Unveiling Enhancer RNAs in Gliomas: A Systematic Review and Qualitative Synthesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
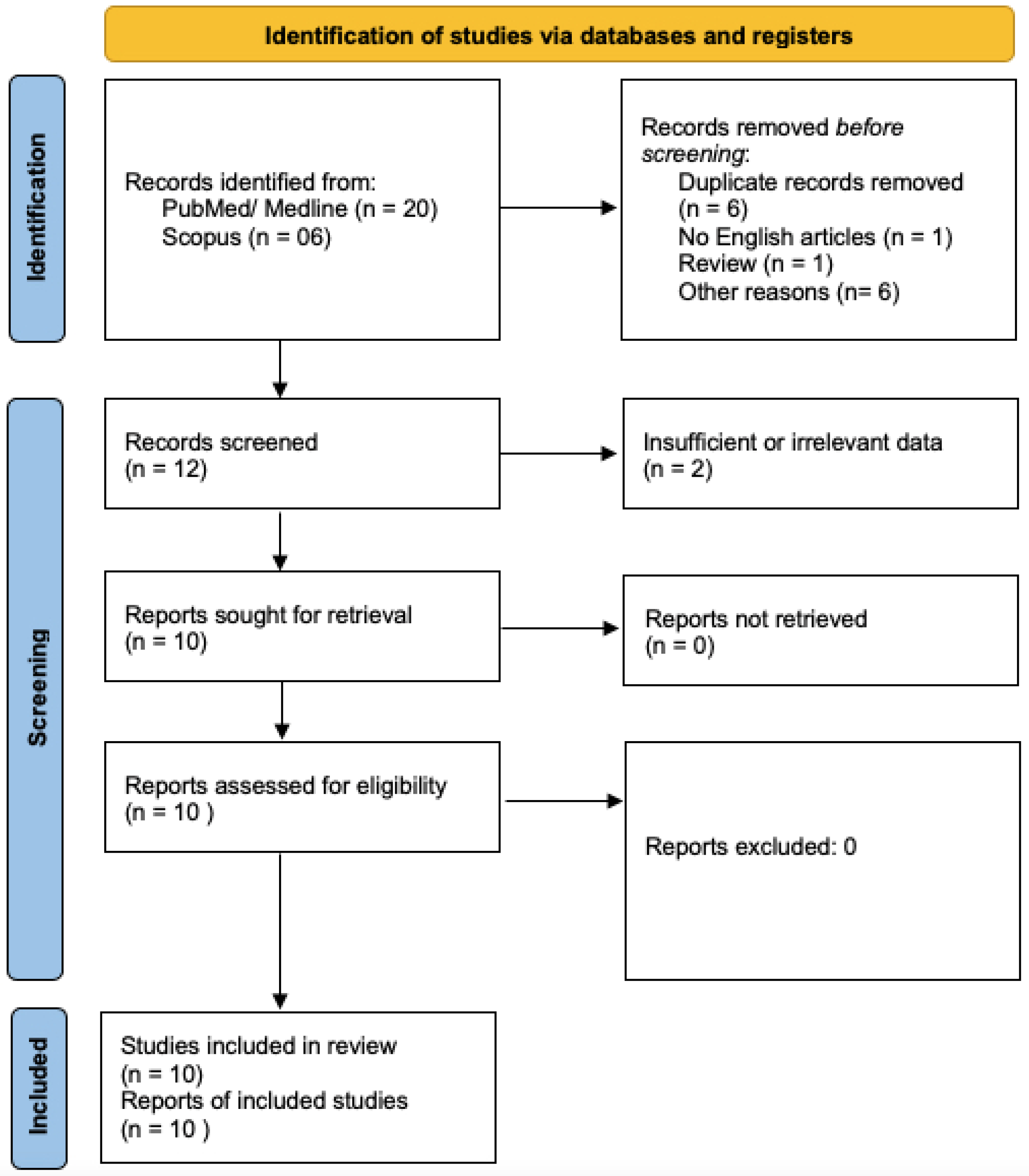
2.1. Search Methods
2.2. Selection of Studies
2.3. Data Extraction
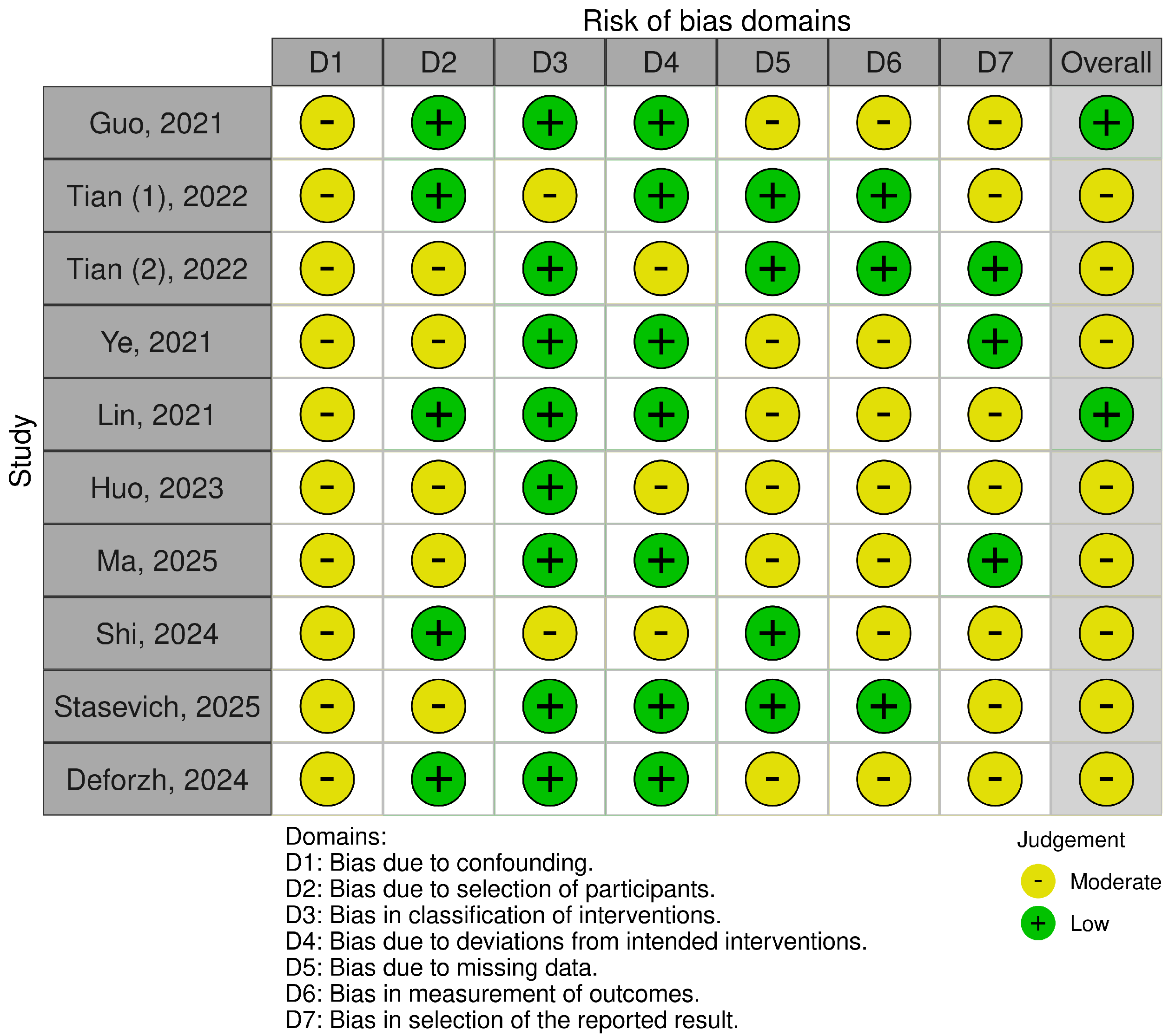
2.4. Risk of Bias
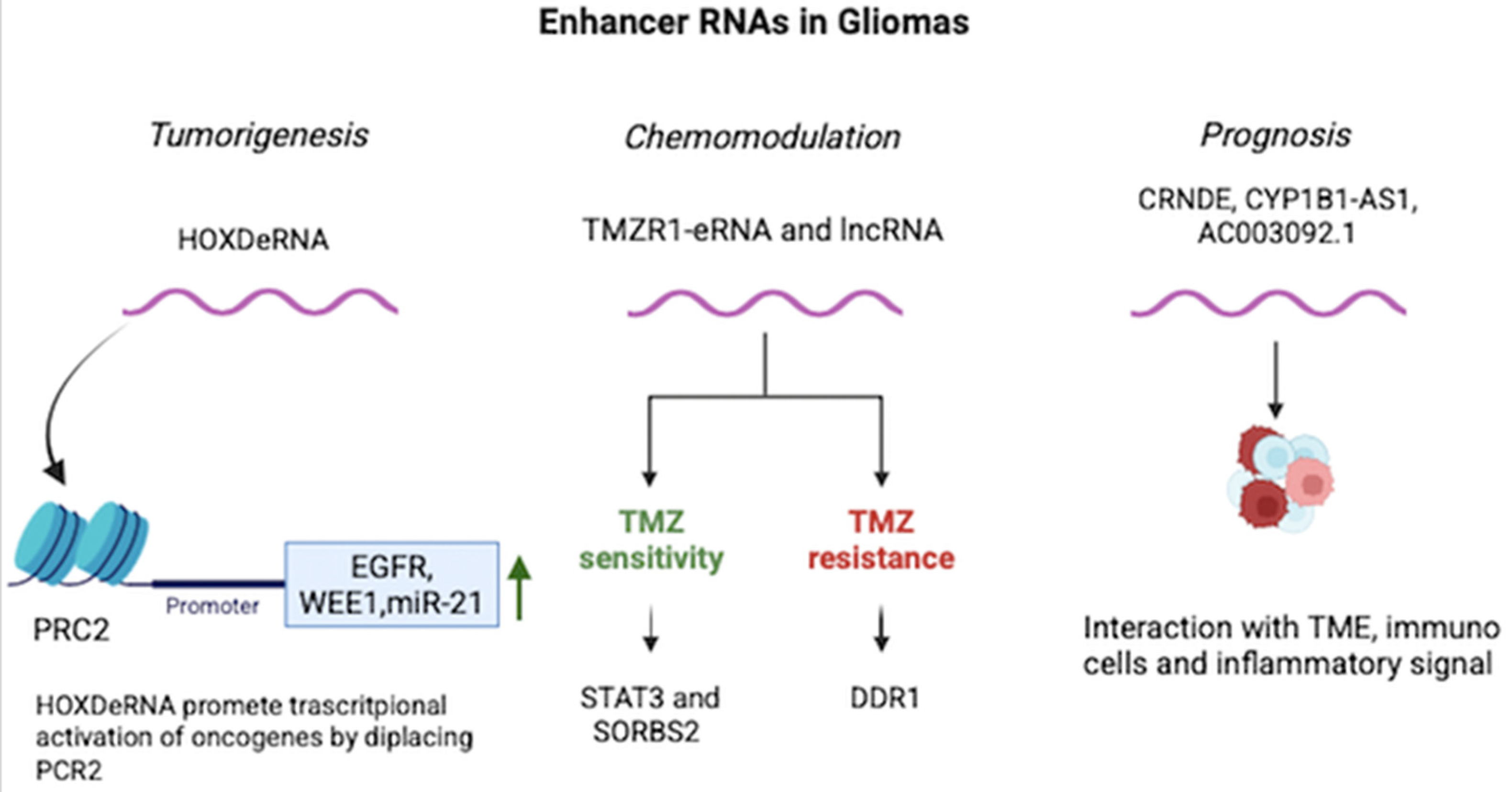
3. Results
Qualitative Analysis (Systematic Review)
4. Discussion
4.1. eRNAs and Chemoresistance
4.2. eRNAs as Prognostic Biomarkers
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CGGA | Chinese Glioma Genome Atlas |
| DDR1 | Discoidin Domain Receptor 1 |
| eRNA/eRNAs | Enhancer RNA/Enhancer RNAs |
| EZH2 | Enhancer of Zeste Homolog 2 (subunit of PRC2) |
| GBM | Glioblastoma |
| GEO | Gene Expression Omnibus |
| HGG | High-Grade Glioma |
| LGG | Low-Grade Glioma |
| lncRNA/lncRNAs | Long non-coding RNA/Long non-coding RNAs |
| PICO | Population, Intervention, Comparator, Outcome |
| PRC2 | Polycomb Repressive Complex 2 |
| seRNA/seRNAs | Super-enhancer RNA/Super-enhancer RNAs |
| SORBS2 | Sorbin and SH3 Domain Containing 2 |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| TCGA | The Cancer Genome Atlas |
| TFPI2 | Tissue Factor Pathway Inhibitor 2 |
| TMZ | Temozolomide |
| USP28 | Ubiquitin Specific Peptidase 28 |
References
- Wapinski, O.; Chang, H.Y. Long Noncoding RNAs and Human Disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Spizzo, R.; Almeida, M.I.; Colombatti, A.; Calin, G.A. Long Non-Coding RNAs and Cancer: A New Frontier of Translational Research? Oncogene 2012, 31, 4577–4587. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The Functions and Unique Features of Long Intergenic Non-Coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Li, H.; Cui, L.; Zhou, M.; Zou, Q.; Zhao, Y.; Lin, H.; Liang, Y.; Chieh Kow, A.W.; Wang, G. Single-Cell Multi-Omics Data Reveal Heterogeneity in Liver Tissue Microenvironment Induced by Hypertension. Mol. Ther. Nucleic Acids 2025, 36, 102696. