Clinical Application of Vision Transformers for Melanoma Classification: A Multi-Dataset Evaluation Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Preparation
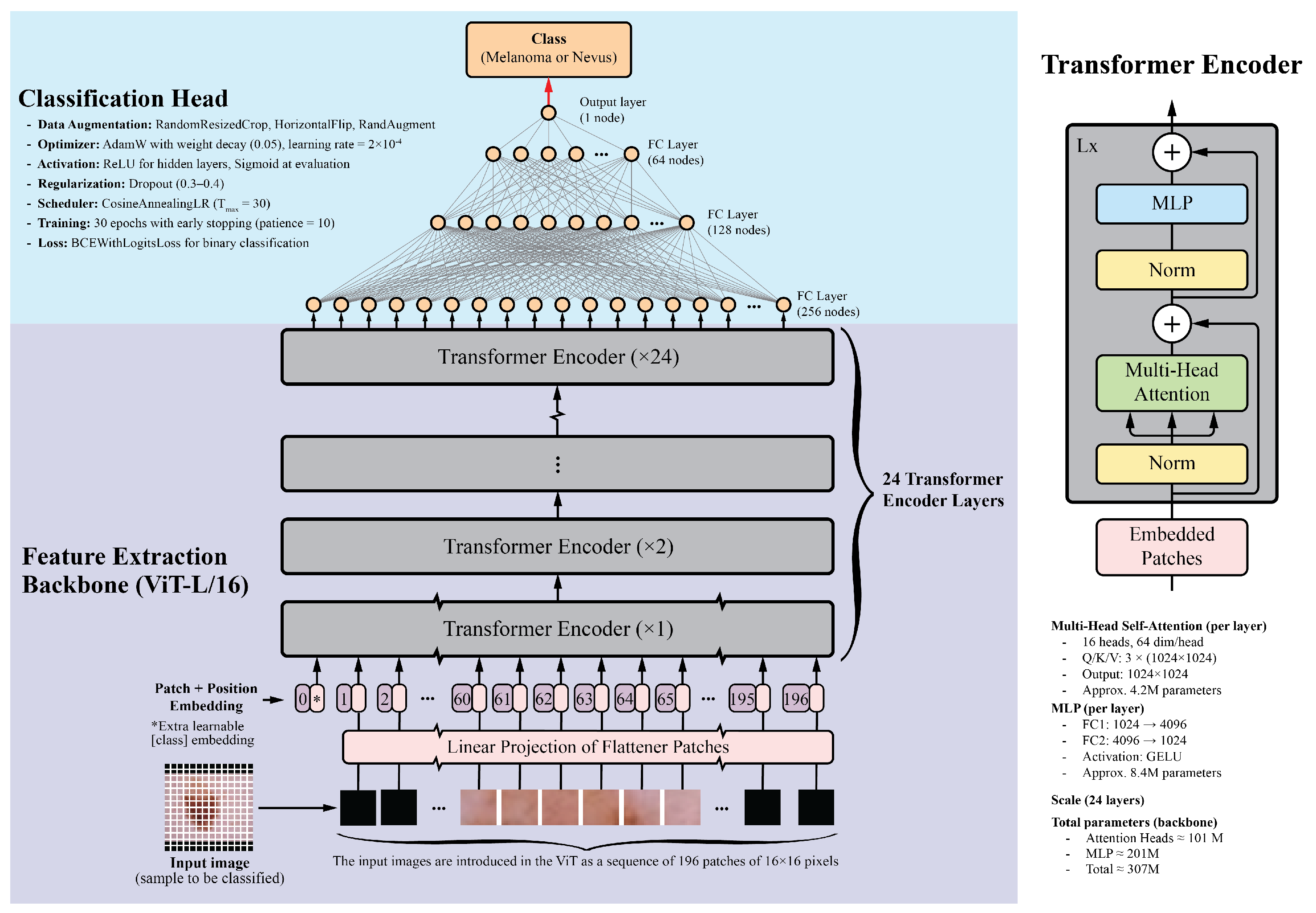
2.2. Model Architecture
2.3. Training Configuration and Optimization
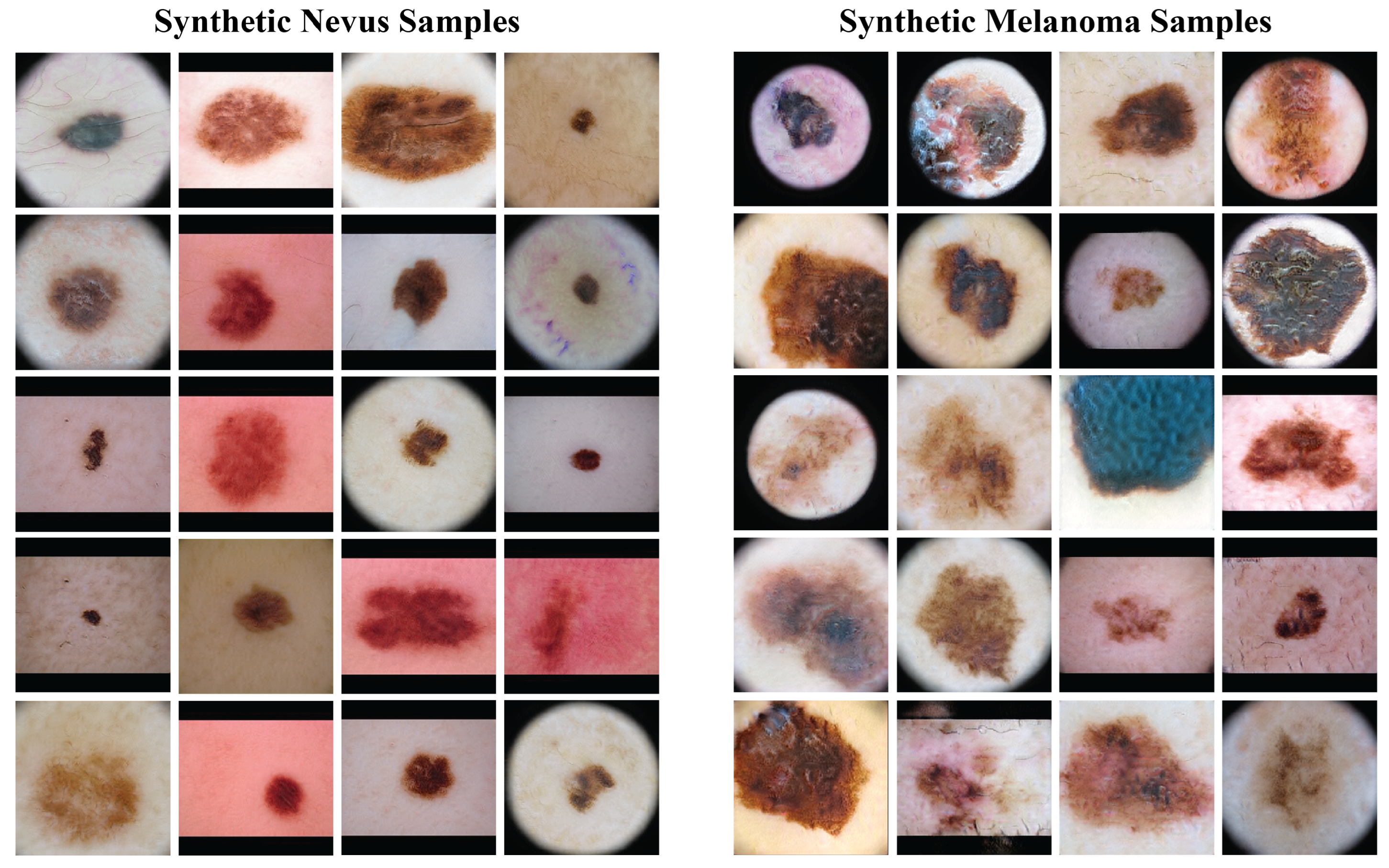
2.4. Data Augmentation
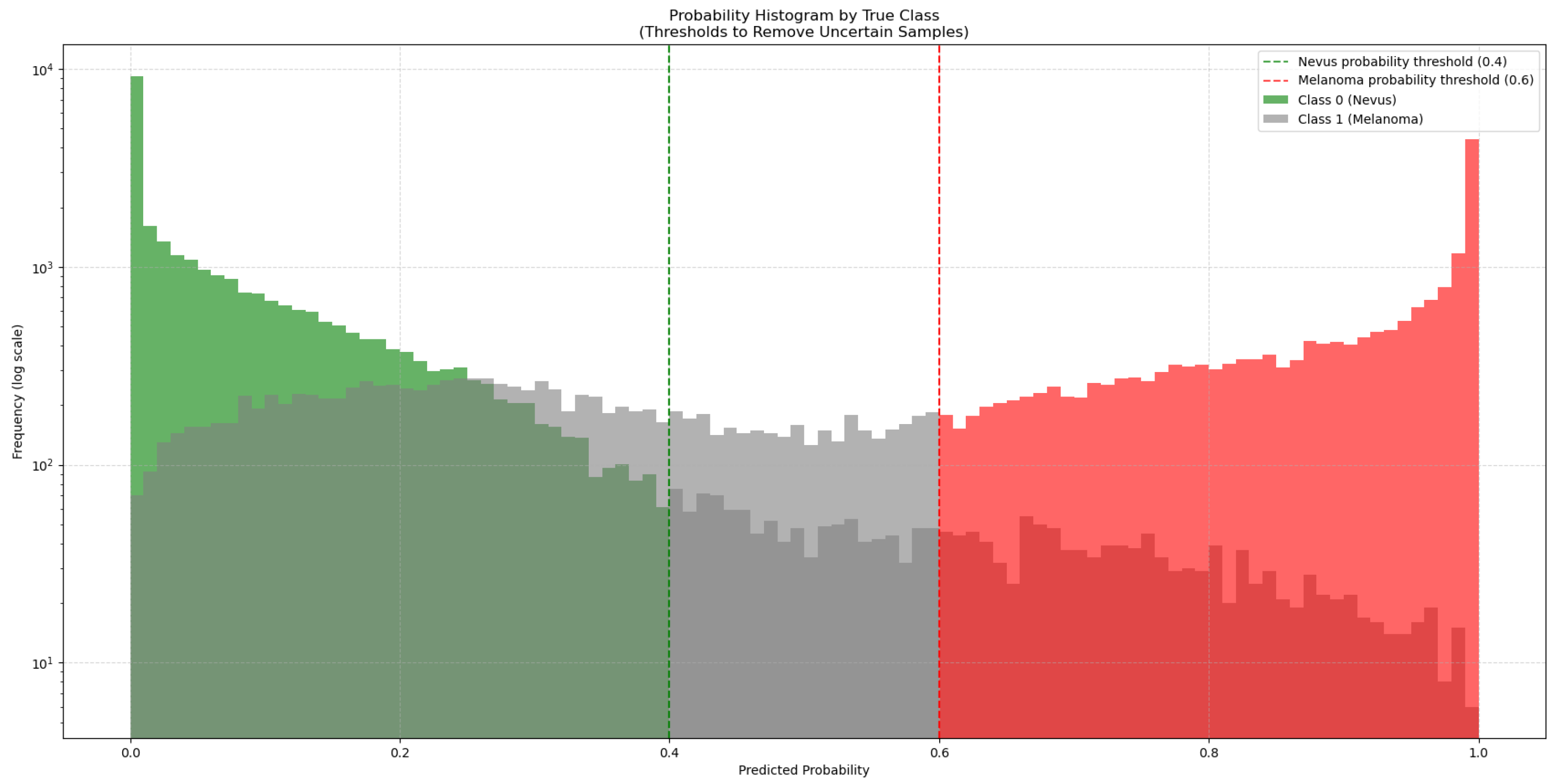
2.5. Model Performance Monitoring and Evaluation
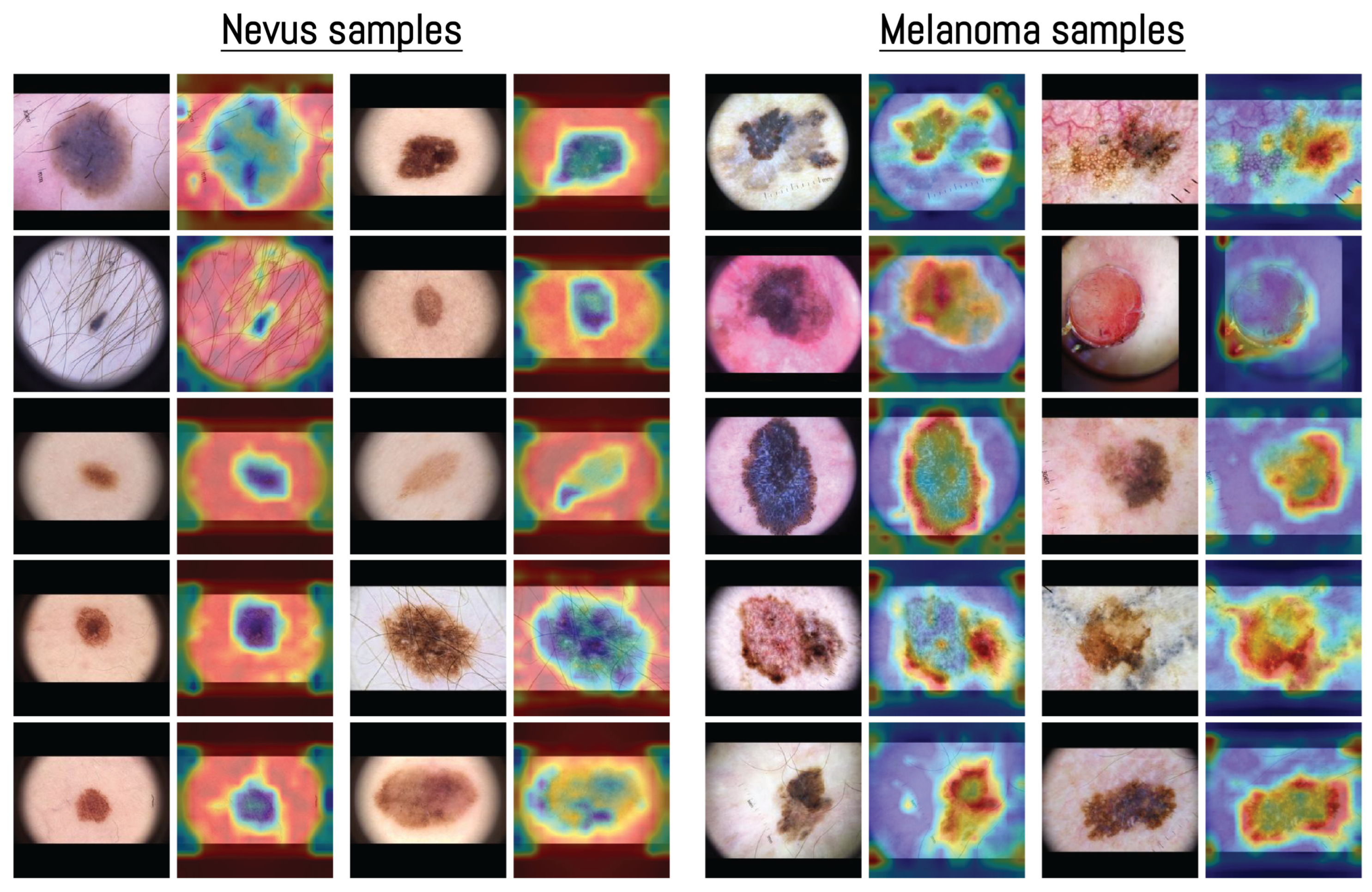
2.6. Model Interpretability
3. Results
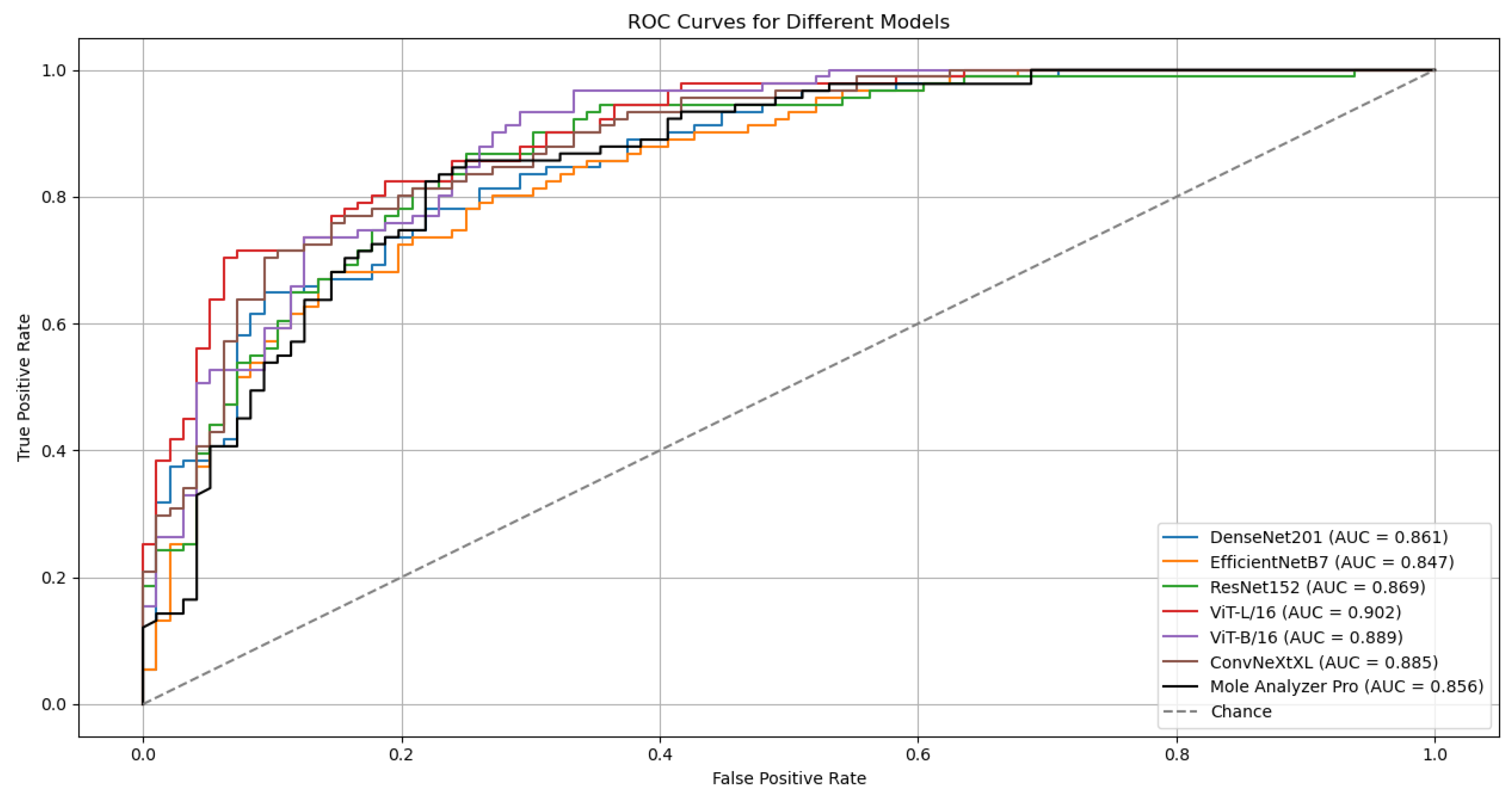
3.1. Model Performance
3.2. Results with Data Augmentation
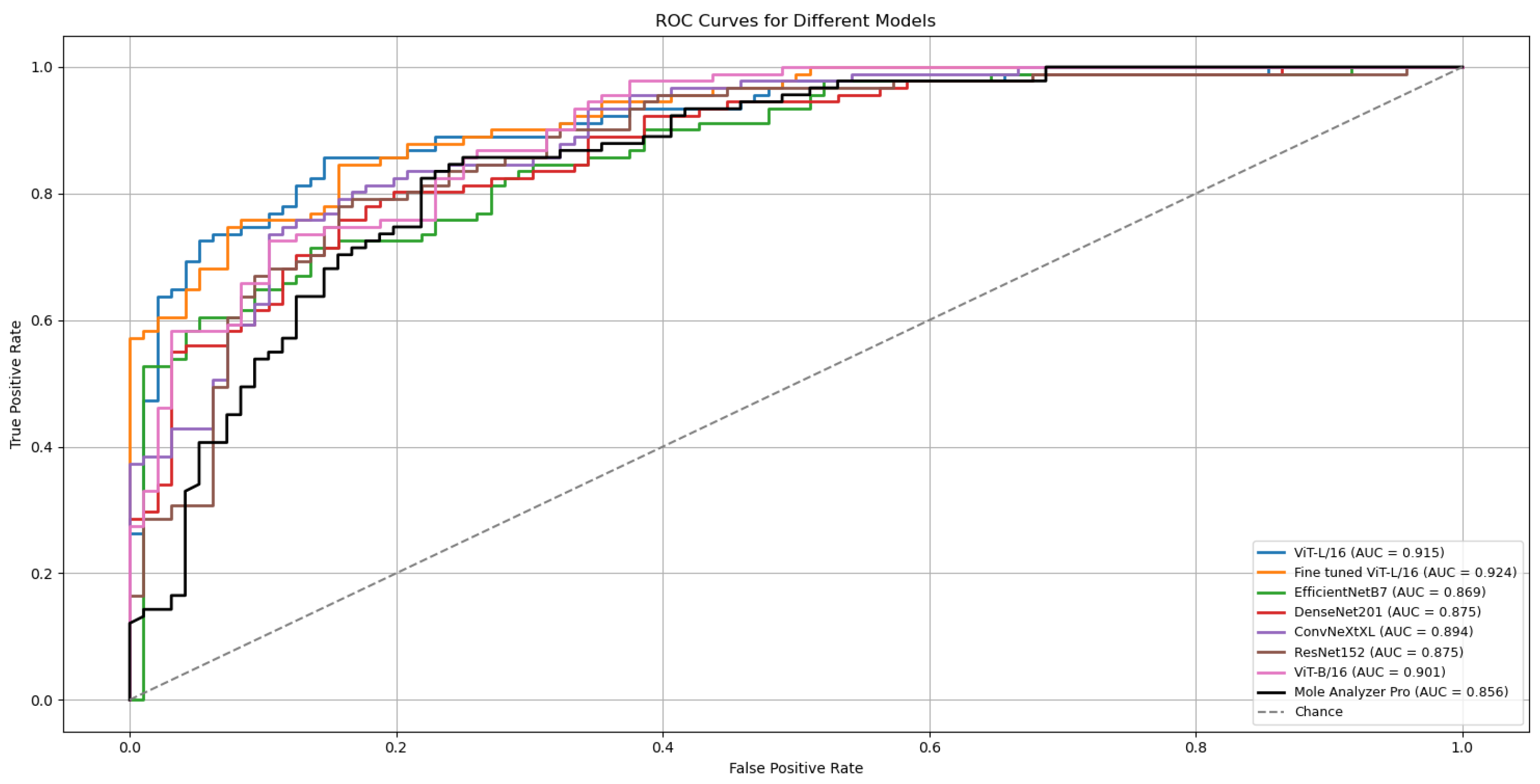
3.3. Model Performance with Data Augmentation
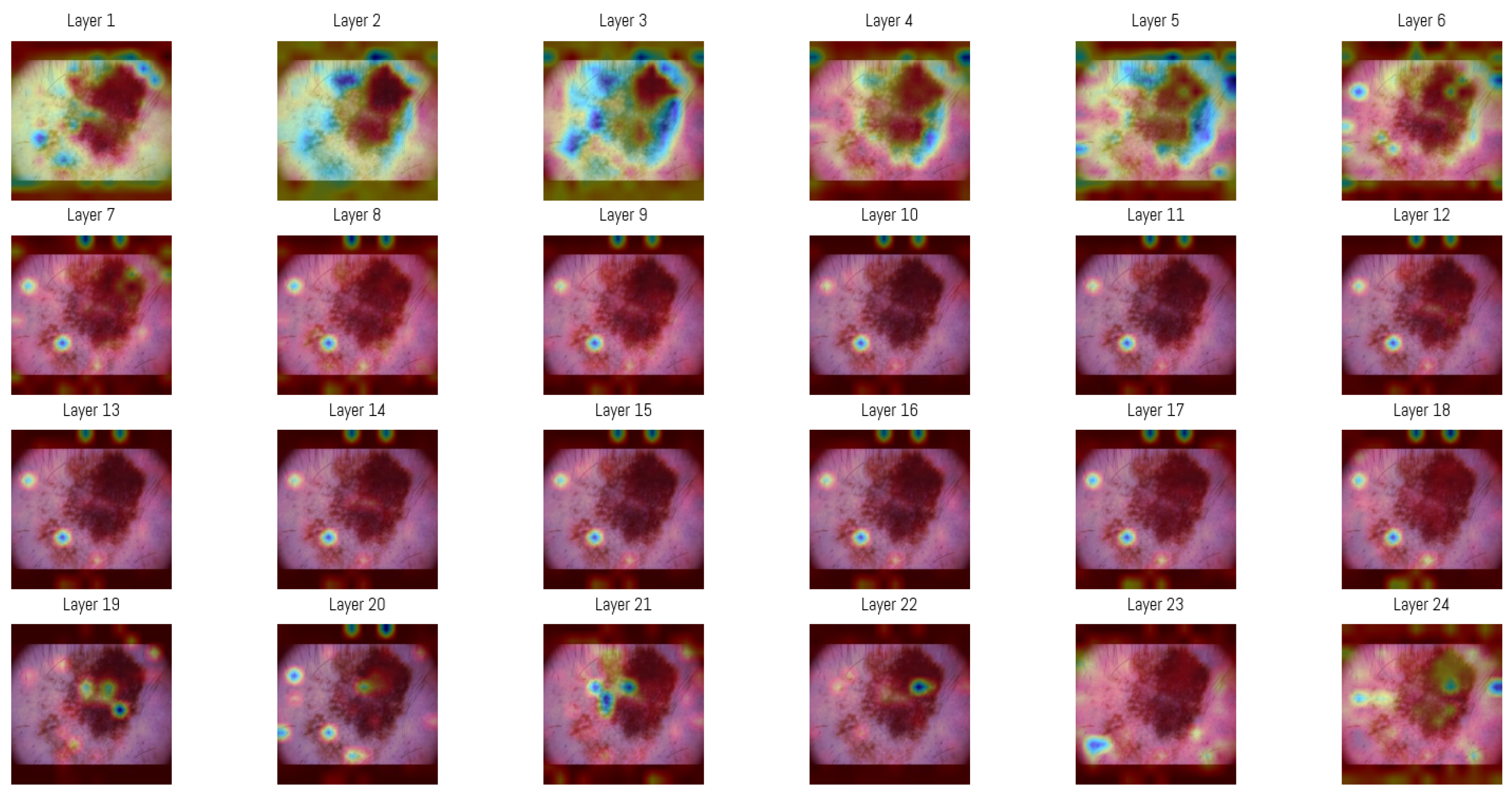
3.4. Attention Map Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, W.; Wang, H.; Li, C. Signal pathways of melanoma and targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 424. [Google Scholar] [CrossRef]
- Platz, A.; Egyhazi, S.; Ringborg, U.; Hansson, J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol. Oncol. 2008, 1, 395–405. [Google Scholar] [CrossRef]
- Thomas, N.E.; Edmiston, S.N.; Alexander, A.; Groben, P.A.; Parrish, E.; Kricker, A.; Armstrong, B.K.; Anton-Culver, H.; Gruber, S.B.; From, L.; et al. Association Between NRAS and BRAF Mutational Status and Melanoma-Specific Survival Among Patients with Higher-Risk Primary Melanoma. JAMA Oncol. 2015, 1, 359. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, Y.H. Molecular Frontiers in Melanoma: Pathogenesis, Diagnosis, and Therapeutic Advances. Int. J. Mol. Sci. 2024, 25, 2984. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Capone, M.; Ascierto, M.L.; Gentilcore, G.; Stroncek, D.F.; Casula, M.; Sini, M.C.; Palla, M.; Mozzillo, N.; Ascierto, P.A. Main roads to melanoma. J. Transl. Med. 2009, 7, 86. [Google Scholar] [CrossRef]
- Craig, S.; Earnshaw, C.H.; Virós, A. Ultraviolet light and melanoma. J. Pathol. 2018, 244, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; De Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495. [Google Scholar] [CrossRef]
- De Pinto, G.; Mignozzi, S.; La Vecchia, C.; Levi, F.; Negri, E.; Santucci, C. Global trends in cutaneous malignant melanoma incidence and mortality. Melanoma Res. 2024, 34, 265–275. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Guy, G.P. Stemming the Rising Incidence of Melanoma: Calling Prevention to Action. JNCI J. Natl. Cancer Inst. 2016, 108, djv381. [Google Scholar] [CrossRef]
- Elder, D.E. Precursors to melanoma and their mimics: Nevi of special sites. Mod. Pathol. 2006, 19, S4–S20. [Google Scholar] [CrossRef]
