Radiomics and Deep Learning Interplay for Predicting MGMT Methylation in Glioblastoma: The Crucial Role of Segmentation Quality
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Analysis Flow
- The first step was the identification of structural and diffusion MRI sequences that contain a relevant amount of information related to the classification of MGMT promoter methylation status. To achieve this goal, the T1, T2, T1Gd, and FLAIR sequences were analyzed using both a radiomic and a deep learning approach, while DTI images have been used only to train and evaluate the radiomic approach.
- Then, we developed different joint models using the Fractional Anisotropy (FA) and the Axial Diffusivity (AD) maps to train a 3D Convolutional Neural Newtork (CNN) to explore whether these volumes contain information about the MGMT methylation status.
- Furthermore, we trained a multi-modal 3D CNN along with radiomic features in a joint fusion approach to maximize the performance of the classifier.
- Finally, since the performance we obtained was not satisfactory to claim for a reliable prediction of the MGMT promoter status, we end our analysis with specific insights into the dependence of the robustness and the predictive power of radiomic features on the quality of provided segmentation masks.
2.3. Mri Data Pre-Processing
2.4. Computation of Radiomic Features
- 18 histogram-based features (also known as First Order Statistics or intensity features) computed on pixel gray-level histograms;
- 14 shape-based features, dependent only on the shape of the mask;
- 75 texture-based features, derived from the gray-level co-occurrence matrix (GLCM), gray-level size zone matrix (GLSZM), gray-level dependence matrix (GLDM), gray-level run length matrix (GLRLM), and neighboring gray tone difference matrix (NGTDM).
2.5. Selection of More Informative Structural MRI Sequences
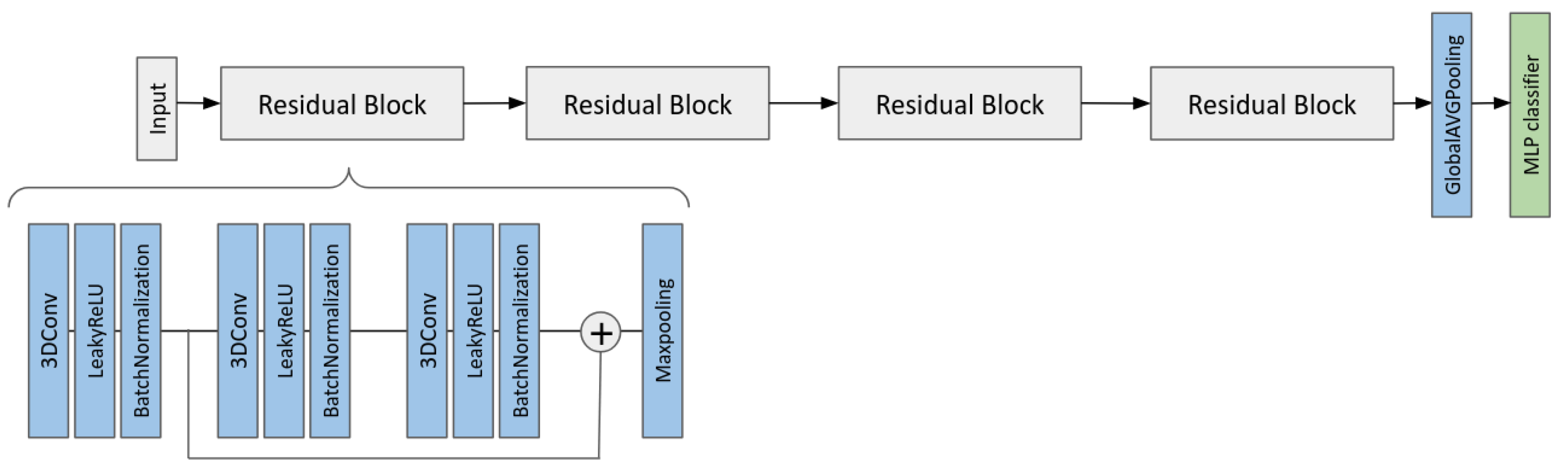
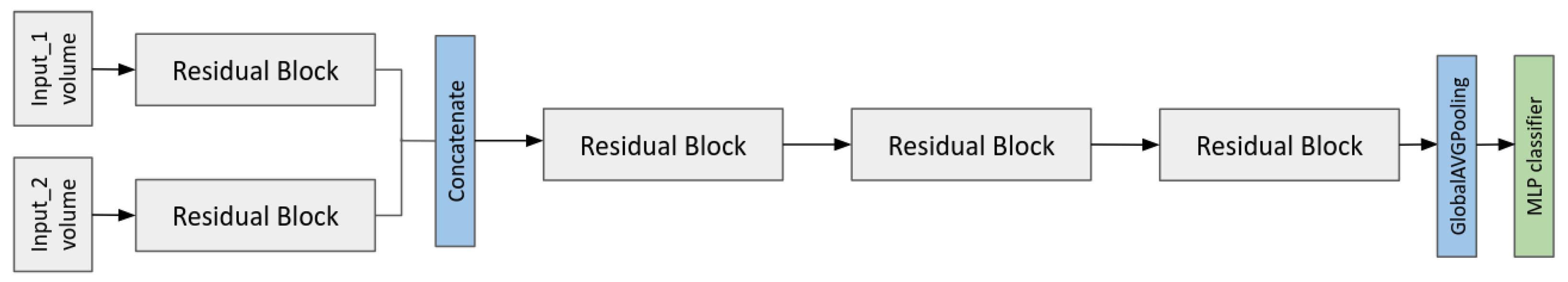
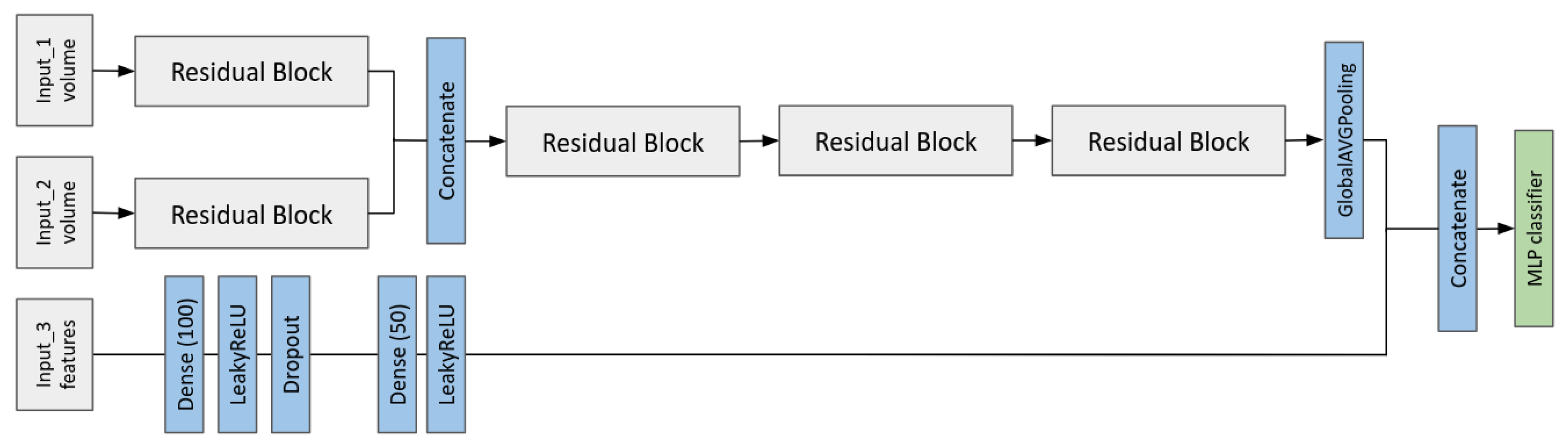
2.6. Multi-Input CNN Models to Predict the MGMT Status
- A multi-branch CNN that takes as input the two MRI volumes;
- A multi-modal CNN-MLP that takes as input two MRI volumes and all the radiomic features.
