Transformer-Based Deep Learning for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
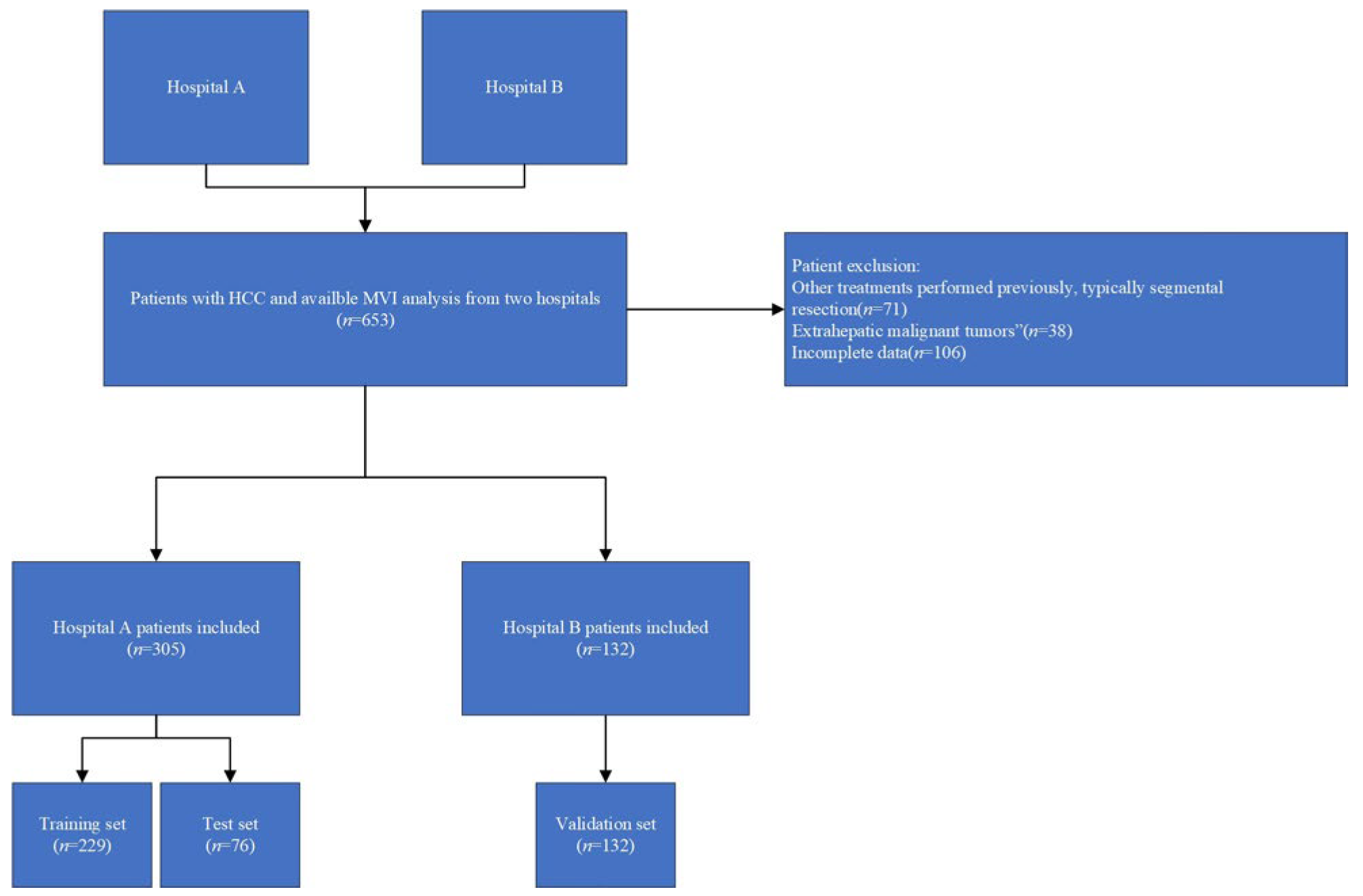
2.1. Patient Enrollment and Clinical Data Collection
2.2. MRI Image Acquisition
2.3. Imaging Analysis
2.4. Pathological Evaluation
2.5. Radiomic Feature Extraction
2.6. Data Preprocessing
- Missing Value Handling: Samples with more than 10% missing values were excluded. For the remaining samples, missing values were imputed separately for each feature using k-nearest neighbor (KNN) imputation (k = 3) implemented in Scikit-learn (version 1.3.1, Python Software Foundation, Wilmington, DE, USA)., based on the Euclidean distance in the feature space [21,22].