- Raluca Jitian (Mihulecea), C.; Frățilă, S.; Rotaru, M. Clinical-dermoscopic similarities between atypical nevi and early stage melanoma. Exp. Ther. Med. 2021, 22, 854. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.T.; Lin, T.L.; Mukundan, A.; Karmakar, R.; Chandrasekar, A.; Chang, W.Y.; Wang, H.C. Skin Cancer: Epidemiology, Screening and Clinical Features of Acral Lentiginous Melanoma (ALM), Melanoma In Situ (MIS), Nodular Melanoma (NM) and Superficial Spreading Melanoma (SSM). J. Cancer 2025, 16, 3972–3990. [Google Scholar] [CrossRef]
- Liu, P.; Su, J.; Zheng, X.; Chen, M.; Chen, X.; Li, J.; Peng, C.; Kuang, Y.; Zhu, W. A Clinicopathological Analysis of Melanocytic Nevi: A Retrospective Series. Front. Med. 2021, 8, 681668. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C.; Bestvater, F.; et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Fröhling, S.; et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur. J. Cancer 2019, 111, 148–154. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef]
- Jojoa Acosta, M.F.; Caballero Tovar, L.Y.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef]
- Haenssle, H.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Nasr-Esfahani, E.; Samavi, S.; Karimi, N.; Soroushmehr, S.; Jafari, M.; Ward, K.; Najarian, K. Melanoma detection by analysis of clinical images using convolutional neural network. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1373–1376. [Google Scholar] [CrossRef]
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef]
- Yan, S.; Yu, Z.; Primiero, C.; Vico-Alonso, C.; Wang, Z.; Yang, L.; Tschandl, P.; Hu, M.; Ju, L.; Tan, G.; et al. A multimodal vision foundation model for clinical dermatology. Nat. Med. 2025, 31, 2691–2702. [Google Scholar] [CrossRef] [PubMed]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human–computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef]
- Winkler, J.K.; Blum, A.; Kommoss, K.; Enk, A.; Toberer, F.; Rosenberger, A.; Haenssle, H.A. Assessment of Diagnostic Performance of Dermatologists Cooperating with a Convolutional Neural Network in a Prospective Clinical Study: Human with Machine. JAMA Dermatol. 2023, 159, 621. [Google Scholar] [CrossRef]
- Han, S.S.; Kim, Y.J.; Moon, I.J.; Jung, J.M.; Lee, M.Y.; Lee, W.J.; Won, C.H.; Lee, M.W.; Kim, S.H.; Navarrete-Dechent, C.; et al. Evaluation of Artificial Intelligence–Assisted Diagnosis of Skin Neoplasms: A Single-Center, Paralleled, Unmasked, Randomized Controlled Trial. J. Investig. Dermatol. 2022, 142, 2353–2362.e2. [Google Scholar] [CrossRef]