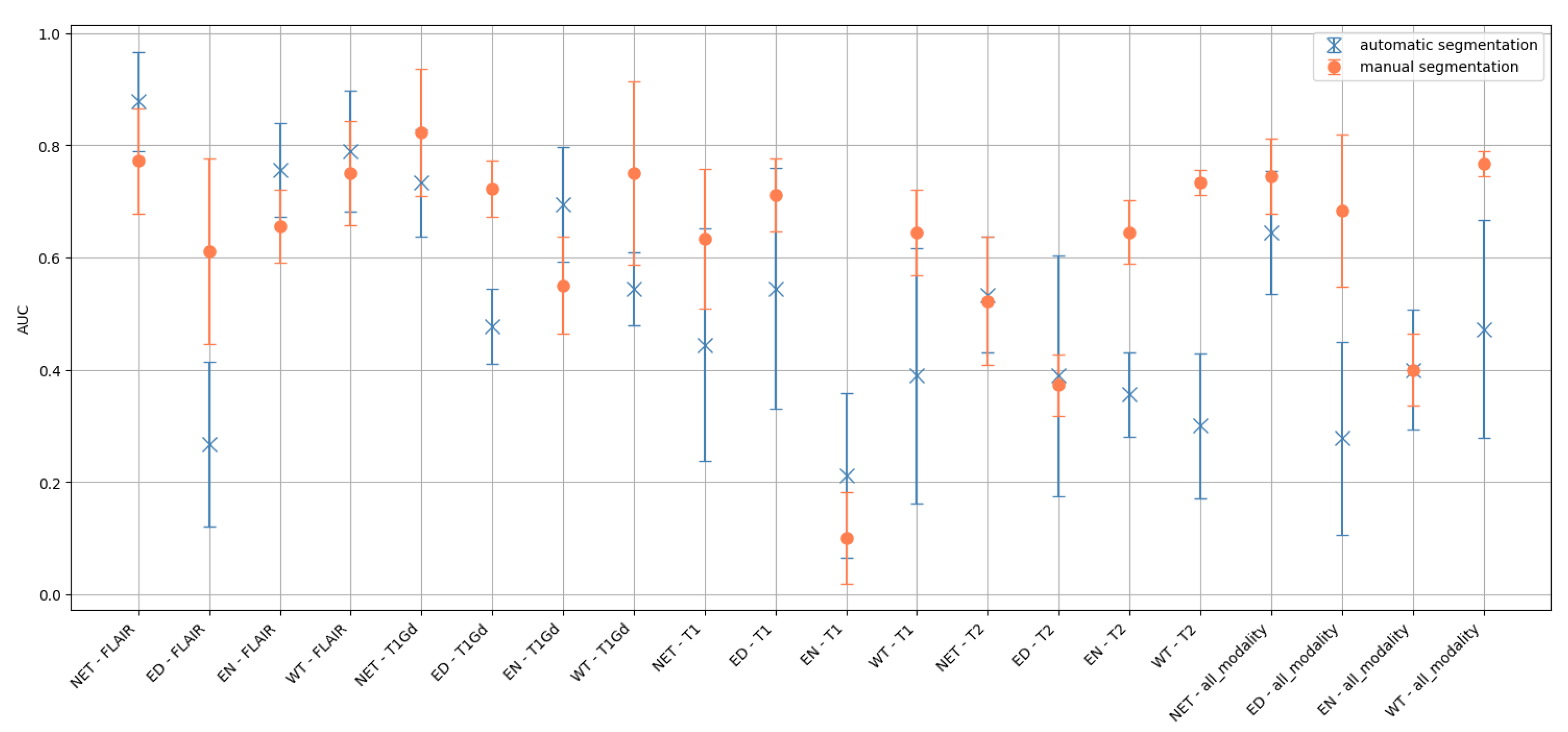
2.7. Robustness of Radiomic Features with Respect to Variations in Segmentation Masks
3. Results
3.1. Analysis to Select the More Informative MRI Sequences
3.2. MGMT Status Prediction by Multi-Input CNN Models
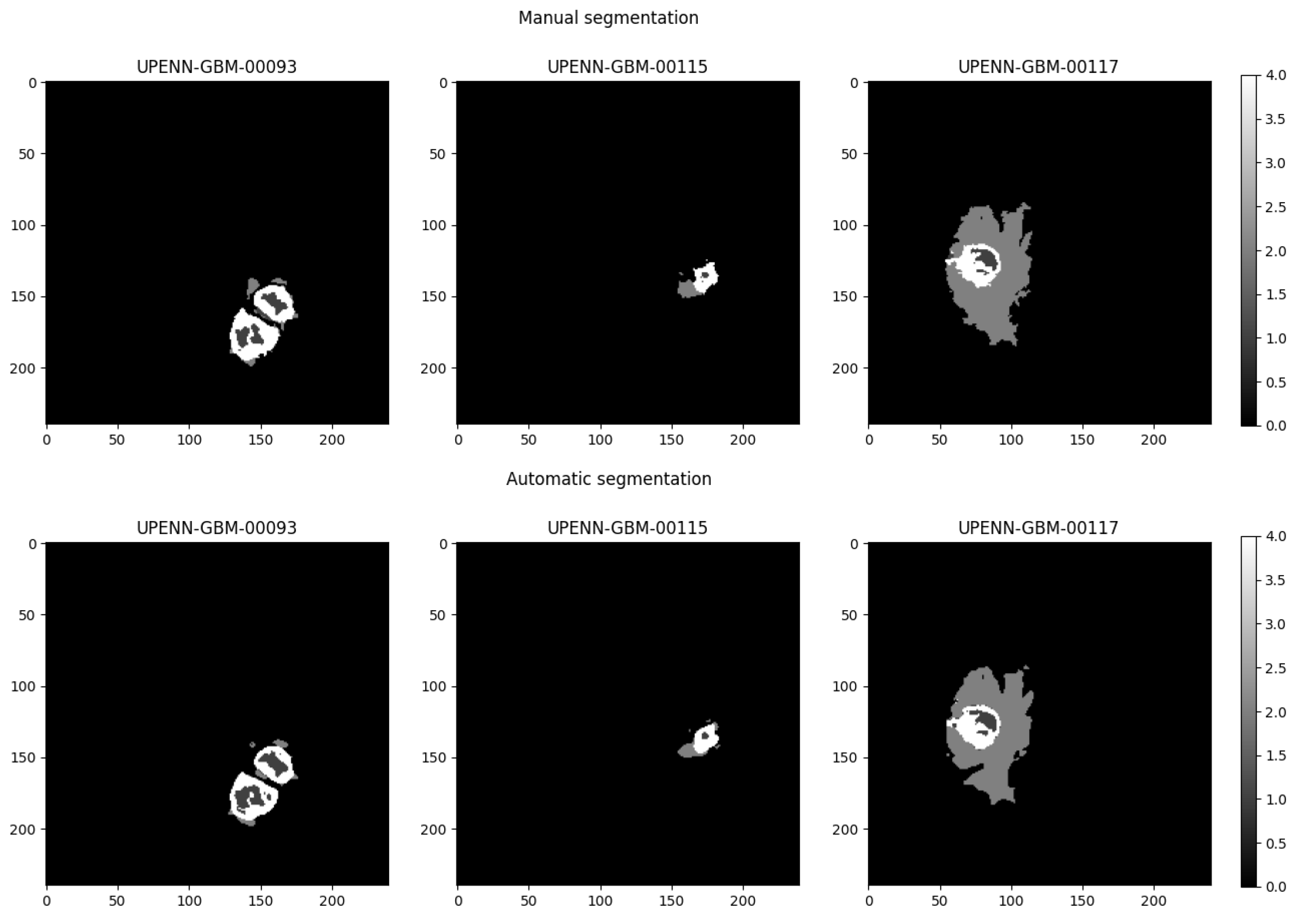
3.3. Dependence of the Classification Performance on Segmentation Quality
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanderi, T.; Munakomi, S.; Gupta, V. Glioblastoma Multiforme; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Do, D.T.; Yang, M.R.; Lam, L.H.T.; Le, N.Q.K.; Wu, Y.W. Improving MGMT methylation status prediction of glioblastoma through optimizing radiomics features using genetic algorithm-based machine learning approach. Sci. Rep. 2022, 12, 13412. [Google Scholar] [CrossRef]
- Zappe, K.; Pühringer, K.; Pflug, S.; Berger, D.; Böhm, A.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Association between MGMT Enhancer Methylation and MGMT Promoter Methylation, MGMT Protein Expression, and Overall Survival in Glioblastoma. Cells 2023, 12, 1639. [Google Scholar] [CrossRef] [PubMed]
- Brandner, S.; McAleenan, A.; Kelly, C.; Spiga, F.; Cheng, H.Y.; Dawson, S.; Schmidt, L.; Faulkner, C.L.; Wragg, C.; Jefferies, S.; et al. MGMT promoter methylation testing to predict overall survival in people with glioblastoma treated with temozolomide: A comprehensive meta-analysis based on a Cochrane Systematic Review. Neuro-Oncology 2021, 23, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Tasci, E.; Shah, Y.; Jagasia, S.; Zhuge, Y.; Shephard, J.; Johnson, M.O.; Elemento, O.; Joyce, T.; Chappidi, S.; Zgela, T.C.; et al. MGMT ProFWise: Unlocking a New Application for Combined Feature Selection and the Rank-Based Weighting Method to Link MGMT Methylation Status to Serum Protein Expression in Patients with Glioblastoma. Int. J. Mol. Sci. 2024, 25, 4082. [Google Scholar] [CrossRef] [PubMed]