- Normalization: Radiomic features were normalized using Z-score transformation. The mean and standard deviation were calculated exclusively from the training set and then applied to the validation and test sets to ensure no information leakage during model evaluation [23]. Normalization parameters were derived from the training set and applied unchanged to the test and validation sets.
2.7. Feature Selection

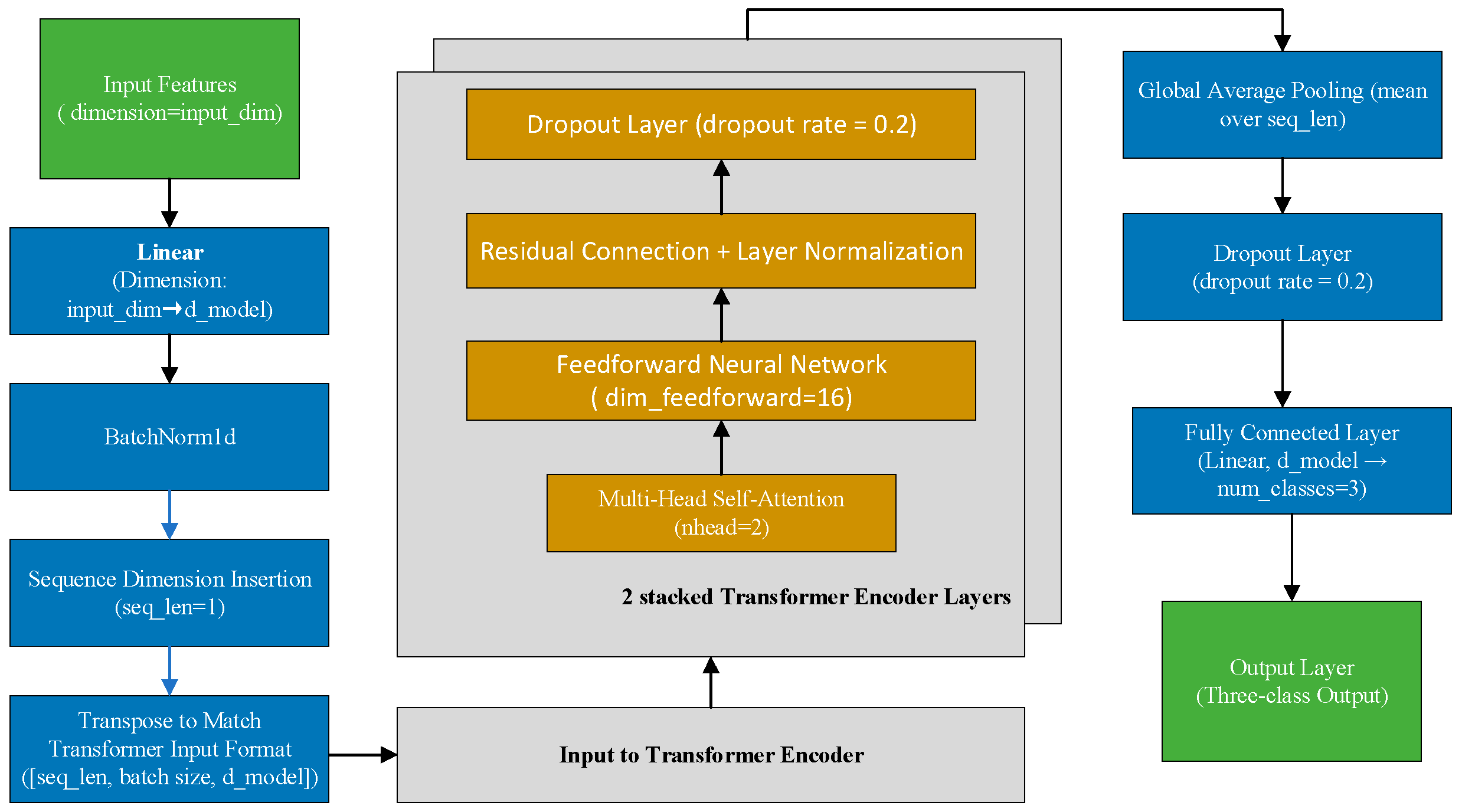
2.8. Model Architecture and Training
2.9. Data Augmentation and Balancing
3. Results
3.1. Baseline Characteristics
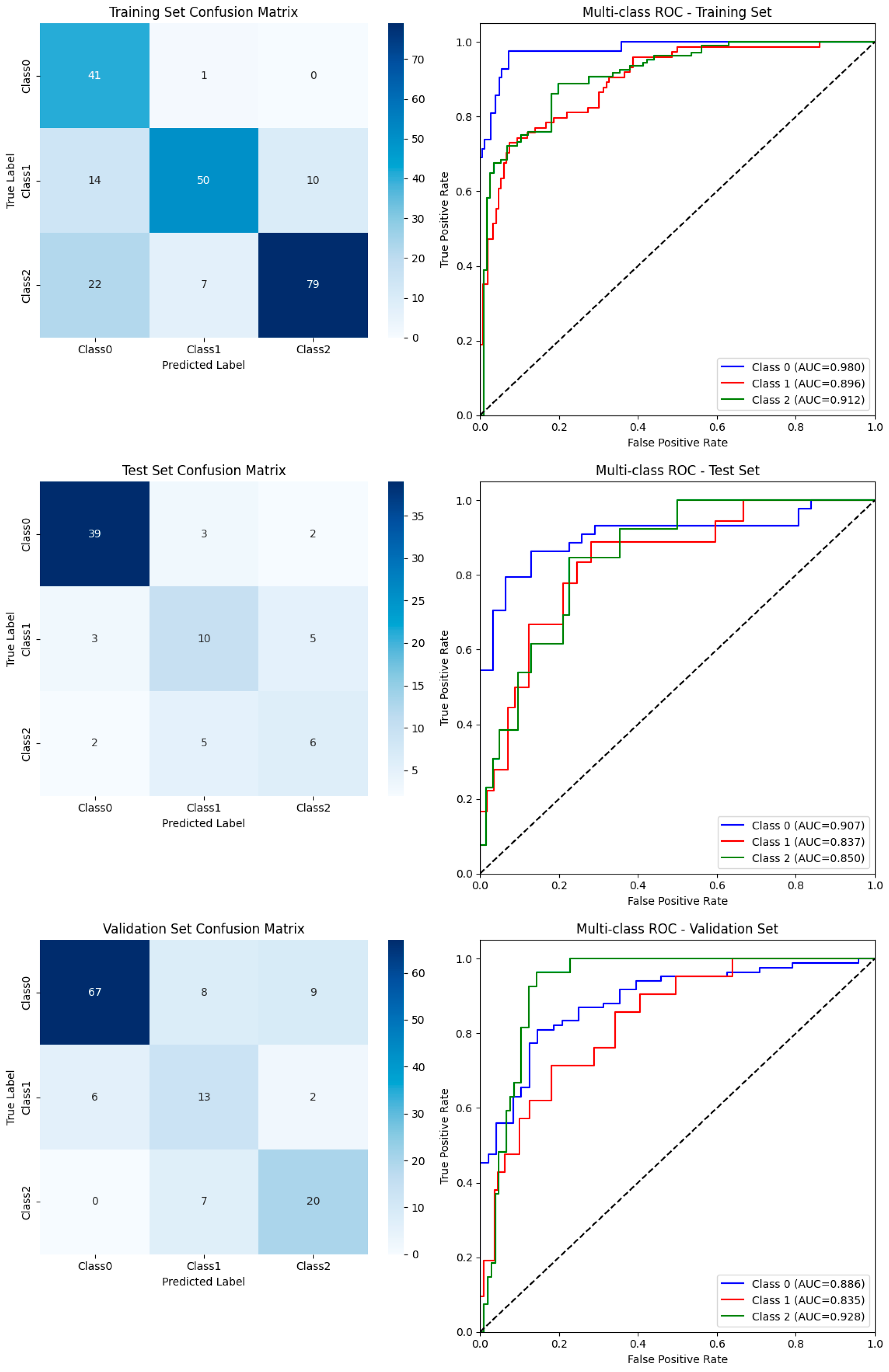
3.2. Overall Model Performance
3.3. Class-Wise Diagnostic Performance
3.4. Visualization of Classification Outcomes
3.5. Comparison with Traditional Machine Learning Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| MVI | Microvascular invasion |
| MRI | Magnetic resonance imaging |
| AUC | Area under the curve |
| Gd-BOPTA | Gadolinium benzyloxypropionictetraacetate |
| RFE | Recursive feature elimination |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Calderaro, J.; Seraphin, T.P.; Luedde, T.; Simon, T.G. Artificial Intelligence for the Prevention and Clinical Management of Hepatocellular Carcinoma. J. Hepatol. 2022, 76, 1348–1361. [Google Scholar] [CrossRef]
- Lei, Z.; Li, J.; Wu, D.; Xia, Y.; Wang, Q.; Si, A.; Wang, K.; Wan, X.; Lau, W.Y.; Wu, M.; et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus–Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg. 2016, 151, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kahn, J.; Terrault, N.A. Liver