- Nigar, N.; Umar, M.; Shahzad, M.K.; Islam, S.; Abalo, D. A Deep Learning Approach Based on Explainable Artificial Intelligence for Skin Lesion Classification. IEEE Access 2022, 10, 113715–113725. [Google Scholar] [CrossRef]
- Barata, C.; Celebi, M.E.; Marques, J.S. Explainable skin lesion diagnosis using taxonomies. Pattern Recognit. 2021, 110, 107413. [Google Scholar] [CrossRef]
- Shorfuzzaman, M. An explainable stacked ensemble of deep learning models for improved melanoma skin cancer detection. Multimed. Syst. 2021, 28, 1309–1323. [Google Scholar] [CrossRef]
- Mridha, K.; Uddin, M.M.; Shin, J.; Khadka, S.; Mridha, M.F. An Interpretable Skin Cancer Classification Using Optimized Convolutional Neural Network for a Smart Healthcare System. IEEE Access 2023, 11, 41003–41018. [Google Scholar] [CrossRef]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin Lesions Classification Into Eight Classes for ISIC 2019 Using Deep Convolutional Neural Network and Transfer Learning. IEEE Access 2020, 8, 114822–114832. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Lu, C.T.; Karmakar, R.; Nampalley, K.; Mukundan, A.; Hsiao, Y.P.; Hsieh, S.C.; Wang, H.C. Assessing the Efficacy of the Spectrum-Aided Vision Enhancer (SAVE) to Detect Acral Lentiginous Melanoma, Melanoma In Situ, Nodular Melanoma, and Superficial Spreading Melanoma. Diagnostics 2024, 14, 1672. [Google Scholar] [CrossRef]
- Yang, G.; Luo, S.; Greer, P. A Novel Vision Transformer Model for Skin Cancer Classification. Neural Process. Lett. 2023, 55, 9335–9351. [Google Scholar] [CrossRef]
- Xin, C.; Liu, Z.; Zhao, K.; Miao, L.; Ma, Y.; Zhu, X.; Zhou, Q.; Wang, S.; Li, L.; Yang, F.; et al. An improved transformer network for skin cancer classification. Comput. Biol. Med. 2022, 149, 105939. [Google Scholar] [CrossRef]
- Khan, R.F.; Lee, B.D.; Lee, M.S. Transformers in medical image segmentation: A narrative review. Quant. Imaging Med. Surg. 2023, 13, 5048–5064. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, Z.; Fang, Z. An effective CNN and Transformer complementary network for medical image segmentation. Pattern Recognit. 2023, 139, 109520. [Google Scholar] [CrossRef]
- Al-hammuri, K.; Gebali, F.; Kanan, A.; Chelvan, I.T. Vision transformer architecture and applications in digital health: A tutorial and survey. Vis. Comput. Ind. Biomed. Art 2023, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Xi, Z.; Yin, S.; Zhao, Z.; Zhao, L. Advantages of transformer and its application for medical image segmentation: A survey. Biomed. Eng. Online 2024, 23, 14. [Google Scholar] [CrossRef]
