AI-Based Characterization of Breast Cancer in Mammography and Tomosynthesis: A Review of Radiomics and Deep Learning for Subtyping, Staging, and Prognosis
Simple Summary
Abstract
1. Introduction
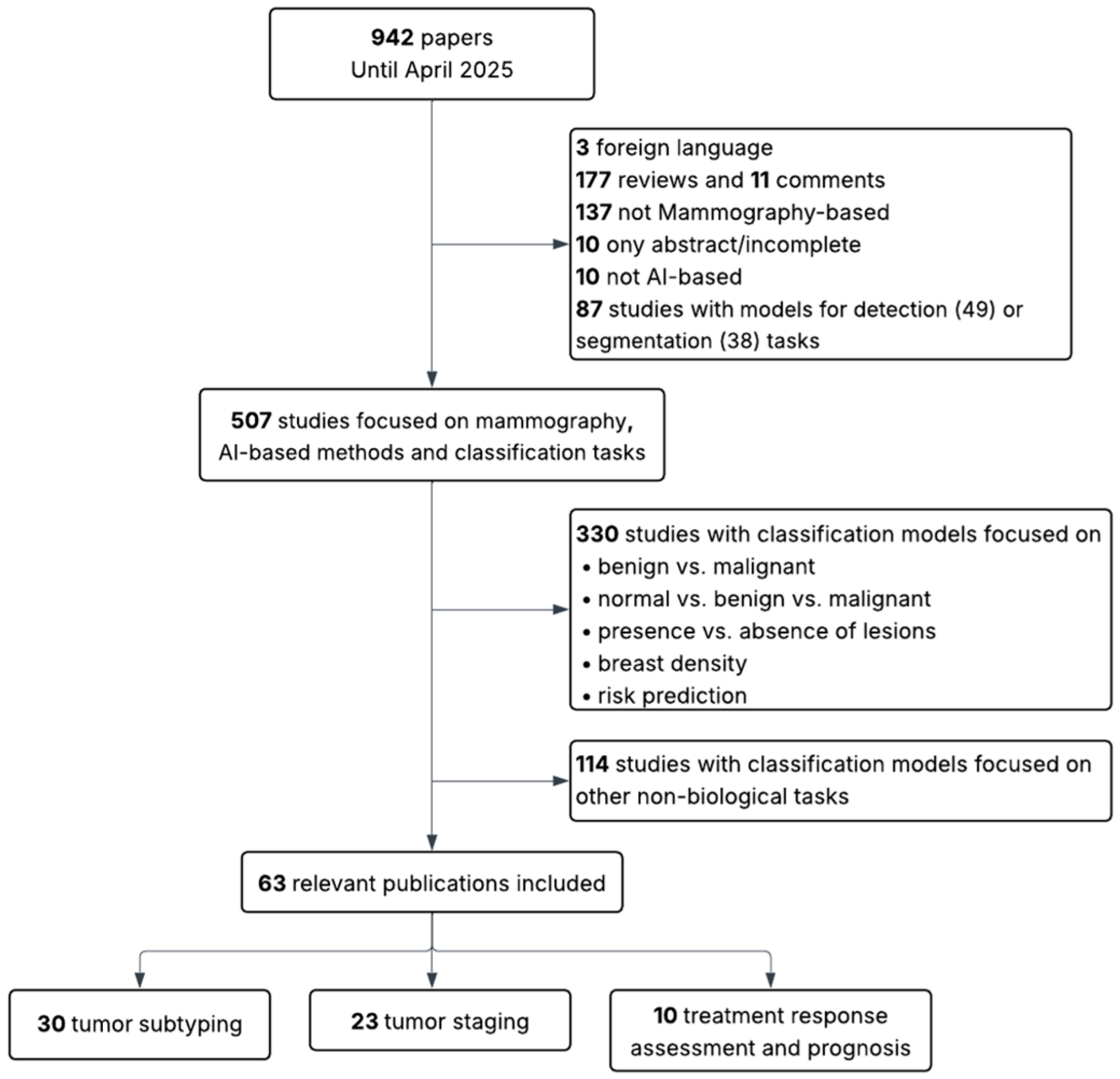
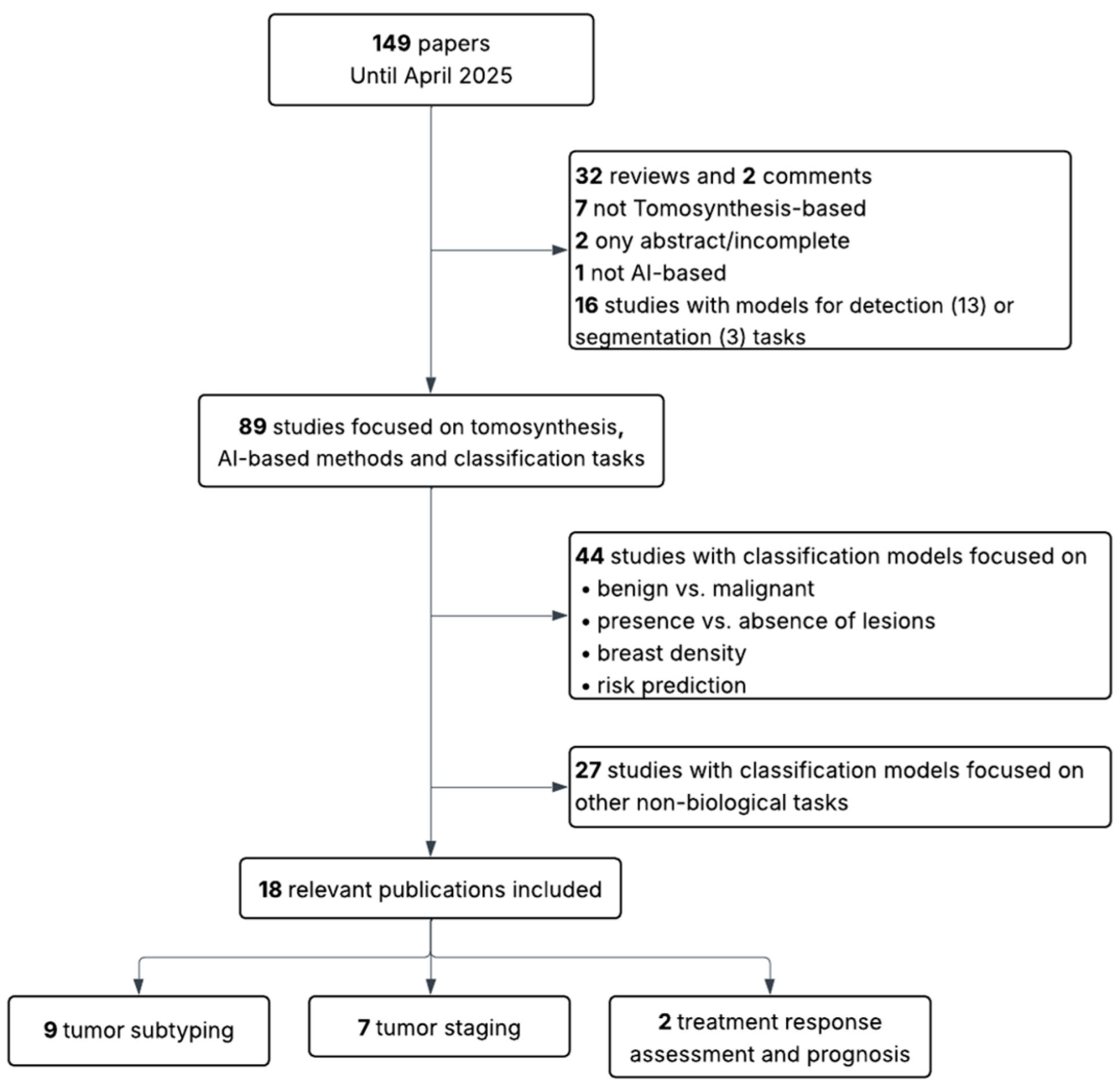
2. Methodology
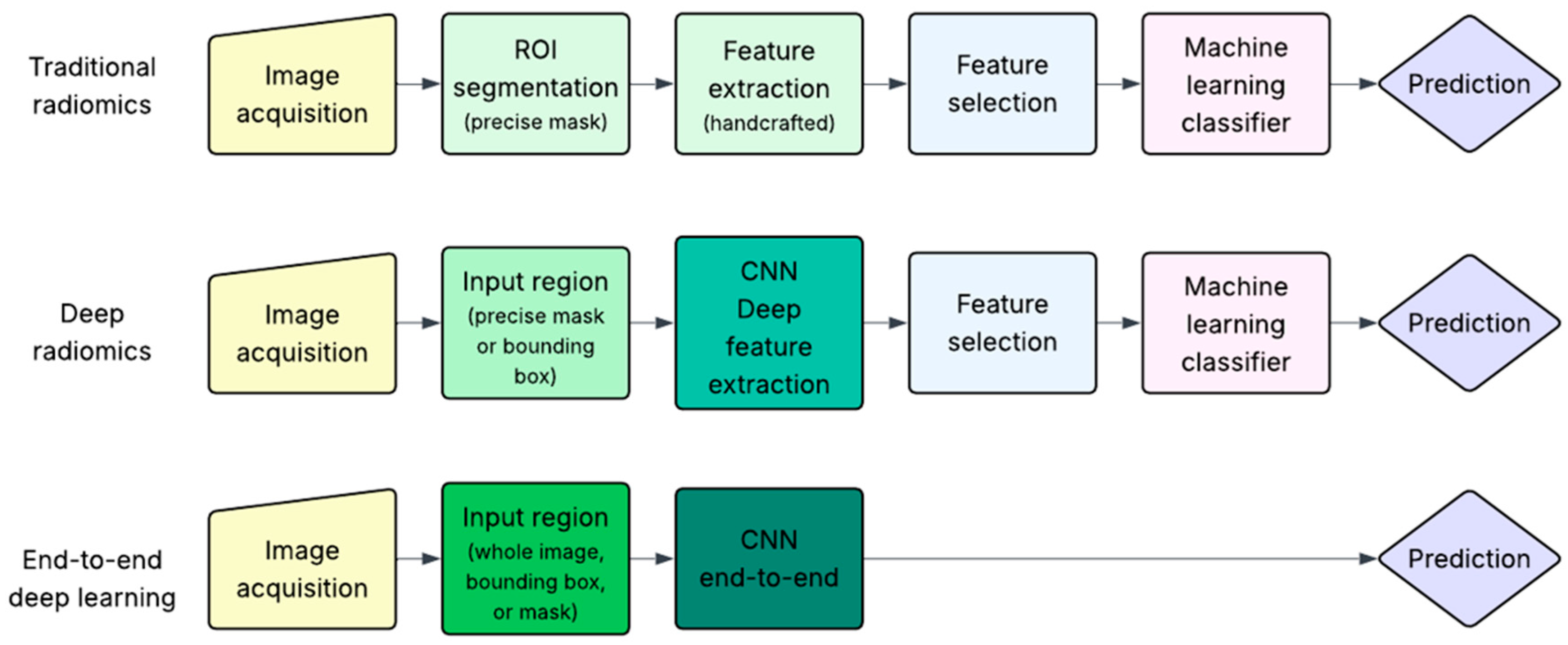
3. AI-Based Pipelines Used in Breast Cancer Characterization
4. AI-Based Characterization of Breast Cancer from Mammography and Tomosynthesis
4.1. Tumor Subtyping
4.2. Tumor Staging
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Zhou et al., 2019 [29] | Receptor status (HER2) | MM | TradRad | 306 patients | private | AUC = 0.787 (95% CI, 0.673–0.885), Acc = 0.770 |
| Ma et al., 2019 [30] | Molecular subtypes | MM | TradRad | 182 patients | private | AUC = 0.865 (Acc = 0.796) TN vs. non-TN AUC = 0.784 (Acc = 0.748) HER2-enriched vs. non-HER2-enriched AUC = 0.752 (Acc = 0.788) luminal vs. non-luminal |
| Tagliafico et al., 2019 [31] | ki-67 | BT (2D synthetic) | TradRad | 70 patients | private | AUC = 0.698 |
| La Forgia et al., 2020 [17] | Receptor status, tumor grade and ki-67 | CESM | TradRad | 52 patients | private | AUC = 0.8276 ER-positive vs. ER-negative AUC = 0.8213 PR-positive vs. PR-negative AUC = 0.8348 HER2-positive vs. HER2-negative AUC = 0.7985 ki-67+ vs. ki-67- AUC = 0.7680 High-grade vs. Low-grade AUC = 0.9089 TN vs. non-TN |
| Wang et al., 2020 [32] | Molecular subtype (TN vs. non-TN) | MM | TradRad | 112 patients | private | AUC = 0.84 (95% CI, 0.73–0.96), Acc = 0.783 |
| Marino et al., 2020 [33] | Receptor status and tumor grade | CESM | TradRad | 100 patients | private | Acc = 0.784 HR-positive vs. HR-negative Acc = 0.972 HR-positive/HER2-negative vs. HR-negative/HER2-positive Acc = 1.00 HR-positive/HER2-positive (triple positive) vs. HR-negative/HER2-negative (TN) Acc = 0.821 TN vs. HR-positive Acc = 0.900 G1 vs. G2 + G3 for invasive cancers Acc = 1.00 G1 vs. G2 + G3 for non-invasive cancers |
| Son et al., 2020 [34] | Molecular subtypes | BT (2D synthetic) | TradRad | 221 patients | private | AUC = 0.677 (95% CI, 0.552–0.802) Luminal vs. non-luminal AUC = 0.582 (95% CI, 0.361–0.804) HER2 vs. non-HER2 AUC = 0.868 (95% CI, 0.730–1.000) TN vs. non-TN |
| Kanbayti et al., 2021 [35] | Receptor status (PR) | MM | TradRad | 184 patients | private | AUC = 0.652 (95% CI, 0.560–0.744) PR status |
| Ueda et al., 2021 [36] | Receptor status | MM | End-to-end DL | 1373 images | private | AUC = 0.67 (95% CI, 0.58–0.76) ER-positive vs. ER-negative AUC = 0.61 (95% CI, 0.53–0.68) PR-positive vs. PR-negative AUC = 0.75 (95% CI, 0.68–0.82) HER2-positive vs. HER2-negative |
| Zhang et al., 2021 [21] | Molecular subtype (TN vs. non-TN) | CESM | TradRad | 367 patients | private | AUC = 0.90 (95% CI, 0.85–0.96) |
| Mao et al., 2021 [26] | Tumor grade | CESM | TradRad | 205 patients | private | AUC = 0.80 (95%CI, 0.67–0.92), Acc = 0.85 |
| Ge et al., 2022 [37] | Molecular subtype (TN vs. non-TN) | MM | TradRad | 319 patients | private | AUC = 0.809 (95% CI, 0.711–0.907), Acc = 0.806 |
| Petrillo et al., 2022 [38] | Receptor status and tumor grade | CESM | TradRad | 182 patients | private | AUC = 0.8237, Acc = 0.9167 G1 vs. G2 + G3 AUC = 0.7081, Acc = 0.8929 HER2-positive vs. HER2-negative AUC = 0.7500, Acc = 0.8500 HR-positive vs. HR-negative |
| Dominique et al., 2022 [39] | Tumor grade, ki-67, receptor status | CESM | End-to-end DL | 389 patients | private | AUC = 0.621, Acc = 0.6028 G1 + G2 vs. G3 AUC = 0.858, Acc = 0.8125 ER-positive vs. ER-negative AUC = 0.630, Acc = 0.5958 PR-positive vs. PR-negative AUC = 0.644, Acc = 0.5544 HER2-positive vs. HER2-negative AUC = 0.610, Acc = 0.5645 Ki-67 (high or not) AUC = 0.908, Acc = 0.8468 TN |
| Jiang et al., 2022 [40] | ki-67 | BT (slice-based) | TradRad and DeepRad | 266 patients | private | AUC = 0.792 (95% CI, 0.647–0.897), Acc = 0.652 (95% CI, 0.404–0.913) |
| Zhang et al., 2023 [18] | Receptor status (ER and PR) | MM | End-to-end DL | 2321 patients | private | AUC = 0.921, Acc = 0.802 |
| Zhu et al., 2023 [22] | Molecular subtypes | CESM | TradRad | 356 patients | private | AUC = 0.82 (95% CI, 0.69–0.92), Acc = 0.72 Luminal vs. Non-Luminal AUC = 0.83 (95% CI, 0.70–0.94), Acc = 0.83 HER2-enriched vs. Non-HER2-enriched AUC = 0.68 (95% CI, 0.47–0.86), Acc = 0.75 TN vs. Non-TN |
| Rong et al., 2023 [41] | Tumor grade | MM | TradRad | 534 patients | private | AUC = 0.75 (95% CI, 0.66–0.83), Acc = 0.68 G3 vs. G1 + G2 |
| Deng et al., 2024 [42] | Receptor status (HER2) | MM | TradRad | 1512 patients | private | AUC = 0.724 (95% CI, 0.637–0.811), Acc = 0.722 |
| Duan et al., 2024 [19] | Receptor status (ER) | MM | End-to-end DL | 358 patients | private | AUC = 0.886 (95% CI, 0.809–0.934), Acc = 0.831 |
| Petrillo et al., 2024 [27] | Receptor status, molecular subtype (luminal vs. non-luminal) and tumor grade | CESM | TradRad | 169 patients | private | Acc = 0.6071 HER2-positive Acc = 0.6786 HR-positive Acc = 0.8205 grade Acc = 0.9375 luminal vs. non-luminal |
| Mota et al., 2024 [43] | Molecular subtypes | MM | End-to-end DL | 660 patients | semiPublic—OPTIMAM | AUC = 0.6599, Acc = 0.7372 Luminal A vs. non-Luminal A AUC = 0.6545, Acc = 0.7491 Luminal B1 vs. non-Luminal B1 AUC = 0.6530, Acc = 0.6284 HER2 vs. non-HER2, AUC = 0.6445, Acc = 0.7706 TN vs. non-TN Multiclassification: AUC = 0.5992 Luminal A AUC = 0.5981 Luminal B1 AUC = 0.5920 Luminal B2 AUC = 0.6440 HER2 AUC = 0.6089 TN |