Transplantation as Therapy for Hepatocellular Carcinoma. Liver Int. 2020, 40, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Tampaki, M.; Papatheodoridis, G.V.; Cholongitas, E. Intrahepatic Recurrence of Hepatocellular Carcinoma after Resection: An Update. Clin. J. Gastroenterol. 2021, 14, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Sumie, S.; Nakashima, O.; Okuda, K.; Kuromatsu, R.; Kawaguchi, A.; Nakano, M.; Satani, M.; Yamada, S.; Okamura, S.; Hori, M.; et al. The Significance of Classifying Microvascular Invasion in Patients with Hepatocellular Carcinoma. Ann. Surg. Oncol. 2014, 21, 1002–1009. [Google Scholar] [CrossRef]
- Cai, Y.; Fu, Y.; Liu, C.; Wang, X.; You, P.; Li, X.; Song, Y.; Mu, X.; Fang, T.; Yang, Y.; et al. Stathmin 1 Is a Biomarker for Diagnosis of Microvascular Invasion to Predict Prognosis of Early Hepatocellular Carcinoma. Cell Death Dis. 2022, 13, 176. [Google Scholar] [CrossRef]
- Sheng, X.; Ji, Y.; Ren, G.-P.; Lu, C.-L.; Yun, J.-P.; Chen, L.-H.; Meng, B.; Qu, L.-J.; Duan, G.-J.; Sun, Q.; et al. A Standardized Pathological Proposal for Evaluating Microvascular Invasion of Hepatocellular Carcinoma: A Multicenter Study by LCPGC. Hepatol. Int. 2020, 14, 1034–1047. [Google Scholar] [CrossRef]
- Rodríguez-Perálvarez, M.; Luong, T.V.; Andreana, L.; Meyer, T.; Dhillon, A.P.; Burroughs, A.K. A Systematic Review of Microvascular Invasion in Hepatocellular Carcinoma: Diagnostic and Prognostic Variability. Ann. Surg. Oncol. 2013, 20, 325–339. [Google Scholar] [CrossRef]
