Correction: Simbulan-Rosenthal et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490
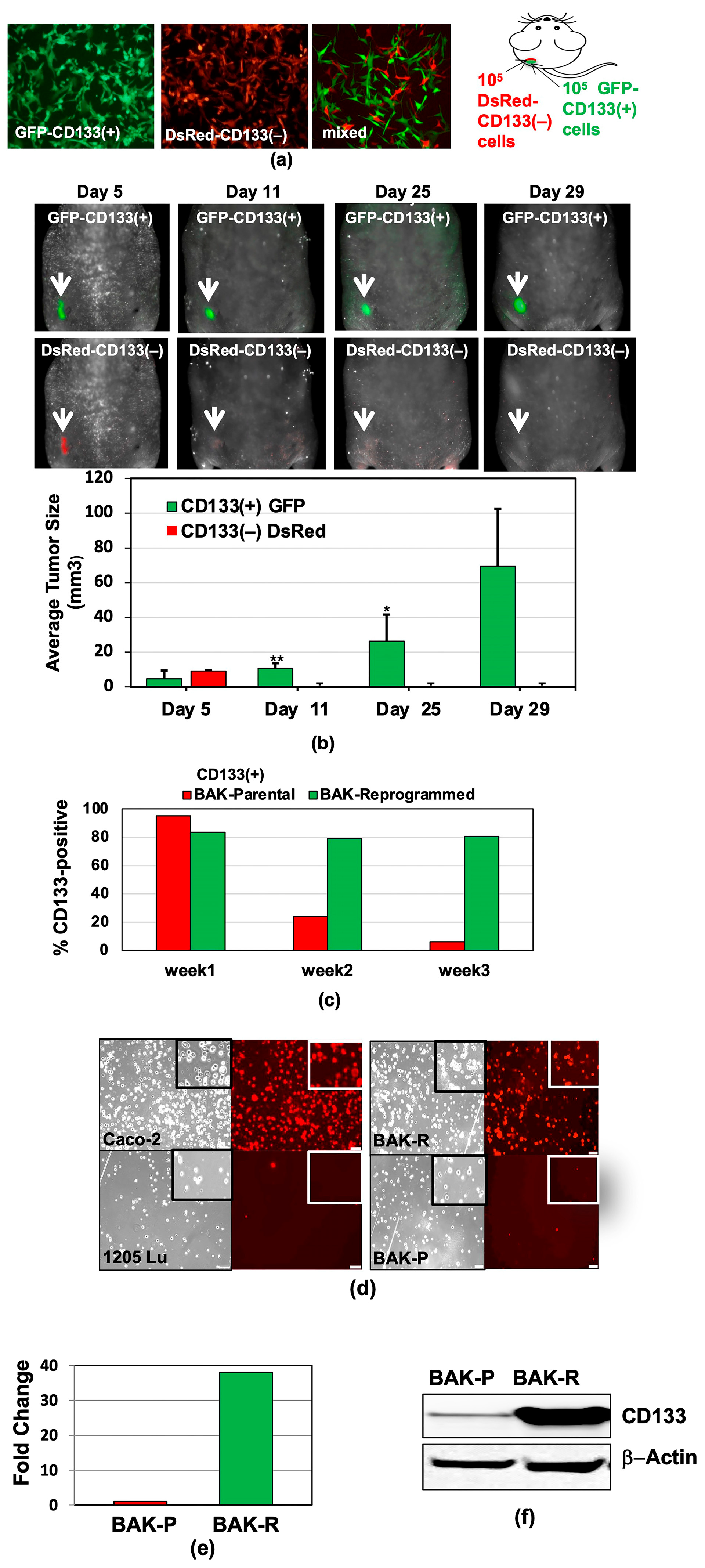
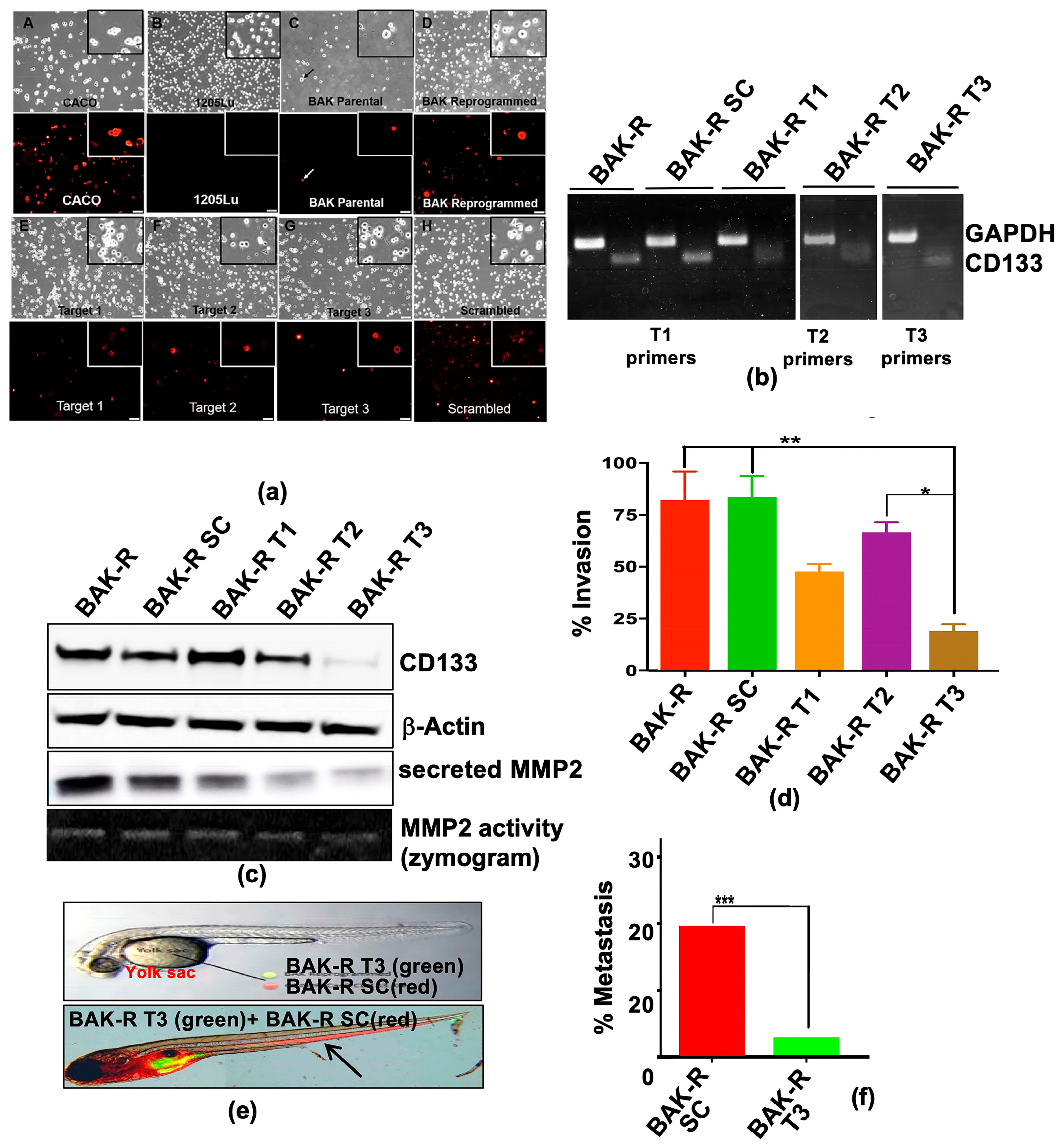
Error in Figures
Error in Supplementary Materials
- Supplementary Materials: The following are available online at https://www.mdpi.com/2072-6694/11/10/1490/s1, Figure S1: (a) CRISPR Cas9 pLenti plasmid from ABM and Gene Targets, (b) Frameshift mutation analysis; Figure S2: (a) Frameshift mutation analysis from Next Generation Sequencing and (b) visualization of allelic mutations at CRISPR target sites; Figure S3: Scans of whole gel western blots showing molecular weight sizes of relevant proteins (indicated in arrows) for Figures 1f, 2a, 3a, 4c, 5a, 6b, 6d, 6f, and 6g; Figure S4: Densitometry analysis of western blots; Figure S5: RT-PCR gel images for Figure 4b.
Reference
- Simbulan-Rosenthal, C.M.; Dougherty, R.; Vakili, S.; Ferraro, A.M.; Kuo, L.-W.; Alobaidi, R.; Aljehane, L.; Gaur, A.; Sykora, P.; Glasgow, E.; et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simbulan-Rosenthal, C.M.; Dougherty, R.; Vakili, S.; Ferraro, A.M.; Kuo, L.