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wei, M.; Tian, Q.; Guo, W. CEBPB as a Prognostic Biomarker and Its Association with Immune Cells in Clear Cell Renal Cell Carcinoma. Biochem. Biophys. Rep. 2025, 44, 102231. [Google Scholar] [CrossRef] [PubMed]
- Speicher, D.J.; Rose, J.; McKernan, K. Quantification of Residual Plasmid DNA and SV40 Promoter-Enhancer Sequences in Pfizer/BioNTech and Moderna modRNA COVID-19 Vaccines from Ontario, Canada. Autoimmunity 2025, 58, 2551517. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, K.; Lu, C.; Yao, C.; Xie, F. YY1-Induced Long Non-Coding RNA HOXA11-AS Activates Oxidative Stress and Inflammation by Epigenetic Modification of Nrf2 Pathway to Promote Keloid Formation. Redox Rep. 2025, 30, 2539030. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, X.; Zhang, Y.; Yu, M.; Yuan, X.; Xu, Y.; Ma, L.; Wang, X.; Xing, H. Gut Microbial Metabolite Butyrate Suppresses Hepatocellular Carcinoma Growth via CXCL11-Dependent Enhancement of Natural Killer Cell Infiltration. Gut Microbes 2025, 17, 2519706. [Google Scholar] [CrossRef]
- Xie, B.; Dean, A. Noncoding Function of Super Enhancer Derived Cpox Pre-mRNA in Modulating Neighbouring Gene Expression and Chromatin Interactions. RNA Biol. 2025, 22, 1–17. [Google Scholar] [CrossRef]
- Stirling, D.C.; de Miguel Ferrer, M.; Kim, S.; Wane, M.; Kysh, D.; Caproni, L.J.; Tregoning, J.S. Modifying Non-Coding Regions of Linear DNA Vaccines to Explore the Interplay of Expression and Inflammation in Immunogenicity. Hum. Vaccin Immunother. 2025, 21, 2430826. [Google Scholar] [CrossRef]