- Wang, J.; Lin, Z.; Lin, Z.; Yang, L.; Yu, M.; Xie, R.; Lin, W.; Yang, Y.; Tu, H. Predicting Microvascular Invasion in Solitary Hepatocellular Carcinoma: A Multi-Center Study Integrating Clinical, MRI Assessments, and Radiomics Indicators. Front. Oncol. 2025, 15, 1511260. [Google Scholar] [CrossRef]
- Hu, G.; Qu, J.; Gao, J.; Chen, Y.; Wang, F.; Zhang, H.; Zhang, H.; Wang, X.; Ma, H.; Xie, H.; et al. Radiogenomics Nomogram Based on MRI and microRNAs to Predict Microvascular Invasion of Hepatocellular Carcinoma. Front. Oncol. 2024, 14, 1371432. [Google Scholar] [CrossRef]
- Li, J.; Song, W.; Li, J.; Cai, L.; Jiang, Z.; Wei, M.; Nong, B.; Lai, M.; Jiang, Y.; Zhao, E.; et al. A Clinical Study Exploring the Prediction of Microvascular Invasion in Hepatocellular Carcinoma through the Use of Combined Enhanced CT and MRI Radiomics. PLoS ONE 2025, 20, e0318232. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning Methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 September 2017. [Google Scholar]
- Huang, X.; Khetan, A.; Cvitkovic, M.; Karnin, Z. TabTransformer: Tabular Data Modeling Using Contextual Embedding. arXiv 2020, arXiv:2012.06678. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Parmar, C.; Rios Velazquez, E.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing Value Estimation Methods for DNA Microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A Study of the Behavior of Several Methods for Balancing Machine Learning Training Data. ACM SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]