-W.; Alobaidi, R.; Aljehane, L.; Gaur, A.; Sykora, P.; Glasgow, E.; et al. Correction: Simbulan-Rosenthal et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. Cancers 2025, 17, 3436. https://doi.org/10.3390/cancers17213436
Simbulan-Rosenthal CM, Dougherty R, Vakili S, Ferraro AM, Kuo L-W, Alobaidi R, Aljehane L, Gaur A, Sykora P, Glasgow E, et al. Correction: Simbulan-Rosenthal et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. Cancers. 2025; 17(21):3436. https://doi.org/10.3390/cancers17213436
Chicago/Turabian StyleSimbulan-Rosenthal, Cynthia M., Ryan Dougherty, Sahar Vakili, Alexandra M. Ferraro, Li-Wei Kuo, Ryyan Alobaidi, Leala Aljehane, Anirudh Gaur, Peter Sykora, Eric Glasgow, and et al. 2025. "Correction: Simbulan-Rosenthal et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490" Cancers 17, no. 21: 3436. https://doi.org/10.3390/cancers17213436
APA StyleSimbulan-Rosenthal, C. M., Dougherty, R., Vakili, S., Ferraro, A. M., Kuo, L.-W., Alobaidi, R., Aljehane, L., Gaur, A., Sykora, P., Glasgow, E., Agarwal, S., & Rosenthal, D. S. (2025). Correction: Simbulan-Rosenthal et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. Cancers, 17(21), 3436. https://doi.org/10.3390/cancers17213436