| Bakker et al., 2024 [44] | Molecular subtypes | MM | TradRad | 186 patients | semipublic-OPTIMAM | AUC = 0.855 (95% CI, 0.779–0.930), Acc = 0.815 luminal A AUC = 0.812 (95% CI, 0.736–0.889), Acc = 0.734 luminal B AUC = 0.755 (95% CI, 0.644–0.867), Acc = 0.637 HER2 AUC = 0.789 (95% CI, 0.701–0.878), Acc = 0.718 TN |
| Nissar et al., 2024 [45] | Molecular subtypes | MM | End-to-end DL | 749 patients | public—CMMD | Subtype classification (4-class: Luminal A, Luminal B, HER2-enriched, TN): Acc = 0.90 (mass-based) Acc = 0.90 (calcification-based) Acc = 0.92 (combined mass + calcification) |
| Oba et al., 2024 [24] | ki-67 | BT (slice-based) | End-to-end DL | 126 patients | private | AUC = 0.883, Acc = 0.912 AUC = 0.890 mass AUC = 0.750 calcification AUC = 0.870 distortion AUC = 0.660 focal asymmetric density |
| Zeng et al., 2025 [46] | Receptor status | MM | End-to-end DL | 352 patients | private | AUC = 0.708 (95% CI, 0.609–0.808), Acc = 0.651 HER2 status AUC = 0.785 (95% CI, 0.673–0.897), Acc = 0.845 ER status AUC = 0.706 (95% CI, 0.603–0.809), Acc = 0.678 PR status |
| Rabah et al., 2025 [47] | Molecular subtypes | MM | End-to-end DL | 1775 patients (public) | public—CMMD | AUC = 0.8887, Acc = 0.6379 (multiclass classification) AUC = 0.67 Luminal A vs. Others AUC = 0.74 Luminal B vs. Others AUC = 0.78 HER2 vs. Others AUC = 0.78 TN vs. Others |
| Hu et al., 2025 [48] | ki-67 | BT (slice-based) | TradRad | 289 patients | private | AUC = 0.755 (95% CI, 0.629–0.880), Acc = 0.782 |
| Mota et al., 2025 [49] | Molecular subtypes | BT (slice-based) | End-to-end DL | 453 images | semiPublic—OPTIMAM | AUC = 0.5928 Luminal B2 vs. non-Luminal B2 AUC = 0.7317 HER2+ vs. non-HER2+ AUC = 0.6522 TN vs. non-TN |
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Marino et al., 2020 [28] | Receptor status and tumor grade | CESM and MRI | TradRad | 48 patients | private | CESM: Acc = 0.956 HR positive vs. HR negative Acc = 0.778 G1 + G2 vs. G3 invasive cancers MRI: Acc = 0.826 HR positive vs. HR negative Acc = 0.778 G1 + G2 vs. G3 cancers invasive cancers |
| Jiang et al., 2022 [25] | ki-67 | MM and MRI and BT (slice-based) | TradRad | 209 patients | private | AUC = 0.866 |
| Niu et al., 2022 [23] | Molecular subtypes | MM and MRI and BT (slice-based) | TradRad | 241 patients | private | AUC = 0.773 (95% CI, 0.650–0.895) Luminal A (MM + BT) AUC = 0.747 (0.578–0.917) Luminal A (DW + DCE) AUC = 0.807 (0.706–0.908) Luminal B (MM + BT) AUC = 0.784 (0.675–0.894) Luminal B (DW + DCE) AUC = 0.802 (0.658–0.946) HER2 (MM + BT) AUC = 0.877 (0.795–0.959) HER2 (DW + DCE) AUC = 0.874 (0.759–0.990) TN (MM + BT) AUC = 0.938 (0.883–0.992) TN (DW + DCE) |
| Liu et al., 2023 [50] | Molecular subtype (Luminal A vs. non-Luminal A) | MM and MRI | End-to-end DL | 158 patients | private | AUC = 0.802 (95% CI, 0.657–0.906), Acc = 0.711 (MM + MRI) AUC = 0.593 (95%CI, 0.436–0.737), Acc = 0.533 (only MM) |
| Zhang et al., 2023 [51] | Molecular subtypes | MM and US | End-to-end DL | 4162 images | private | AUC = 0.929 (95% CI, 0.903, 0.951), Acc = 88.5 (86.0–90.9) Luminal (Luminal A and Luminal B) vs. Non-Luminal (HER2-enriched and TN) |
| Liu et al., 2024 [52] | ki-67 | BT (slice-based) and US | TradRad | 149 patients | private | AUC = 0.818 (95% CI, 0.685–0.950) |
| Wang et al., 2025 [20] | Receptor status (HER2) | MM and MRI | TradRad and DeepRad | 550 patients (private) | private and public (4validation)—Duke, CMMD | AUC = 0.824 (95% CI, 0.749–0.884), Acc = 0.8074, HER2-Positive vs. HER2 Zero/Low AUC = 0.811 (95% CI, 0.735–0.874), Acc = 0. 7926, HER2-Zero vs. HER2-Low/Positive Acc = 0.8444 (HER2-zero) Acc = 0.8000 (HER2-low) Acc = 0.8815 (HER2-positive) |
| Yang et al., 2025 [53] | Molecular subtypes | MM and MRI | TradRad | 243 patients | private | AUC = 0.648, Acc = 0.627 Luminal A vs. Luminal B AUC = 0.819, Acc = 0.793 luminal A vs. HER2-enriched AUC = 0.725, Acc = 0.696 luminal A vs. TN AUC = 0.644, Acc = 0.560 luminal B vs. HER2-enriched AUC = 0.625, Acc = 0.636 luminal B vs. TN AUC = 0.598, Acc = 0.500 TN vs. HER2-enriched |
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Shi et al., 2018 [63] | Pure DCIS vs. DCIS with invasion | MM | TradRad and DeepRad | 99 patients | private | TradRad: AUC = 0.68 (95% CI, 0.66–0.71) DeepRad: AUC = 0.70 (95% CI, 0.68–0.73) |
| Yang et al., 2019 [64] | ALN metastasis | MM | TradRad | 147 patients | private | AUC = 0.875 (95% CI, 0.698–0.891), Acc = 0.800 (95% CI, 66.4–83.2%) |
| Li et al., 2019 [65] | Pure DCIS vs. DCIS with invasion | MM | TradRad | 362 patients | private | AUC = 0.72 (95% CI, 0.63–0.81) |
| Tan et al., 2020 [66] | ALN metastasis | MM | TradRad | 216 patients | private | AUC = 0.863 (95% CI, 0.821–0.897), Acc = 0.7917 |
| Mao et al., 2020 [67] | ALN metastasis | CESM | TradRad | 394 patients | private | AUC = 0.767 (95% CI, 0.583–0.857) internal validation AUC = 0.790 (95% CI, 0.63–0.94) external validation |
| Marino et al., 2020 [33] | Pure DCIS vs. DCIS with invasion | CESM | TradRad | 100 patients | private | Acc = 0.874 invasive vs. noninvasive cancer |
| Kanbayti et al., 2021 [35] | ALN metastasis | MM | TradRad | 184 patients | private | AUC = 0.681 (95% CI, 0.559–0.804) |