- Sammut, C.; Webb, G.I. (Eds.) Encyclopedia of Machine Learning and Data Mining; Springer: New York, NY, USA, 2017; ISBN 978-1-4899-7685-7. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene Selection for Cancer Classification Using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Somepalli, G.; Goldblum, M.; Schwarzschild, A.; Bruss, C.B.; Goldstein, T. SAINT: Improved Neural Networks for Tabular Data via Row Attention and Contrastive Pre-Training. arXiv 2021, arXiv:2106.01342. [Google Scholar] [CrossRef]
- Feng, S.-T.; Jia, Y.; Liao, B.; Huang, B.; Zhou, Q.; Li, X.; Wei, K.; Chen, L.; Li, B.; Wang, W.; et al. Preoperative Prediction of Microvascular Invasion in Hepatocellular Cancer: A Radiomics Model Using Gd-EOB-DTPA-Enhanced MRI. Eur. Radiol. 2019, 29, 4648–4659. [Google Scholar] [CrossRef]
- He, M.; Zhang, P.; Ma, X.; He, B.; Fang, C.; Jia, F. Radiomic Feature-Based Predictive Model for Microvascular Invasion in Patients with Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 574228. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, Y.; Jin, Q. Radiomics and Its Feature Selection: A Review. Symmetry 2023, 15, 1834. [Google Scholar] [CrossRef]
- Riley, R.D.; Ensor, J.; Snell, K.I.E.; Debray, T.P.A.; Altman, D.G.; Moons, K.G.M.; Collins, G.S. External Validation of Clinical Prediction Models Using Big Datasets from E-Health Records or IPD Meta-Analysis: Opportunities and Challenges. BMJ 2016, 353, i3140. [Google Scholar] [CrossRef]
- Riley, R.D.; Archer, L.; Snell, K.I.E.; Ensor, J.; Dhiman, P.; Martin, G.P.; Bonnett, L.J.; Collins, G.S. Evaluation of Clinical Prediction Models (Part 2): How to Undertake an External Validation Study. BMJ 2024, 384, e074820. [Google Scholar] [CrossRef]