- Khan, S.; Ali, H.; Shah, Z. Identifying the role of vision transformer for skin cancer—A scoping review. Front. Artif. Intell. 2023, 6, 1202990. [Google Scholar] [CrossRef]
- Arshed, M.A.; Mumtaz, S.; Ibrahim, M.; Ahmed, S.; Tahir, M.; Shafi, M. Multi-Class Skin Cancer Classification Using Vision Transformer Networks and Convolutional Neural Network-Based Pre-Trained Models. Information 2023, 14, 415. [Google Scholar] [CrossRef]
- Khan, S.; Khan, A. SkinViT: A transformer based method for Melanoma and Nonmelanoma classification. PLoS ONE 2023, 18, e0295151. [Google Scholar] [CrossRef]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar] [CrossRef]
- Jeong, J.J.; Tariq, A.; Adejumo, T.; Trivedi, H.; Gichoya, J.W.; Banerjee, I. Systematic Review of Generative Adversarial Networks (GANs) for Medical Image Classification and Segmentation. J. Digit. Imaging 2022, 35, 137–152. [Google Scholar] [CrossRef]
- Goceri, E. GAN based augmentation using a hybrid loss function for dermoscopy images. Artif. Intell. Rev. 2024, 57, 234. [Google Scholar] [CrossRef]
- Behara, K.; Bhero, E.; Agee, J.T. Skin Lesion Synthesis and Classification Using an Improved DCGAN Classifier. Diagnostics 2023, 13, 2635. [Google Scholar] [CrossRef] [PubMed]
- Yazdanparast, T.; Shamsipour, M.; Ayatollahi, A.; Delavar, S.; Ahmadi, M.; Samadi, A.; Firooz, A. Comparison of the Diagnostic Accuracy of Teledermoscopy, Face-to-Face Examinations and Artificial Intelligence in the Diagnosis of Melanoma. Indian J. Dermatol. 2024, 69, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Cerminara, S.E.; Cheng, P.; Kostner, L.; Huber, S.; Kunz, M.; Maul, J.T.; Böhm, J.S.; Dettwiler, C.F.; Geser, A.; Jakopović, C.; et al. Diagnostic performance of augmented intelligence with 2D and 3D total body photography and convolutional neural networks in a high-risk population for melanoma under real-world conditions: A new era of skin cancer screening? Eur. J. Cancer 2023, 190, 112954. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.E.; Kamali, K.; Dorey, R.A.; MacIntyre, O.C.; Cleminson, K.; MacGillivary, M.L.; Green, P.J.; Langley, R.G.; Purdy, K.S.; DeCoste, R.C.; et al. Using Artificial Intelligence as a Melanoma Screening Tool in Self-Referred Patients. J. Cutan. Med. Surg. 2024, 28, 37–43. [Google Scholar] [CrossRef]
- Hartman, R.I.; Trepanowski, N.; Chang, M.S.; Tepedino, K.; Gianacas, C.; McNiff, J.M.; Fung, M.; Braghiroli, N.F.; Grant-Kels, J.M. Multicenter prospective blinded melanoma detection study with a handheld elastic scattering spectroscopy device. JAAD Int. 2024, 15, 24–31. [Google Scholar] [CrossRef]