| Wu et al., 2022 [68] | ALN metastasis | CESM | DeepRad | 182 patients | private | for non-sentinel lymph node metastasis status: AUC = 0.85 (95% CI, 0.71–0.99), Acc = 0.81 (95% CI, 0.63–0.93) testing AUC = 0.82 (95% CI, 0.67–0.97), Acc = 0.74 (95% CI, 0.55–0.88) temporal validation axillary tumor burden: AUC = 0.82 (95% CI, 0.66–0.97), Acc = 0.75 (95% CI, 0.55–0.88) testing AUC = 0.77 (95% CI, 0.62–0.93), Acc = 0.74 (95% CI, 0.55–0.88) temporal validation |
| Abel et al., 2022 [57] | ALN metastasis | MM | End-to-end DL | 74 patients | private | Acc = 0.9596 |
| Hou et al., 2022 [69] | Pure DCIS vs. DCIS with invasion | MM | TradRad | 700 patients | private | AUC = 0.71 (95% CI, 0.62–0.79) |
| Wang et al., 2022 [61] | Lymphovascular Invasion | BT (volume-based) | TradRad | 135 patients | private | AUC = 0.835 (95% CI, 0.712–0.958) |
| Lin et al., 2023 [70] | ALN metastasis | CESM | TradRad | 365 patients | private | AUC = 0.7567 (95% CI, 0.6717–0.8678), Acc = 0.7551 (0.6113–0.8666) |
| Wang et al., 2023 [71] | ALN metastasis | CESM | TradRad | 809 patients | private | AUC = 0.732, Acc = 0.681 |
| Xu et al., 2023 [58] | ALN metastasis | BT (slice-based) | TradRad | 120 patients | private | AUC = 0.920 (95% CI, 0.806–1.000), Acc = 0.833 |
| Shimokawa et al., 2023 [72] | Stromal invasion/pure DCIS vs. DCIS with invasion | BT (slice-based) | End-to-end DL | 140 patients | private | AUC = 0.75 (95% CI, 0.69–0.81) |
| Tsai et al., 2024 [73] | Pure DCIS vs. DCIS with invasion | MM | End-to-end DL | 1436 patients | private | AUC = 0.747 (95% CI, 0.677–0.813) |
| Alaeikhanehshir et al., 2024 [54] | Pure DCIS vs. DCIS with invasion | MM | End-to-end DL | 464 patients | private | AUC = 0.72 low-risk DCIS vs. high risk AUC = 0.76 low-risk DCIS vs. high-risk and/or upstaged DCIS (occult IBC) |
| Xu et al., 2024 [62] | Lymphovascular Invasion | BT (slice-based) | TradRad | 178 patients | private | AUC = 0.905 (95% CI, 0.823–0.986), Acc = 0.815 |
| He et al., 2025 [74] | ALN metastasis | BT (volume-based) | TradRad | 536 patients | private | AUC = 0.792, Acc = 0.726 |
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Marino et al., 2020 [28] | Pure DCIS vs. DCIS with invasion | CESM + MRI | TradRad | 48 patients | private | CESM Acc = 0.92 MRI Acc = 0.90 |
| Cheng et al., 2022 [75] | ALN metastasis | MM + MRI + BT (slice-based) | TradRad | 208 patients | private | AUC = 0.786, Acc = 0.771 (MM + BT) AUC = 0.826, Acc = 0.829 (DCE-MRI + DWI) |
| Haraguchi et al., 2023 [76] | ALN metastasis | MM + BT (2D synthetic) | TradRad | 77 patients | private | AUC = 0.738 (95% CI, 0.608–0.867), MM AUC = 0.742 (95% CI, 0.613–0.871), BT (2D synthetic) |
| Wang et al., 2024 [59] | ALN metastasis | MM + MRI | TradRad | 485 patients | private | AUC = 0.892 (95% CI, 0.826–0.939), Acc = 0.8182 |
| Hua et al., 2024 [60] | ALN metastasis | MM + MRI | TradRad | 492 patients | private | AUC = 0.902 (95% CI, 0.833–0.972), Acc = 0.847 |
| Guo et al., 2024 [77] | ALN metastasis | MM + MRI | TradRad and DeepRad | 270 patients | private | AUC = 0.846, Acc = 0.765 |
| Wu et al., 2024 [55] | Pure DCIS vs. DCIS with invasion | MM + US | End-to-end DL | 733 patients | private | AUC = 0.859–0.907, Acc = 0.752–0.766 in low-grade DCIS vs. intermediate-to-high-grade DCIS vs. upstaged DCIS AUC = 0.829–0.861, Acc = 0.751–0.780 in DCIS vs. upstaged DCIS AUC = 0.769–0.987, Acc = 0.818–0.939 in low-grade DCIS vs. upstaged low-grade DCIS |
| Cheng et al., 2025 [78] | ALN metastasis | MM + MRI | TradRad and End-to-end DL | 326 patients | private | AUC = 0.877, Acc = 0.727 |
| Wu et al., 2025 [56] | Pure DCIS vs. DCIS with invasion | MM + US | TradRad | 237 patients | private | AUC = 0.92 (95% CI, 0.84–0.96), Acc = 0.88 low nuclear grade DCIS vs. Intermediate to high nuclear grade |
4.3. Treatment Response and Prognosis
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Jiang et al., 2020 [82] | Prognostic/recurrence prediction | MM | TradRad | 200 patients | private | C-index = 0.944 (95% CI, 0.883–1.004) for predicting iDFS (invasive disease-free survival) |
| Wang et al., 2021 [86] | pCR to neoadjuvant chemotherapy | CESM | TradRad | 117 patients | private | AUC = 0.81 (95% CI, 0.575–0.948), Acc = 0.80 |
| Mao et al., 2021 [83] | Prognostic/recurrence prediction | MM | TradRad | 304 patients | private | AUC = 0.88 (95% CI, 0.75–1.00) internal test set AUC = 0.84 (95% CI, 0.69–0.99) external test set |
| Yu et al., 2021 [85] | Tumor-infiltrating lymphocyte | MM | TradRad | 121 patients | private | AUC = 0.79 (95% CI, 0.615–0.964), Acc = 0.639 |
| Skarping et al., 2022 [87] | pCR to neoadjuvant chemotherapy | MM | End-to-end DL | 453 patients | private | AUC = 0.71 (95% CI, 0.53–0.90) |
| Mao et al., 2022 [79] | pCR to neoadjuvant chemotherapy | CESM | TradRad | 118 patients | private | AUC = 0.85 (95% CI, 0.72–0.98) |
| Zhang et al., 2023 [88] | pCR to neoadjuvant chemotherapy | CESM | TradRad | 118 patients | private | AUC = 0.790 (95% CI, 0.554–0.952), Acc = 0.861 |
| Xing et al., 2024 [80] | pCR to neoadjuvant chemotherapy | CESM | End-to-end DL | 265 patients | private | AUC = 0.95 (95% CI, 0.847–1.0), Acc = 0.94 |
| Förnvik et al., 2024 [81] | pCR to neoadjuvant chemotherapy | BT (volume-based) | End-to-end DL | 149 patients | private | AUC = 0.83 (95% CI, 0.63–1.00) |