- Kickingereder, P.; Bonekamp, D.; Nowosielski, M.; Kratz, A.; Sill, M.; Burth, S.; Wick, A.; Eidel, O.; Schlemmer, H.-P.; Radbruch, A.; et al. Radiogenomics of Glioblastoma: Machine Learning–Based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology 2016, 281, 907–918. [Google Scholar] [CrossRef]
- Wehrle, C.J.; Hong, H.; Kamath, S.; Schlegel, A.; Fujiki, M.; Hashimoto, K.; Kwon, D.C.H.; Miller, C.; Walsh, R.M.; Aucejo, F. Tumor Mutational Burden From Circulating Tumor DNA Predicts Recurrence of Hepatocellular Carcinoma After Resection: An Emerging Biomarker for Surveillance. Ann. Surg. 2024, 280, 504–513. [Google Scholar] [CrossRef]
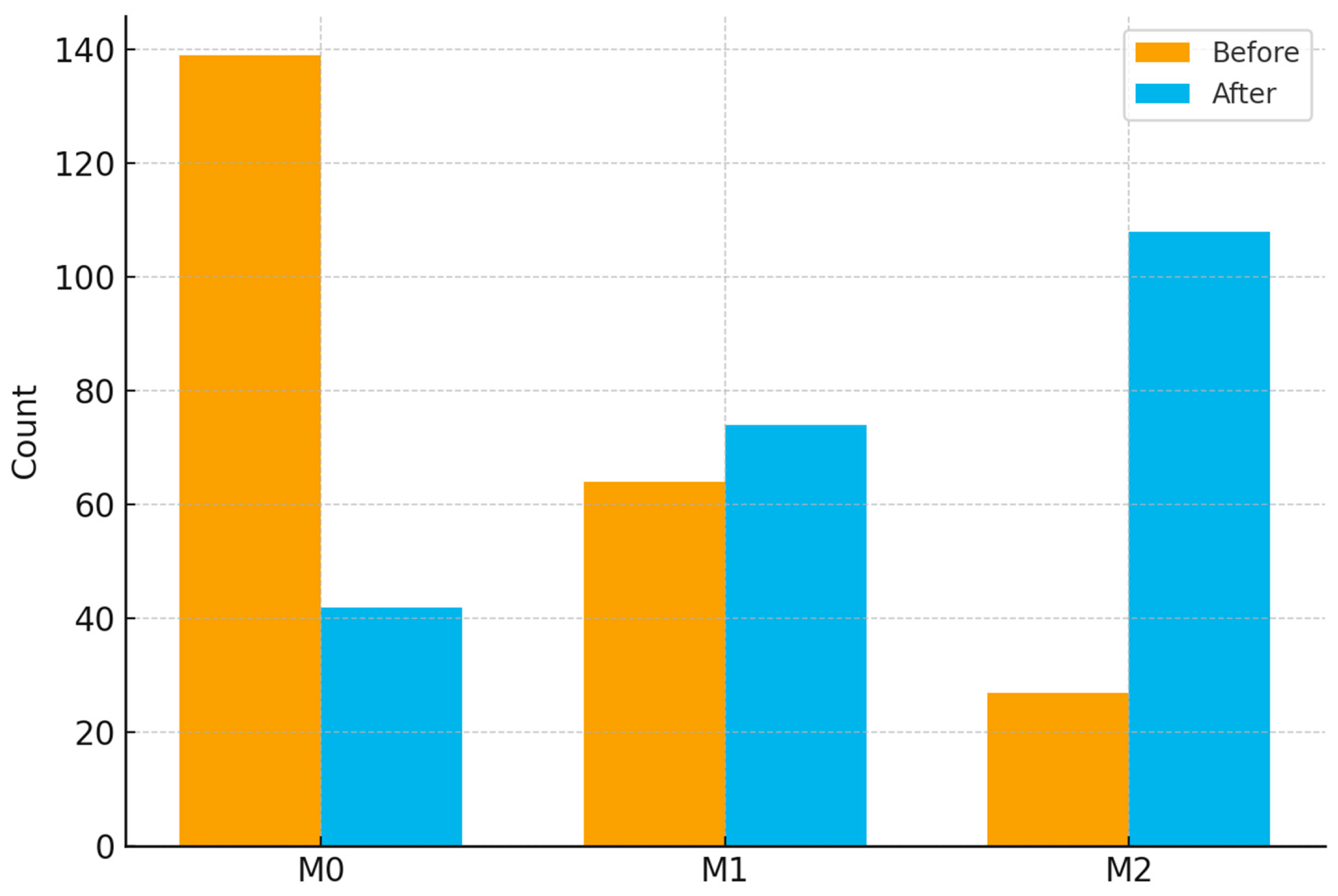
| Characteristic | M0 | M1 | M2 | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | 56.49 ± 10.43 | 55.21 ± 10.95 | 55.51 ± 9.56 | 0.52 |
| Sex | 227 (85.0%) | 85 (82.5%) | 55 (82.1%) | 0.758 |
| Tumor characteristics | ||||
| Tumor number | 1.00 [1.00–1.00] | 1.00 [1.00–1.00] | 1.00 [1.00–1.00] | 0.822 |
| BCLC stage | 156 (58.4%) | 63 (61.2%) | 35 (52.2%) | 0.145 |
| Child–Pugh stage | 247 (93.2%) | 100 (97.1%) | 60 (89.6%) | 0.122 |
| AFP (ng/mL) | 12.79 [3.54–163.90] | 40.70 [9.54–544.92] | 198.80 [28.70–1210.00] | <0.001 |
| PIVKA-II (mAU/mL) | 100.00 [31.00–553.25] | 163.00 [35.75–839.25] | 234.00 [81.00–2204.00] | <0.001 |
| Laboratory indicators | ||||
| CA19-9 (U/mL) | 16.40 [8.65–30.06] | 14.20 [9.20–24.80] | 12.45 [8.60–26.74] | 0.323 |
| CEA (ng/mL) | 2.40 [1.60–3.42] | 2.80 [1.50–4.07] | 2.70 [1.80–3.60] | 0.346 |