- MacLellan, A.N.; Price, E.L.; Publicover-Brouwer, P.; Matheson, K.; Ly, T.Y.; Pasternak, S.; Walsh, N.M.; Gallant, C.J.; Oakley, A.; Hull, P.R.; et al. The use of noninvasive imaging techniques in the diagnosis of melanoma: A prospective diagnostic accuracy study. J. Am. Acad. Dermatol. 2021, 85, 353–359. [Google Scholar] [CrossRef]
- Miller, I.; Rosic, N.; Stapelberg, M.; Hudson, J.; Coxon, P.; Furness, J.; Walsh, J.; Climstein, M. Performance of Commercial dermoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review. Cancers 2024, 16, 1443. [Google Scholar] [CrossRef]
- Wei, M.L.; Tada, M.; So, A.; Torres, R. Artificial intelligence and skin cancer. Front. Med. 2024, 11, 1331895. [Google Scholar] [CrossRef]
- International Skin Imaging Collaboration (ISIC) Team. ISIC 2019 Challenge: Skin Lesion Classification. 2019. Available online: https://challenge.isic-archive.com/landing/2019/ (accessed on 15 October 2025).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the International Conference on Learning Representations (ICLR), Online, 3–7 May 2021. [Google Scholar]
- Emara, T.; Afify, H.M.; Ismail, F.H.; Hassanien, A.E. A Modified Inception-v4 for Imbalanced Skin Cancer Classification Dataset. In Proceedings of the 2019 14th International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 17 December 2019; pp. 28–33. [Google Scholar] [CrossRef]
- Alzamel, M.; Iliopoulos, C.; Lim, Z. Deep learning approaches and data augmentation for melanoma detection. Neural Comput. Appl. 2024, 37, 10591–10604. [Google Scholar] [CrossRef]
- Safdar, K.; Akbar, S.; Shoukat, A. A Majority Voting based Ensemble Approach of Deep Learning Classifiers for Automated Melanoma Detection. In Proceedings of the 2021 International Conference on Innovative Computing (ICIC), Lahore, Pakistan, 9–10 November 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Karras, T.; Aittala, M.; Hellsten, J.; Laine, S.; Lehtinen, J.; Aila, T. Training generative adversarial networks with limited data. In Proceedings of the Advances in Neural Information Processing Systems, Virtual, 6–12 December 2020; pp. 12104–12114. [Google Scholar]
- NVlabs. StyleGAN2-ADA PyTorch. 2020. Available online: https://github.com/NVlabs/stylegan2-ada-pytorch (accessed on 24 October 2025).
- Obuchowski, N.A.; Bullen, J.A. Receiver operating characteristic (ROC) curves: Review of methods with applications in diagnostic medicine. Phys. Med. Biol. 2018, 63, 07TR01. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- He, Z.; Zhang, Q.; Song, M.; Tan, X.; Wang, W. Four overlooked errors in ROC analysis: How to prevent and avoid. BMJ Evid.-Based Med. 2025, 30, 208–211. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837. [Google Scholar] [CrossRef]