| Study | Clinical Target | Modality | AI Approach | No. of Cases | Dataset | Performance Metric |
|---|---|---|---|---|---|---|
| Han et al., 2024 [89] | Prognostic/recurrence prediction | MM + US | End-to-end DL | 1242 patients | private | AUC = 0.739 (95% CI, 0.608–0.871), Acc = 0.798 (95% CI, 0.797–0.800) disease-free survival |
| Ma et al., 2024 [84] | Prognostic/recurrence prediction | MM + MRI | TradRad | 131 patients | private | AUC = 0.95 (95% CI, 0.92–0.98), Acc = 0.923 disease-free survival |
| Cai et al., 2024 [90] | pCR to neoadjuvant chemotherapy | BT (slice-based) + US | TradRad | 720 patients | private | AUC = 0.81 (95% CI, 0.75–0.87), Acc =0. 764 |
5. Conclusions
Funding
Conflicts of Interest
Abbreviations
| MM | mammography |
| BT | breast tomosynthesis |
| US | breast ultrasound |
| MRI | magnetic resonance imaging |
| AI | artificial intelligence |
| CESM | contrast-enhanced spectral mammography |
| DL | deep learning |
| CNN | convolutional neural networks |
| ROI | region of interest |
| ALN | axillary lymph node |
| pCR | pathological complete response |
| ER | estrogen receptor |
| PR | progesterone receptor |
| HER2 | human epidermal growth factor receptor 2 |
| IHC | immunohistochemistry |
| TN | triple-negative |
| DCIS | ductal carcinoma in situ |
References
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2021, 95, 20211033. [Google Scholar] [CrossRef]
- Tabár, L.; Dean, P.B.; Chen, T.H.-H.; Yen, A.M.-F.; Chen, S.L.-S.; Fann, J.C.-Y.; Chiu, S.Y.-H.; Ku, M.M.-S.; Wu, W.Y.-Y.; Hsu, C.-Y.; et al. The incidence of fatal breast cancer measures the increased effectiveness of therapy in women participating in mammography screening. Cancer 2019, 125, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Stanislaw, S.; Spain, L.; Gallegos, L.L.; Rowan, A.; Schnidrig, D.; Rosenbaum, H.; Harle, A.; Au, L.; Hill, S.M.; et al. Representative Sequencing: Unbiased Sampling of Solid Tumor Tissue. Cell Rep. 2020, 31, 107550. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, H.; Sun, Y. Effect of acute inflammatory reaction induced by biopsy on tumor microenvironment. J. Cancer Res. Clin. Oncol. 2024, 150, 177. [Google Scholar] [CrossRef]
- Crouigneau, R.; Li, Y.-F.; Auxillos, J.; Goncalves-Alves, E.; Marie, R.; Sandelin, A.; Pedersen, S.F. Mimicking and analyzing the tumor microenvironment. Cell Rep. Methods 2024, 4, 100866. [Google Scholar] [CrossRef]
- Bareche, Y.; Buisseret, L.; Gruosso, T.; Girard, E.; Venet, D.; Dupont, F.; Desmedt, C.; Larsimont, D.; Park, M.; Rothé, F.; et al. Unraveling Triple-Negative Breast Cancer Tumor Microenvironment Heterogeneity: Towards an Optimized Treatment Approach. J. Natl. Cancer Inst. 2020, 112, 708–719. [Google Scholar] [CrossRef]
- Kumar Yadav, S.; Sharma, D.; Bala Sharma, D.; Kintu-Luwaga, R.; Jha, C.K.; Shekhar, S. Barriers and challenges in providing standard breast cancer care in low resource settings. Trop. Dr. 2022, 52, 532–537. [Google Scholar] [CrossRef]
- Zackrisson, S.; Lång, K.; Rosso, A.; Johnson, K.; Dustler, M.; Förnvik, D.; Förnvik, H.; Sartor, H.; Timberg, P.; Tingberg, A.; et al. One-view breast tomosynthesis versus two-view mammography in the Malmö Breast Tomosynthesis Screening Trial (MBTST): A prospective, population-based, diagnostic accuracy study. Lancet Oncol. 2018, 19, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Hofvind, S.; Hovda, T.; Holen, Å.S.; Lee, C.I.; Albertsen, J.; Bjørndal, H.; Brandal, S.H.B.; Gullien, R.; Lømo, J.; Park, D.; et al. Digital Breast Tomosynthesis and Synthetic 2D Mammography versus Digital Mammography: Evaluation in a Population-based Screening Program. Radiology 2018, 287, 787–794. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Al-Karawi, D.; Al-Zaidi, S.; Helael, K.A.; Obeidat, N.; Mouhsen, A.M.; Ajam, T.; Alshalabi, B.A.; Salman, M.; Ahmed, M.H. A Review of Artificial Intelligence in Breast Imaging. Tomography 2024, 10, 705–726. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.M.; Yin, M.; Yu, M.H.; Yu, J.; Zeng, S.E.; Lv, W.Z.; Li, J.; Ye, H.R.; Cui, X.W.; Dietrich, C.F. Artificial Intelligence in Medical Imaging of the Breast. Front. Oncol. 2021, 11, 600557. [Google Scholar] [CrossRef] [PubMed]
- Elahi, R.; Nazari, M. An updated overview of radiomics-based artificial intelligence (AI) methods in breast cancer screening and diagnosis. Radiol. Phys. Technol. 2024, 17, 795–818. [Google Scholar] [CrossRef]
- Hussain, S.; Lafarga-Osuna, Y.; Ali, M.; Naseem, U.; Ahmed, M.; Tamez-Peña, J.G. Deep learning, radiomics and radiogenomics applications in the digital breast tomosynthesis: A systematic review. BMC Bioinform. 2023, 24, 401. [Google Scholar] [CrossRef]
- La Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic Analysis in Contrast-Enhanced Spectral Mammography for Predicting Breast Cancer Histological Outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, C.; Cai, L.; Zhao, J.; Xu, Y.; Xing, J.; Sun, J.; Zhang, Y. Developing a weakly supervised deep learning framework for breast cancer diagnosis with HR status based on mammography images. Comput. Struct. Biotechnol. J. 2023, 22, 17–26. [Google Scholar] [CrossRef]