- Doniselli, F.M.; Pascuzzo, R.; Mazzi, F.; Padelli, F.; Moscatelli, M.; D’Antonoli, T.A.; Cuocolo, R.; Aquino, D.; Cuccarini, V.; Sconfienza, L.M. Quality assessment of the MRI-radiomics studies for MGMT promoter methylation prediction in glioma: A systematic review and meta-analysis. Eur. Radiol. 2024, 34, 5802–5815. [Google Scholar] [CrossRef]
- Saeed, N.; Ridzuan, M.; Alasmawi, H.; Sobirov, I.; Yaqub, M. MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models. Med. Image Anal. 2023, 90, 102989. [Google Scholar] [CrossRef]
- Han, L.; Kamdar, M.R. MRI to MGMT: Predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Biocomputing 2018, 2018, 331–342. [Google Scholar] [CrossRef]
- Lu, Y.; Patel, M.; Natarajan, K.; Ughratdar, I.; Sanghera, P.; Jena, R.; Watts, C.; Sawlani, V. Machine learning-based radiomic, clinical and semantic feature analysis for predicting overall survival and MGMT promoter methylation status in patients with glioblastoma. Magn. Reson. Imaging 2020, 74, 161–170. [Google Scholar] [CrossRef]
- Restini, F.C.F.; Torfeh, T.; Aouadi, S.; Hammoud, R.; Al-Hammadi, N.; Starling, M.T.M.; Sousa, C.F.P.M.; Mancini, A.; Brito, L.H.; Yoshimoto, F.H.; et al. AI tool for predicting MGMT methylation in glioblastoma for clinical decision support in resource limited settings. Sci. Rep. 2024, 14, 27995. [Google Scholar] [CrossRef]
- Faghani, S.; Khosravi, B.; Moassefi, M.; Conte, G.M.; Erickson, B.J. A Comparison of Three Different Deep Learning-Based Models to Predict the MGMT Promoter Methylation Status in Glioblastoma Using Brain MRI. J. Digit. Imaging 2023, 36, 837–846. [Google Scholar] [CrossRef]
- Saeed, N.; Hardan, S.; Abutalip, K.; Yaqub, M. Is it Possible to Predict MGMT Promoter Methylation from Brain Tumor MRI Scans using Deep Learning Models? In Proceedings of the 5th International Conference on Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022. [Google Scholar]