- Chefer, H.; Gur, S.; Wolf, L. Transformer Interpretability Beyond Attention Visualization. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 782–791. [Google Scholar] [CrossRef]
- Wu, J.; Kang, W.; Tang, H.; Hong, Y.; Yan, Y. On the Faithfulness of Vision Transformer Explanations. In Proceedings of the 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 17–21 June 2024; pp. 10936–10945. [Google Scholar] [CrossRef]
- Martín, J.; Bella-Navarro, R.; Jordá, E. Vascular Patterns in Dermoscopy. Actas-Dermo-Sifiliográficas (Engl. Ed.) 2012, 103, 357–375. [Google Scholar] [CrossRef]
- Longo, C.; Pampena, R.; Moscarella, E.; Chester, J.; Starace, M.; Cinotti, E.; Piraccini, B.M.; Argenziano, G.; Peris, K.; Pellacani, G. Dermoscopy of melanoma according to different body sites: Head and neck, trunk, limbs, nail, mucosal and acral. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 1718–1730. [Google Scholar] [CrossRef]
- Hedegaard, L.; Alok, A.; Jose, J.; Iosifidis, A. Structured pruning adapters. Pattern Recognit. 2024, 156, 110724. [Google Scholar] [CrossRef]
- Na, G.S. Efficient learning rate adaptation based on hierarchical optimization approach. Neural Netw. 2022, 150, 326–335. [Google Scholar] [CrossRef]
- Razuvayevskaya, O.; Wu, B.; Leite, J.A.; Heppell, F.; Srba, I.; Scarton, C.; Bontcheva, K.; Song, X. Comparison between parameter-efficient techniques and full fine-tuning: A case study on multilingual news article classification. PLoS ONE 2024, 19, e0301738. [Google Scholar] [CrossRef] [PubMed]
| Category | Parameter | Value / Setting | Description / Notes |
|---|---|---|---|
| Hardware | –gpus | 2 | Number of GPUs used (2× NVIDIA RTX A5000) |
| –workers | 3 | Number of DataLoader workers per process | |
| Dataset | –data | dataset_nevus.zip | Path to training dataset (datasets are provided to the model in zip compressed files. Both datasets, nevus and melanoma, are trained with the same parameters, only changing the dataset file) |
| –mirror | True | Enables x-flip augmentation (data doubling) | |
| –subset | None | Uses full dataset (no subset restriction) | |
| Resolution | 256×256 | Input image resolution for generator and discriminator | |
| Base Configuration | –cfg | auto | Auto-selects model depth, learning rate, and |
| –kimg | 1000 | Total training duration (1000 k images = 1 M samples) | |
| –batch | 16 | Global batch size | |
| –gamma | Auto (≈) | R1 regularization weight (≈ at ) | |
| –snap | 5 | Snapshot / FID computation interval (in ticks) | |
| Optimizer | Adam | Used for both Generator and Discriminator | |