- Duan, W.; Wu, Z.; Zhu, H.; Zhu, Z.; Liu, X.; Shu, Y.; Zhu, X.; Wu, J.; Peng, D. Deep learning modeling using mammography images for predicting estrogen receptor status in breast cancer. Am. J. Transl. Res. 2024, 16, 2411–2422. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Z.Q.; Huang, C.C.; Xue, H.W.; Zhang, H.; Bo, F.; Guan, W.T.; Zhou, W.; Bai, G.J. Dual-Modality Virtual Biopsy System Integrating MRI and MG for Noninvasive Predicting HER2 Status in Breast Cancer. Acad Radiol. 2025, 32, 3858–3869. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Zhang, H.; Ma, H.; Sun, J.; Zhang, R.; Song, L.; Shi, H. Diagnostic Value of Radiomics Analysis in Contrast-Enhanced Spectral Mammography for Identifying Triple-Negative Breast Cancer. Front. Oncol. 2021, 11, 773196. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, S.; Guo, S.; Wu, R.; Zhang, J.; Kong, M.; Pan, L.; Gu, Y.; Yu, S. Contrast-Enhanced Mammography Radiomics Analysis for Preoperative Prediction of Breast Cancer Molecular Subtypes. Acad. Radiol. 2023, 31, 2228–2238. [Google Scholar] [CrossRef]
- Niu, S.; Jiang, W.; Zhao, N.; Jiang, T.; Dong, Y.; Luo, Y.; Yu, T.; Jiang, X. Intra- and peritumoral radiomics on assessment of breast cancer molecular subtypes based on mammography and MRI. J. Cancer Res. Clin. Oncol. 2022, 148, 97–106. [Google Scholar] [CrossRef]
- Oba, K.; Adachi, M.; Kobayashi, T.; Takaya, E.; Shimokawa, D.; Fukuda, T.; Takahashi, K.; Yagishita, K.; Ueda, T.; Tsunoda, H. Deep learning model to predict Ki-67 expression of breast cancer using digital breast tomosynthesis. Breast Cancer 2024. [Google Scholar] [CrossRef]
- Jiang, T.; Song, J.; Wang, X.; Niu, S.; Zhao, N.; Dong, Y.; Wang, X.; Luo, Y.; Jiang, X. Intratumoral and Peritumoral Analysis of Mammography, Tomosynthesis, and Multiparametric MRI for Predicting Ki-67 Level in Breast Cancer: A Radiomics-Based Study. Mol. Imaging Biol. 2022, 24, 550–559. [Google Scholar] [CrossRef]
- Mao, N.; Jiao, Z.; Duan, S.; Xu, C.; Xie, H. Preoperative prediction of histologic grade in invasive breast cancer by using contrast-enhanced spectral mammography-based radiomics. J. X-Ray Sci. Technol. 2021, 29, 763–772. [Google Scholar] [CrossRef]
- Petrillo, A.; Fusco, R.; Petrosino, T.; Vallone, P.; Granata, V.; Rubulotta, M.R.; Pariante, P.; Raiano, N.; Scognamiglio, G.; Fanizzi, A.; et al. A multicentric study of radiomics and artificial intelligence analysis on contrast-enhanced mammography to identify different histotypes of breast cancer. La Radiol. Medica 2024, 129, 864–878. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.A.; Leithner, D.; Sung, J.; Avendano, D.; Morris, E.A.; Pinker, K.; Jochelson, M.S. Radiomics for Tumor Characterization in Breast Cancer Patients: A Feasibility Study Comparing Contrast-Enhanced Mammography and Magnetic Resonance Imaging. Diagnostics 2020, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tan, H.; Bai, Y.; Li, J.; Lu, Q.; Chen, R.; Zhang, M.; Feng, Q.; Wang, M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019, 121, 108718. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhao, Y.; Ji, Y.; Guo, X.; Jian, X.; Liu, P.; Wu, S. Breast Cancer Molecular Subtype Prediction by Mammographic Radiomic Features. Acad Radiol. 2019, 26, 196–201. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Bignotti, B.; Rossi, F.; Matos, J.; Calabrese, M.; Valdora, F.; Houssami, N. Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur. Radiol. Exp. 2019, 3, 36. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, W.; Xie, X.; Liu, W.; Wang, H.; Shen, J.; Ding, Y.; Zhang, B.; Song, B. Application of digital mammography-based radiomics in the differentiation of benign and malignant round-like breast tumors and the prediction of molecular subtypes. Gland Surg. 2020, 9, 2005–2016. [Google Scholar] [CrossRef]
- Marino, M.A.; Pinker, K.; Leithner, D.; Sung, J.; Avendano, D.; Morris, E.A.; Jochelson, M. Contrast-Enhanced Mammography and Radiomics Analysis for Noninvasive Breast Cancer Characterization: Initial Results. Mol. Imaging Biol. 2020, 22, 780–787. [Google Scholar] [CrossRef]
- Son, J.; Lee, S.E.; Kim, E.-K.; Kim, S. Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from digital breast tomosynthesis. Sci. Rep. 2020, 10, 21566. [Google Scholar] [CrossRef]
- Kanbayti, I.H.; Rae, W.I.D.; McEntee, M.F.; Gandomkar, Z.; Ekpo, E.U. Clinicopathologic breast cancer characteristics: Predictions using global textural features of the ipsilateral breast mammogram. Radiol. Phys. Technol. 2021, 14, 248–261. [Google Scholar] [CrossRef]
- Ueda, D.; Yamamoto, A.; Takashima, T.; Onoda, N.; Noda, S.; Kashiwagi, S.; Morisaki, T.; Honjo, T.; Shimazaki, A.; Miki, Y. Training, Validation, and Test of Deep Learning Models for Classification of Receptor Expressions in Breast Cancers From Mammograms. JCO Precis. Oncol. 2021, 5, 543–551. [Google Scholar] [CrossRef]
- Ge, S.; Yixing, Y.; Jia, D.; Ling, Y. Application of mammography-based radiomics signature for preoperative prediction of triple-negative breast cancer. BMC Med. Imaging 2022, 22, 166. [Google Scholar] [CrossRef]
- Petrillo, A.; Fusco, R.; Di Bernardo, E.; Petrosino, T.; Barretta, M.L.; Porto, A.; Granata, V.; Di Bonito, M.; Fanizzi, A.; Massafra, R.; et al. Prediction of Breast Cancer Histological Outcome by Radiomics and Artificial Intelligence Analysis in Contrast-Enhanced Mammography. Cancers 2022, 14, 2132. [Google Scholar] [CrossRef]
