 | 0.83 | 1.82 | 0.09 |
| Advanced Stage | 1.25 | 1.06 | 1.48 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bushara, O.; Devaro, D.; Ahn, S.S.; Dourlain, J.; Farooq, M.S.; Brunetti, A.; Chen, S.; Feldser, D.; Kucharczuk, J.C.; Singhal, S. Characterizing the Clinical and Molecular Profile of SETD2-Mutated Lung Adenocarcinoma. Cancers 2025, 17, 3540. https://doi.org/10.3390/cancers17213540
Bushara O, Devaro D, Ahn SS, Dourlain J, Farooq MS, Brunetti A, Chen S, Feldser D, Kucharczuk JC, Singhal S. Characterizing the Clinical and Molecular Profile of SETD2-Mutated Lung Adenocarcinoma. Cancers. 2025; 17(21):3540. https://doi.org/10.3390/cancers17213540
Chicago/Turabian StyleBushara, Omar, David Devaro, Shawn S. Ahn, Jordan Dourlain, Mohammad S. Farooq, Alessandro Brunetti, Simon Chen, David Feldser, John C. Kucharczuk, and Sunil Singhal. 2025. "Characterizing the Clinical and Molecular Profile of SETD2-Mutated Lung Adenocarcinoma" Cancers 17, no. 21: 3540. https://doi.org/10.3390/cancers17213540
APA StyleBushara, O., Devaro, D., Ahn, S. S., Dourlain, J., Farooq, M. S., Brunetti, A., Chen, S., Feldser, D., Kucharczuk, J. C., & Singhal, S. (2025). Characterizing the Clinical and Molecular Profile of SETD2-Mutated Lung Adenocarcinoma. Cancers, 17(21), 3540. https://doi.org/10.3390/cancers17213540