| ALT (U/L) | 28.00 [19.00–42.00] | 25.50 [17.00–39.75] | 30.00 [23.00–42.50] | 0.316 |
| AST (U/L) | 26.00 [20.00–38.00] | 26.00 [20.00–37.00] | 26.00 [21.25–41.75] | 0.408 |
| Albumin (g/L) | 42.00 [39.38–44.80] | 41.55 [39.30–44.50] | 42.35 [39.32–44.33] | 0.995 |
| TBIL (µmol/L) | 13.75 [10.85–19.55] | 14.60 [11.00–18.70] | 14.50 [10.93–20.27] | 0.918 |
| DBIL (µmol/L) | 5.35 [4.10–7.62] | 5.70 [4.20–6.80] | 5.80 [3.95–7.97] | 0.839 |
| IBIL (µmol/L) | 8.10 [6.38–11.55] | 9.15 [6.70–11.57] | 8.40 [6.83–11.40] | 0.582 |
| GGT (U/L) | 40.00 [25.75–83.00] | 43.00 [26.00–79.75] | 51.00 [28.50–111.25] | 0.456 |
| Cholinesterase (U/L) | 6906.82 ± 1783.92 | 6997.93 ± 1737.74 | 6885.03 ± 1826.85 | 0.903 |
| Total protein (g/L) | 68.65 ± 5.75 | 68.64 ± 4.95 | 67.03 ± 5.13 | 0.088 |
| Cholesterol (mmol/L) | 3.75 [3.27–4.38] | 3.78 [3.38–4.28] | 4.11 [3.57–4.59] | 0.187 |
| Triglyceride (mmol/L) | 1.01 [0.78–1.31] | 1.02 [0.80–1.38] | 1.04 [0.88–1.33] | 0.596 |
| HDL-C (mmol/L) | 1.15 [0.97–1.38] | 1.05 [0.91–1.32] | 1.04 [0.84–1.23] | 0.015 |
| LDL-C (mmol/L) | 2.33 [1.93–2.84] | 2.44 [2.07–2.71] | 2.63 [2.20–3.18] | 0.02 |
| HBsAg | 220 (82.4%) | 94 (91.3%) | 56 (83.6%) | 0.102 |
| HBsAb | 40 (15.0%) | 13 (12.6%) | 12 (17.9%) | 0.637 |
| HBeAg | 48 (18.0%) | 23 (22.3%) | 13 (19.4%) | 0.635 |
| HBeAb | 185 (69.3%) | 67 (65.0%) | 48 (71.6%) | 0.622 |
| HBcAb | 253 (94.8%) | 101 (98.1%) | 64 (95.5%) | 0.32 |
| HBV DNA | 260 (97.4%) | 100 (97.1%) | 65 (97.0%) | 0.98 |
| Dataset | Accuracy | Weighted F1-Score | Macro-Average Precision | Macro-Average Recall | Macro-Average AUC (95% CI) |