- Chen, Y.; Ke, B.; Qiu, M.; Yan, C.; Zou, H.; Tu, W. Role of E2F1, Taking Account for the Transcriptional Activity of lncRNA HOTAIR, in Vascular Calcification via Epigenetic Silencing of KLF16/Klotho. J. Hypertens. 2025, 43, 1802–1815. [Google Scholar] [CrossRef]
- Namous, H.; Arruri, V.; Galleske, T.; Vemuganti, R. Ischemic Stroke Altered the Expression Profiles of Super Enhancer RNAs in Mouse Brain in a Sexually Dimorphic Manner. Exp. Neurol. 2025, 393, 115372. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and Muscle Regeneration Are Regulated by the LINC00961-Encoded SPAR Polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as Non-Coding RNA Transcription Units: Recent Insights and Future Perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Cardon, T.; Fournier, I.; Salzet, M. Unveiling a Ghost Proteome in the Glioblastoma Non-Coding RNAs. Front. Cell Dev. Biol. 2021, 9, 703583. [Google Scholar] [CrossRef]
- Rion, N.; Rüegg, M.A. LncRNA-Encoded Peptides: More than Translational Noise? Cell Res. 2017, 27, 604–605. [Google Scholar] [CrossRef]
- Deforzh, E.; Kharel, P.; Zhang, Y.; Karelin, A.; El Khayari, A.; Ivanov, P.; Krichevsky, A.M. HOXDeRNA Activates a Cancerous Transcription Program and Super Enhancers via Genome-Wide Binding. Mol. Cell 2024, 84, 3950–3966.e6. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Guo, D.; Zheng, Y.; Wang, W.; Bi, J.; He, A.; Fan, S.; Su, G.; Zhao, X.; Zhao, Z.; et al. Bivalent Activity of Super-Enhancer RNA LINC02454 Controls 3D Chromatin Structure and Regulates Glioma Sensitivity to Temozolomide. Cell Death Dis. 2024, 15, 6. [Google Scholar] [CrossRef]
- Ye, T.; Li, L.; Peng, X.; Li, Q. CYP1B1-AS1 Is a Novel Biomarker in Glioblastoma by Comprehensive Analysis. Dis. Markers 2021, 2021, 8565943. [Google Scholar] [CrossRef]