- Dominique, C.; Callonnec, F.; Berghian, A.; Defta, D.; Vera, P.; Modzelewski, R.; Decazes, P. Deep learning analysis of contrast-enhanced spectral mammography to determine histoprognostic factors of malignant breast tumours. Eur. Radiol. 2022, 32, 4834–4844. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Jiang, W.; Chang, S.; Wang, H.; Niu, S.; Yue, Z.; Yang, H.; Wang, X.; Zhao, N.; Fang, S.; et al. Intratumoral analysis of digital breast tomosynthesis for predicting the Ki-67 level in breast cancer: A multi-center radiomics study. Med. Phys. 2022, 49, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.-C.; Kang, Y.-H.; Shi, G.-F.; Ren, J.-L.; Liu, Y.-H.; Li, Z.-G.; Yang, G. The use of mammography-based radiomics nomograms for the preoperative prediction of the histological grade of invasive ductal carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 11635–11645. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, Y.; Li, X.; Zhu, Y.; Zhao, Y.; Ruan, Z.; Mei, N.; Yin, B.; Liu, L. Prediction of human epidermal growth factor receptor 2 (HER2) status in breast cancer by mammographic radiomics features and clinical characteristics: A multicenter study. Eur. Radiol. 2024, 34, 5464–5476. [Google Scholar] [CrossRef]
- Mota, A.M.; Mendes, J.; Matela, N. Breast Cancer Molecular Subtype Prediction: A Mammography-Based AI Approach. Biomedicines 2024, 12, 1371. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.A.G.; Ovalho, M.d.L.; Matela, N.; Mota, A.M. Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images. J. Imaging 2024, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Nissar, I.; Alam, S.; Masood, S.; Kashif, M. MOB-CBAM: A dual-channel attention-based deep learning generalizable model for breast cancer molecular subtypes prediction using mammograms. Comput. Methods Programs Biomed. 2024, 248, 108121. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Chen, H.; Jing, R.; Yang, W.; He, L.; Zou, T.; Liu, P.; Liang, B.; Shi, D.; Wu, W.; et al. An assessment of breast cancer HER2, ER, and PR expressions based on mammography using deep learning with convolutional neural networks. Sci. Rep. 2025, 15, 4826. [Google Scholar] [CrossRef]
- Ben Rabah, C.; Sattar, A.; Ibrahim, A.; Serag, A. A Multimodal Deep Learning Model for the Classification of Breast Cancer Subtypes. Diagnostics 2025, 15, 995. [Google Scholar] [CrossRef]
- Hu, Z.; Xu, M.; Yang, H.; Hao, H.; Zhao, P.; Yang, Y.; Liu, G. Development of an Intra- and Peritumoral Radiomics Nomogram Using Digital Breast Tomosynthesis for Preoperative Assessment of Ki-67 Expression in Invasive Breast Cancer. Acad. Radiol. 2025, 32, 2465–2476. [Google Scholar] [CrossRef]
- Mota, A.M. AI and Tomosynthesis for Breast Cancer Molecular Subtyping: A Step Toward Precision Medicine. In Proceedings of the 47th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2025), Copenhagen, Denmark, 14–17 July 2025. [Google Scholar]
- Liu, M.; Zhang, S.; Du, Y.; Zhang, X.; Wang, D.; Ren, W.; Sun, J.; Yang, S.; Zhang, G. Identification of Luminal A breast cancer by using deep learning analysis based on multi-modal images. Front. Oncol. 2023, 13, 1243126. [Google Scholar] [CrossRef]
- Zhang, T.; Tan, T.; Han, L.; Appelman, L.; Veltman, J.; Wessels, R.; Duvivier, K.M.; Loo, C.; Gao, Y.; Wang, X.; et al. Predicting breast cancer types on and beyond molecular level in a multi-modal fashion. npj Breast Cancer 2023, 9, 16. [Google Scholar] [CrossRef]
- Liu, J.; Yan, C.; Liu, C.; Wang, Y.; Chen, Q.; Chen, Y.; Guo, J.; Chen, S. Predicting Ki-67 expression levels in breast cancer using radiomics-based approaches on digital breast tomosynthesis and ultrasound. Front. Oncol. 2024, 14, 1403522. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, J.; Sun, H.; Chen, J.; Xie, J.; Peng, Y.; Shang, T.; Pan, T. Radiomics Integration of Mammography and DCE-MRI for Predicting Molecular Subtypes in Breast Cancer Patients. Breast Cancer (Dove Med. Press) 2025, 17, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Alaeikhanehshir, S.; Voets, M.M.; van Duijnhoven, F.H.; Lips, E.H.; Groen, E.J.; van Oirsouw, M.C.J.; Hwang, S.E.; Lo, J.Y.; Wesseling, J.; Mann, R.M.; et al. Application of deep learning on mammographies to discriminate between low and high-risk DCIS for patient participation in active surveillance trials. Cancer Imaging 2024, 24, 48. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Jiang, Y.; Tian, H.; Ye, X.; Cui, C.; Shi, S.; Chen, M.; Ding, Z.; Li, S.; Huang, Z.; et al. Sonography-based multimodal information platform for identifying the surgical pathology of ductal carcinoma in situ. Comput. Methods Programs Biomed. 2024, 245, 108039. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, D.; Zha, Z.; Gu, L.; Chen, J.; Fang, J.; Dou, Z.; Zhang, P.; Zhang, C.; Wang, J. Integrating radiomics into predictive models for low nuclear grade DCIS using machine learning. Sci. Rep. 2025, 15, 7505. [Google Scholar] [CrossRef]
- Abel, F.; Landsmann, A.; Hejduk, P.; Ruppert, C.; Borkowski, K.; Ciritsis, A.; Rossi, C.; Boss, A. Detecting Abnormal Axillary Lymph Nodes on Mammograms Using a Deep Convolutional Neural Network. Diagnostics 2022, 12, 1347. [Google Scholar] [CrossRef]