- Koska, İ.Ö.; Koska, Ç. Deep learning classification of MGMT status of glioblastomas using multiparametric MRI with a novel domain knowledge augmented mask fusion approach. Sci. Rep. 2025, 15, 3273. [Google Scholar] [CrossRef]
- Bakas, S.; Sako, C.; Akbari, H.; Bilello, M.; Sotiras, A.; Shukla, G.; Rudie, J.D.; Santamaría, N.F.; Kazerooni, A.F.; Pati, S.; et al. The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: Advanced MRI, clinical, genomics, & radiomics. Sci. Data 2022, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.W.; Vendt, B.A.; Smith, K.E.; Freymann, J.B.; Kirby, J.S.; Koppel, P.; Moore, S.M.; Phillips, S.R.; Maffitt, D.R.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Ubaldi, L.; Saponaro, S.; Giuliano, A.; Talamonti, C.; Retico, A. Deriving quantitative information from multiparametric MRI via Radiomics: Evaluation of the robustness and predictive value of radiomic features in the discrimination of low-grade versus high-grade gliomas with machine learning. Phys. Medica 2023, 107, 102538. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Valliè res, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Metz, C.E. Receiver Operating Characteristic Analysis: A Tool for the Quantitative Evaluation of Observer Performance and Imaging Systems. J. Am. Coll. Radiol. 2006, 3, 413–422. [Google Scholar] [CrossRef]
- Li, Y.; Daho, M.E.H.; Conze, P.H.; Zeghlache, R.; Boité, H.L.; Tadayoni, R.; Cochener, B.; Lamard, M.; Quellec, G. A review of deep learning-based information fusion techniques for multimodal medical image classification. Comput. Biol. Med. 2024, 177, 108635. [Google Scholar] [CrossRef]
- Vallat, R. Pingouin: Statistics in Python. J. Open Source Softw. 2018, 3, 1026. [Google Scholar] [CrossRef]

| MRI Sequence | Area | AUC ± STD |
|---|---|---|
| FLAIR | NET | |
| ED | ||
| EN | ||
| WT | ||
| T1Gd | NET | |
| ED | ||
| EN | ||
| WT | ||
| T1 | NET | |
| ED | ||
| EN | ||
| WT | ||
| T2 | NET | |
| ED | ||
| EN | ||
| WT | ||
| all_modality | NET | |
| ED | ||
| EN | ||
| WT |
| DTI Sequence | Area | AUC ± STD |