- Pennisi, G.; Bruzzaniti, P.; Burattini, B.; Piaser Guerrato, G.; Della Pepa, G.M.; Sturiale, C.L.; Lapolla, P.; Familiari, P.; La Pira, B.; D’Andrea, G.; et al. Advancements in Telomerase-Targeted Therapies for Glioblastoma: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 8700. [Google Scholar] [CrossRef]
- Pennisi, G.; Valeri, F.; Burattini, B.; Bruzzaniti, P.; Sturiale, C.L.; Talacchi, A.; Papacci, F.; Olivi, A.; Della Pepa, G.M. Targeting Macrophages in Glioblastoma: Current Therapies and Future Directions. Cancers 2025, 17, 2687. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Stasevich, E.M.; Simonova, A.V.; Poteryakhina, A.V.; Bogomolova, E.A.; Uvarova, A.N.; Zheremyan, E.A.; Korneev, K.V.; Schwartz, A.M.; Kuprash, D.V.; Demin, D.E. Enhancer RNA from STAT3 Locus Affects Temozolomide Chemoresistance of Glioblastoma Cells. Gene 2025, 944, 149297. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-Y.; Zhong, S.; Wang, Z.-N.; Xie, T.; Duan, H.; Zhang, J.-Y.; Zhang, G.-H.; Liang, L.; Cui, R.; Hu, H.-R.; et al. Immunogenomic Profiling Demonstrate AC003092.1 as an Immune-Related eRNA in Glioblastoma Multiforme. Front. Genet. 2021, 12, 633812. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, K.; Yan, G.; Han, X.; Liu, Y.; Zhang, Q.; Liu, M. A Novel Prognostic Tool for Glioma Based on Enhancer RNA-Regulated Immune Genes. Front. Cell Dev. Biol. 2022, 9, 798445. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Yang, Y.; Hou, C.; Zheng, J.; Lv, G.; Mao, R.; Xu, P.; Chen, S.; Zhou, Y.; Wang, P.; et al. An Integrated Analysis of Enhancer RNAs in Glioma and a Validation of Their Prognostic Values. Am. J. Transl. Res. 2021, 13, 8611–8631. [Google Scholar]
- Huo, Z.; Guo, L.; Shao, W.; Ding, Q.; Guo, Y.; Xu, Q. CRNDE, an Enhancer RNA of Prognostic Value in Glioma, Correlates with Immune Infiltration: A Pan-Cancer Analysis. Eur. J. Inflamm. 2023, 21, 1721727X221138068. [Google Scholar] [CrossRef]