- Xu, M.; Yang, H.; Yang, Q.; Teng, P.; Hao, H.; Liu, C.; Yu, S.; Liu, G. Radiomics nomogram based on digital breast tomosynthesis: Preoperative evaluation of axillary lymph node metastasis in breast carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 9317–9328. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, Y.; Ding, C.; Guan, W.; Zhang, X.; Jia, J.; Zhou, W.; Liu, Z.; Bai, G. Multi-modality radiomics model predicts axillary lymph node metastasis of breast cancer using MRI and mammography. Eur. Radiol. 2024, 34, 6121–6131. [Google Scholar] [CrossRef]
- Hua, Y.; Peng, Q.; Han, J.; Fei, J.; Sun, A. A two-center study of a combined nomogram based on mammography and MRI to predict ALN metastasis in breast cancer. Magn. Reson. Imaging 2024, 110, 128–137. [Google Scholar] [CrossRef]
- Wang, D.; Liu, M.; Zhuang, Z.; Wu, S.; Zhou, P.; Chen, X.; Zhu, H.; Liu, H.; Zhang, L. Radiomics Analysis on Digital Breast Tomosynthesis: Preoperative Evaluation of Lymphovascular Invasion Status in Invasive Breast Cancer. Acad. Radiol. 2022, 29, 1773–1782. [Google Scholar] [CrossRef]
- Xu, M.; Yang, H.; Sun, J.; Hao, H.; Li, X.; Liu, G. Development of an Intratumoral and Peritumoral Radiomics Nomogram Using Digital Breast Tomosynthesis for Preoperative Assessment of Lymphovascular Invasion in Invasive Breast Cancer. Acad. Radiol. 2024, 31, 1748–1761. [Google Scholar] [CrossRef]
- Shi, B.; Grimm, L.J.; Mazurowski, M.A.; Baker, J.A.; Marks, J.R.; King, L.M.; Maley, C.C.; Hwang, E.S.; Lo, J.Y. Prediction of Occult Invasive Disease in Ductal Carcinoma in Situ Using Deep Learning Features. J. Am. Coll. Radiol. 2018, 15, 527–534. [Google Scholar] [CrossRef]
- Yang, J.; Wang, T.; Yang, L.; Wang, Y.; Li, H.; Zhou, X.; Zhao, W.; Ren, J.; Li, X.; Tian, J.; et al. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Cancer Using Mammography-Based Radiomics Method. Sci. Rep. 2019, 9, 4429. [Google Scholar] [CrossRef]
- Li, J.; Song, Y.; Xu, S.; Wang, J.; Huang, H.; Ma, W.; Jiang, X.; Wu, Y.; Cai, H.; Li, L. Predicting underestimation of ductal carcinoma in situ: A comparison between radiomics and conventional approaches. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 709–721. [Google Scholar] [CrossRef]
- Tan, H.; Wu, Y.; Bao, F.; Zhou, J.; Wan, J.; Tian, J.; Lin, Y.; Wang, M. Mammography-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in breast cancer. Br. J. Radiol. 2020, 93, 20191019. [Google Scholar] [CrossRef]
- Mao, N.; Yin, P.; Li, Q.; Wang, Q.; Liu, M.; Ma, H.; Dong, J.; Che, K.; Wang, Z.; Duan, S.; et al. Radiomics nomogram of contrast-enhanced spectral mammography for prediction of axillary lymph node metastasis in breast cancer: A multicenter study. Eur. Radiol. 2020, 30, 6732–6739. [Google Scholar] [CrossRef]
- Wu, X.; Guo, Y.; Sa, Y.; Song, Y.; Li, X.; Lv, Y.; Xing, D.; Sun, Y.; Cong, Y.; Yu, H.; et al. Contrast-Enhanced Spectral Mammography-Based Prediction of Non-Sentinel Lymph Node Metastasis and Axillary Tumor Burden in Patients With Breast Cancer. Front. Oncol. 2022, 12, 823897. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Grimm, L.J.; Mazurowski, M.A.; Marks, J.R.; King, L.M.; Maley, C.C.; Lynch, T.; van Oirsouw, M.; Rogers, K.; Stone, N.; et al. Prediction of Upstaging in Ductal Carcinoma in Situ Based on Mammographic Radiomic Features. Radiology 2022, 303, 54–62. [Google Scholar] [CrossRef]
- Lin, F.; Li, Q.; Wang, Z.; Shi, Y.; Ma, H.; Zhang, H.; Zhang, K.; Yang, P.; Zhang, R.; Duan, S.; et al. Intratumoral and peritumoral radiomics for preoperatively predicting the axillary non-sentinel lymph node metastasis in breast cancer on the basis of contrast-enhanced mammography: A multicenter study. Br. J. Radiol. 2023, 96, 20220068. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, H.; Lin, F.; Zhang, R.; Ma, H.; Shi, Y.; Yang, P.; Zhang, K.; Zhao, F.; Mao, N.; et al. Intra- and Peritumoral Radiomics of Contrast-Enhanced Mammography Predicts Axillary Lymph Node Metastasis in Patients With Breast Cancer: A Multicenter Study. Acad. Radiol. 2023, 30, S133–S142. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, D.; Takahashi, K.; Oba, K.; Takaya, E.; Usuzaki, T.; Kadowaki, M.; Kawaguchi, K.; Adachi, M.; Kaneno, T.; Fukuda, T.; et al. Deep learning model for predicting the presence of stromal invasion of breast cancer on digital breast tomosynthesis. Radiol. Phys. Technol. 2023, 16, 406–413. [Google Scholar] [CrossRef]
- Tsai, H.Y.; Kao, Y.W.; Wang, J.C.; Tsai, T.Y.; Chung, W.S.; Hsu, J.S.; Hou, M.F.; Weng, S.F. Multitask deep learning on mammography to predict extensive intraductal component in invasive breast cancer. Eur. Radiol. 2024, 34, 2593–2604. [Google Scholar] [CrossRef]
- He, S.; Deng, B.; Chen, J.; Li, J.; Wang, X.; Li, G.; Long, S.; Wan, J.; Zhang, Y. Preoperative DBT-based radiomics for predicting axillary lymph node metastasis in breast cancer: A multi-center study. BMC Med. Imaging 2025, 25, 169. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xu, S.; Wang, H.; Wang, X.; Niu, S.; Luo, Y.; Zhao, N. Intra- and peri-tumoral radiomics for predicting the sentinel lymph node metastasis in breast cancer based on preoperative mammography and MRI. Front. Oncol. 2022, 12, 1047572. [Google Scholar] [CrossRef]