|---|---|---|
| AD | NET | |
| ED | ||
| EN | ||
| WT | ||
| FA | NET | |
| ED | ||
| EN | ||
| WT | ||
| RD | NET | |
| ED | ||
| EN | ||
| WT | ||
| TR | NET | |
| ED | ||
| EN | ||
| WT | ||
| all_modality | NET | |
| ED | ||
| EN | ||
| WT |
| MRI Sequence | Area | AUC ± STD over the Folds |
|---|---|---|
| T1 | Whole Tumor | |
| No segmentation | ||
| T2 | Whole Tumor | |
| No segmentation | ||
| T1Gd | Whole Tumor | |
| No segmentation | ||
| FLAIR | Whole Tumor | |
| No segmentation |
| Input Data | AUC ± STD over the Folds |
|---|---|
| Multi-input CNN with FA and AD | |
| Multi-input CNN + MLP with FA, | |
| AD and radiomic features |
| Tumor Region | Dice Score |
|---|---|
| Whole Tumor | |
| Necrosis | |
| Edema | |
| Enhancing Tumor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lizzi, F.; Saponaro, S.; Giuliano, A.; Talamonti, C.; Ubaldi, L.; Retico, A. Radiomics and Deep Learning Interplay for Predicting MGMT Methylation in Glioblastoma: The Crucial Role of Segmentation Quality. Cancers 2025, 17, 3417. https://doi.org/10.3390/cancers17213417
Lizzi F, Saponaro S, Giuliano A, Talamonti C, Ubaldi L, Retico A. Radiomics and Deep Learning Interplay for Predicting MGMT Methylation in Glioblastoma: The Crucial Role of Segmentation Quality. Cancers. 2025; 17(21):3417. https://doi.org/10.3390/cancers17213417
Chicago/Turabian StyleLizzi, Francesca, Sara Saponaro, Alessia Giuliano, Cinzia Talamonti, Leonardo Ubaldi, and Alessandra Retico. 2025. "Radiomics and Deep Learning Interplay for Predicting MGMT Methylation in Glioblastoma: The Crucial Role of Segmentation Quality" Cancers 17, no. 21: 3417. https://doi.org/10.3390/cancers17213417
APA StyleLizzi, F., Saponaro, S., Giuliano, A., Talamonti, C., Ubaldi, L., & Retico, A. (2025). Radiomics and Deep Learning Interplay for Predicting MGMT Methylation in Glioblastoma: The Crucial Role of Segmentation Quality. Cancers, 17(21), 3417. https://doi.org/10.3390/cancers17213417