- Tian, W.; Yan, G.; Chen, K.; Han, X.; Zhang, W.; Sun, L.; Zhang, Q.; Zhang, Y.; Li, Y.; Liu, M.; et al. Development and Validation of a Novel Prognostic Model for Lower-Grade Glioma Based on Enhancer RNA-Regulated Prognostic Genes. Front. Oncol. 2022, 12, 714338. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, S.; Jia, Y.; Wang, S.; Liu, W.; Yu, J.; Wang, J.; Geng, J.; Zhao, J.; Zhao, Z.; et al. Enhancer RNA-Mediated Transcriptional Regulatory Programs Reveal the Malignant Progression of Glioma. Sci. Adv. 2025, 11, eadu9487. [Google Scholar] [CrossRef]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A Tool for Assessing Risk of Bias in Non-Randomised Studies of Interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef]
| Author | Year | eRNA | Regulated Protein | [TMZ] | Time to TMZ Addition | Resistance to TMZ |
|---|---|---|---|---|---|---|
| Stasevich et al. [23] | 2025 | TMZR1-eRNA | STAT3 | 20 μg/m | 1 h after tranfection | ↑ |
| Shi et al. [18] | 2024 | LINC02454 * | SORBS2 + DDR1 | 1 mM | 72 h | ↑ |
| Author | Year | eRNA | Target | Sample Size | Validation Size | Tested on | Prognosis | ||
|---|---|---|---|---|---|---|---|---|---|
| HHG | LGG | GBM | |||||||
| Guo [24] | 2021 | AC003092.1 | TFPI2 | 151 | 23 | X | X | X | ↓ |
| Tian [25] | 2022 | ENSR00000032650 * | SEMA4G | 428 (TCGA) | 399 (CGGA) | X | ↑ | ||
| ENSR00000032651 * | SEMA4G | X | ↑ | ||||||
| ENSR00000261154 * | SEMA4G | X | ↑ | ||||||
| ENSR00000030804 | NRG3 | X | ↑ | ||||||
| ENSR00000161287 | PPM1L | X | ↑ | ||||||
| ENSR00000260547 | RGR | X | ↑ | ||||||
| ENSR00000203159 | TBPL1 | X | ↑ | ||||||
| ENSR00000265929 | USP28 | X | ↓ | ||||||