| Metrics | [FID50k_full] | Frechet Inception Distance computed every snapshot | |
| Augmentation | –aug | noaug | No adaptive discriminator augmentation applied |
| –augpipe | Default (bgc) | Pipeline defined but inactive due to noaug | |
| –target | N/A | ADA target (0.6 default) unused in this configuration | |
| Optimization | Learning rate | 0.002–0.0025 | Determined automatically by resolution and batch size |
| Betas | (0, 0.99) | Adam momentum parameters | |
| 1 × 10−8 | Adam numerical stability constant | ||
| EMA ramp-up | Enabled (10–20 kimg) | Stabilizes generator weights early in training | |
| Mixed precision | Enabled (FP16) | Reduces GPU memory usage, improves throughput | |
| TF32 / cuDNN | Enabled | Allows TensorFloat32 for convolutions; benchmarking on | |
| Model Architecture | Latent dimensions | 512 | StyleGAN2 latent and mapping vector size (z, w) |
| Mapping layers | 8 | Depth of style mapping network (MLP) | |
| Channel base / max | 32,768/512 | Controls feature-map width scaling | |
| –freezed | None | No frozen discriminator layers | |
| EMA decay | Auto (0.999–0.9999) | Exponential moving average rate for generator weights | |
| Evaluation | Metric | FID50k_full | Computed on 50k generated images using Inception-V3 |
| Stopping criterion | FID < 50 | Early stopping once quality threshold achieved |
| Model | Validation (ISIC-2019) | Testing (MN187) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | |
| ViT-L/16 | 0.921 | 0.919 | 0.919 | 0.920 | 0.919 | 0.902 | 0.896 | 0.896 | 0.902 | 0.902 |
| ViT-B/16 | 0.919 | 0.916 | 0.915 | 0.914 | 0.916 | 0.887 | 0.889 | 0.877 | 0.886 | 0.882 |
| ResNet-152 | 0.909 | 0.908 | 0.907 | 0.908 | 0.908 | 0.857 | 0.861 | 0.869 | 0.860 | 0.861 |
| EfficientNet-B7 | 0.889 | 0.887 | 0.887 | 0.889 | 0.889 | 0.847 | 0.835 | 0.838 | 0.834 | 0.839 |
| DenseNet-201 | 0.915 | 0.916 | 0.914 | 0.914 | 0.914 | 0.842 | 0.858 | 0.855 | 0.861 | 0.845 |
| ConvNeXt-XL | 0.926 | 0.927 | 0.929 | 0.926 | 0.928 | 0.881 | 0.877 | 0.881 | 0.884 | 0.885 |
| Model | Deep Learning Model AUC | MoleAnalyzer Pro AUC | DeLong Test p-Value |
|---|---|---|---|
| ViT-L/16 | 0.901 | 0.856 | 0.070 |
| ConvNeXt-XL | 0.884 | 0.856 | 0.226 |
| ViT-B/16 | 0.888 | 0.856 | 0.240 |
| ResNet-152 | 0.868 | 0.856 | 0.654 |
| EfficientNet-B7 | 0.846 | 0.856 | 0.709 |
| DenseNet-201 | 0.859 | 0.856 | 0.910 |
| Model | Validation (ISIC-2019) | Testing (MN187) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | Run 1 | Run 2 | Run 3 | Run 4 | Run 5 | |
| ViT-L/16 | 0.938 | 0.936 | 0.936 | 0.937 | 0.935 | 0.914 | 0.909 | 0.908 | 0.915 | 0.908 |