- Haraguchi, T.; Goto, Y.; Furuya, Y.; Nagai, M.T.; Kanemaki, Y.; Tsugawa, K.; Kobayashi, Y. Use of machine learning with two-dimensional synthetic mammography for axillary lymph node metastasis prediction in breast cancer: A preliminary study. Transl. Cancer Res. 2023, 12, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Sun, S.; Deng, X.; Wang, Y.; Yao, W.; Yue, P.; Wu, S.; Yan, J.; Zhang, X.; Zhang, Y. Predicting axillary lymph node metastasis in breast cancer using a multimodal radiomics and deep learning model. Front. Immunol. 2024, 15, 1482020. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Wu, C.G.; Lin, Y.Y.; Zou, J.C.; Wang, D.Q.; Haffty, B.G.; Wang, K. Development and validation of a multivariable risk model based on clinicopathological characteristics, mammography, and MRI imaging features for predicting axillary lymph node metastasis in patients with upgraded ductal carcinoma in situ. Gland Surg. 2025, 14, 738–753. [Google Scholar] [CrossRef] [PubMed]
- Mao, N.; Shi, Y.; Lian, C.; Wang, Z.; Zhang, K.; Xie, H.; Zhang, H.; Chen, Q.; Cheng, G.; Xu, C.; et al. Intratumoral and peritumoral radiomics for preoperative prediction of neoadjuvant chemotherapy effect in breast cancer based on contrast-enhanced spectral mammography. Eur. Radiol. 2022, 32, 3207–3219. [Google Scholar] [CrossRef]
- Xing, D.; Lv, Y.; Sun, B.; Chu, T.; Bao, Q.; Zhang, H. Develop and Validate a Nomogram Combining Contrast-Enhanced Spectral Mammography Deep Learning with Clinical-Pathological Features to Predict Neoadjuvant Chemotherapy Response in Patients with ER-Positive/HER2-Negative Breast Cancer. Acad. Radiol. 2024, 31, 3524–3534. [Google Scholar] [CrossRef]
- Förnvik, D.; Borgquist, S.; Larsson, M.; Zackrisson, S.; Skarping, I. Deep learning analysis of serial digital breast tomosynthesis images in a prospective cohort of breast cancer patients who received neoadjuvant chemotherapy. Eur. J. Radiol. 2024, 178, 111624. [Google Scholar] [CrossRef]
- Jiang, X.; Zou, X.; Sun, J.; Zheng, A.; Su, C. A Nomogram Based on Radiomics with Mammography Texture Analysis for the Prognostic Prediction in Patients with Triple-Negative Breast Cancer. Contrast Media Mol. Imaging 2020, 2020, 5418364. [Google Scholar] [CrossRef]
- Mao, N.; Yin, P.; Zhang, H.; Zhang, K.; Song, X.; Xing, D.; Chu, T. Mammography-based radiomics for predicting the risk of breast cancer recurrence: A multicenter study. Br. J. Radiol. 2021, 94, 20210348. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, L.; Xiao, Q.; Huang, Y.; Lin, L.; Peng, W.; Gong, J.; Gu, Y. Predicting Prognosis of Phyllodes Tumors Using a Mammography- and Magnetic Resonance Imaging-Based Radiomics Model: A Preliminary Study. Clin. Breast Cancer 2024, 24, e571–e582.e571. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Meng, X.; Chen, H.; Liu, J.; Gao, W.; Du, L.; Chen, Y.; Wang, Y.; Liu, X.; Liu, B.; et al. Predicting the Level of Tumor-Infiltrating Lymphocytes in Patients With Breast Cancer: Usefulness of Mammographic Radiomics Features. Front. Oncol. 2021, 11, 628577. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, F.; Ma, H.; Shi, Y.; Dong, J.; Yang, P.; Zhang, K.; Guo, N.; Zhang, R.; Cui, J.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for the Prediction of Neoadjuvant Chemotherapy-Insensitive Breast Cancers. Front. Oncol. 2021, 11, 605230. [Google Scholar] [CrossRef]
- Skarping, I.; Larsson, M.; Förnvik, D. Analysis of mammograms using artificial intelligence to predict response to neoadjuvant chemotherapy in breast cancer patients: Proof of concept. Eur. Radiol. 2022, 32, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lin, J.; Lin, F.; Wang, Z.; Zhang, H.; Zhang, S.; Mao, N.; Qiao, G. Radiomics of contrast-enhanced spectral mammography for prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer. J. X-Ray Sci. Technol. 2023, 31, 669–683. [Google Scholar] [CrossRef]
- Han, J.; Hua, H.; Fei, J.; Liu, J.; Guo, Y.; Ma, W.; Chen, J. Prediction of Disease-Free Survival in Breast Cancer using Deep Learning with Ultrasound and Mammography: A Multicenter Study. Clin. Breast Cancer 2024, 24, 215–226. [Google Scholar] [CrossRef]
- Cai, L.; Sidey-Gibbons, C.; Nees, J.; Riedel, F.; Schäfgen, B.; Togawa, R.; Killinger, K.; Heil, J.; Pfob, A.; Golatta, M. Can multi-modal radiomics using pretreatment ultrasound and tomosynthesis predict response to neoadjuvant systemic treatment in breast cancer? Eur. Radiol. 2024, 34, 2560–2573. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mota, A.M. AI-Based Characterization of Breast Cancer in Mammography and Tomosynthesis: A Review of Radiomics and Deep Learning for Subtyping, Staging, and Prognosis. Cancers 2025, 17, 3387. https://doi.org/10.3390/cancers17203387
Mota AM. AI-Based Characterization of Breast Cancer in Mammography and Tomosynthesis: A Review of Radiomics and Deep Learning for Subtyping, Staging, and Prognosis. Cancers. 2025; 17(20):3387. https://doi.org/10.3390/cancers17203387
Chicago/Turabian StyleMota, Ana M. 2025. "AI-Based Characterization of Breast Cancer in Mammography and Tomosynthesis: A Review of Radiomics and Deep Learning for Subtyping, Staging, and Prognosis" Cancers 17, no. 20: 3387. https://doi.org/10.3390/cancers17203387
APA StyleMota, A. M. (2025). AI-Based Characterization of Breast Cancer in Mammography and Tomosynthesis: A Review of Radiomics and Deep Learning for Subtyping, Staging, and Prognosis. Cancers, 17(20), 3387. https://doi.org/10.3390/cancers17203387