| Ye [19] | 2021 | CYP1B1-AS1 | CYP1B1 | 167 | NR | X | X | ↓ | |
| Lin [26] | 2021 | CRNDE | IRX5 | 40 (TCGA, CGGA) | NR | X | X | ↓ | |
| MRPS31P5 | ATP7B, NEK3 | X | X | ↓ | |||||
| LINC00844 | PHYHIPL | X | X | ↑ | |||||
| Huo [27] | 2023 | CRNDE | IRX5 | 1013 (CGGA, GEO) | N/A | X | X | ↓ | |
| Tian [28] | 2022 | ENSR00000210436 | ADCYAP1R1 | 525 (TCGA) | 513 (CCGA) | X | X | ↓ | |
| ENSR00000249159 | FGF13 | X | X | ↓ | |||||
| ENSR00000195717 | PSMB8 | X | X | ↓ | |||||
| Ma [29] | 2025 | chr20:1868395–1868476 | GATA3 | 710 (CGGA) | NR | X | X | X | ↓ |
| chr13:95310907–95311049 | E2F6 | 710 (CGGA) | NR | X | X | X | ↓ | ||
| chr19:55451280–55451568 | NFKB1 | 710 (CGGA) | NR | X | X | X | ↓ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palermo, M.; Pennisi, G.; Burattini, B.; Bruzzaniti, P.; Talacchi, A.; Olivi, A.; Sturiale, C.L. Unveiling Enhancer RNAs in Gliomas: A Systematic Review and Qualitative Synthesis. Cancers 2025, 17, 3326. https://doi.org/10.3390/cancers17203326
Palermo M, Pennisi G, Burattini B, Bruzzaniti P, Talacchi A, Olivi A, Sturiale CL. Unveiling Enhancer RNAs in Gliomas: A Systematic Review and Qualitative Synthesis. Cancers. 2025; 17(20):3326. https://doi.org/10.3390/cancers17203326
Chicago/Turabian StylePalermo, Matteo, Giovanni Pennisi, Benedetta Burattini, Placido Bruzzaniti, Andrea Talacchi, Alessandro Olivi, and Carmelo Lucio Sturiale. 2025. "Unveiling Enhancer RNAs in Gliomas: A Systematic Review and Qualitative Synthesis" Cancers 17, no. 20: 3326. https://doi.org/10.3390/cancers17203326
APA StylePalermo, M., Pennisi, G., Burattini, B., Bruzzaniti, P., Talacchi, A., Olivi, A., & Sturiale, C. L. (2025). Unveiling Enhancer RNAs in Gliomas: A Systematic Review and Qualitative Synthesis. Cancers, 17(20), 3326. https://doi.org/10.3390/cancers17203326