| ViT-B/16 | 0.924 | 0.927 | 0.927 | 0.928 | 0.927 | 0.896 | 0.892 | 0.891 | 0.901 | 0.899 |
| ResNet-152 | 0.911 | 0.909 | 0.912 | 0.912 | 0.911 | 0.875 | 0.870 | 0.873 | 0.872 | 0.872 |
| EfficientNet-B7 | 0.895 | 0.894 | 0.893 | 0.894 | 0.893 | 0.860 | 0.866 | 0.863 | 0.866 | 0.869 |
| DenseNet-201 | 0.921 | 0.921 | 0.920 | 0.920 | 0.921 | 0.870 | 0.868 | 0.872 | 0.875 | 0.861 |
| ConvNeXt-XL | 0.934 | 0.936 | 0.933 | 0.933 | 0.934 | 0.891 | 0.894 | 0.885 | 0.889 | 0.894 |
| Model | Deep Learning Model AUC | MoleAnalyzer Pro AUC | DeLong Test p-Value |
|---|---|---|---|
| ViT-L/16 | 0.920 | 0.856 | 0.032 |
| Fine tuned ViT-L/16 | 0.926 | 0.856 | 0.006 |
| EfficientNet-B7 | 0.870 | 0.856 | 0.645 |
| DenseNet-201 | 0.880 | 0.856 | 0.466 |
| ConvNeXt-XL | 0.897 | 0.856 | 0.154 |
| ResNet-152 | 0.877 | 0.856 | 0.498 |
| ViT-B/16 | 0.905 | 0.856 | 0.098 |
| Rank | Layer | Average Entropy |
|---|---|---|
| 1 | Layer 3 | 0.8615 |
| 2 | Layer 4 | 0.8463 |
| 3 | Layer 5 | 0.8393 |
| 4 | Layer 1 | 0.8166 |
| 5 | Layer 6 | 0.8065 |
| 6 | Layer 2 | 0.7685 |
| 7 | Layer 7 | 0.7272 |
| 8 | Layer 24 | 0.7241 |
| 9 | Layer 23 | 0.7036 |
| 10 | Layer 8 | 0.6853 |
| 11 | Layer 9 | 0.6004 |
| 12 | Layer 21 | 0.5624 |
| 13 | Layer 18 | 0.5598 |
| 14 | Layer 17 | 0.5418 |
| 15 | Layer 11 | 0.5405 |
| 16 | Layer 16 | 0.5379 |
| 17 | Layer 13 | 0.5339 |
| 18 | Layer 12 | 0.5331 |
| 19 | Layer 15 | 0.5322 |
| 20 | Layer 10 | 0.5187 |
| 21 | Layer 14 | 0.5141 |
| 22 | Layer 22 | 0.4742 |
| 23 | Layer 20 | 0.4740 |
| 24 | Layer 19 | 0.4354 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, A.; Zhou, J.; Pinero-Crespo, G.; Beachkofsky, T.; Huang, X. Clinical Application of Vision Transformers for Melanoma Classification: A Multi-Dataset Evaluation Study. Cancers 2025, 17, 3447. https://doi.org/10.3390/cancers17213447
Garcia A, Zhou J, Pinero-Crespo G, Beachkofsky T, Huang X. Clinical Application of Vision Transformers for Melanoma Classification: A Multi-Dataset Evaluation Study. Cancers. 2025; 17(21):3447. https://doi.org/10.3390/cancers17213447
Chicago/Turabian StyleGarcia, Antony, Jixing Zhou, Gabriela Pinero-Crespo, Thomas Beachkofsky, and Xinming Huang. 2025. "Clinical Application of Vision Transformers for Melanoma Classification: A Multi-Dataset Evaluation Study" Cancers 17, no. 21: 3447. https://doi.org/10.3390/cancers17213447
APA StyleGarcia, A., Zhou, J., Pinero-Crespo, G., Beachkofsky, T., & Huang, X. (2025). Clinical Application of Vision Transformers for Melanoma Classification: A Multi-Dataset Evaluation Study. Cancers, 17(21), 3447. https://doi.org/10.3390/cancers17213447