|---|---|---|---|---|---|
| Training Set | 0.759 | 0.766 | 0.761 | 0.794 | 0.920 [0.884–0.955] |
| Test Set | 0.733 | 0.733 | 0.635 | 0.635 | 0.880 [0.807–0.953] |
| Validation Set | 0.758 | 0.769 | 0.676 | 0.719 | 0.886 [0.833–0.940] |
| Dataset | Class | Support | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|
| Training | M0 | 42 | 0.976 [0.87–1.00] | 0.802 [0.74–0.86] |
| Training | M1 | 74 | 0.676 [0.56–0.78] | 0.9467 [0.90–0.98] |
| Training | M2 | 108 | 0.731 [0.64–0.81] | 0.914 [0.85–0.96] |
| Test | M0 | 44 | 0.886 [0.75–0.96] | 0.839 [0.66–0.95] |
| Test | M1 | 18 | 0.556 [0.31–0.78] | 0.860 [0.74–0.94] |
| Test | M2 | 13 | 0.462 [0.19–0.75] | 0.887 [0.78–0.95] |
| Validation | M0 | 84 | 0.798 [0.70–0.88] | 0.875 [0.75–0.95] |
| Validation | M1 | 21 | 0.619 [0.38–0.82] | 0.865 [0.79–0.92] |
| Validation | M2 | 27 | 0.895 [0.54–0.95] | 0.8952 [0.82–0.95] |
| Model | Validation Accuracy | Validation F1-Score | Validation AUC | Test Accuracy | Test F1-Score | Test AUC |
|---|---|---|---|---|---|---|
| Random Forest | 0.682 | 0.694 | 0.834 | 0.640 | 0.660 | 0.827 |
| Logistic Regression | 0.629 | 0.638 | 0.837 | 0.547 | 0.575 | 0.802 |
| XGBoost | 0.545 | 0.568 | 0.807 | 0.533 | 0.557 | 0.804 |
| LightGBM | 0.523 | 0.547 | 0.800 | 0.573 | 0.596 | 0.823 |
| Transformer (ours) | 0.758 | 0.769 | 0.886 | 0.733 | 0.733 | 0.880 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, R.; Chen, H.; Zou, W.; Gu, M.; Zhao, X.; Jia, N.; Liu, W. Transformer-Based Deep Learning for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Cancers 2025, 17, 3314. https://doi.org/10.3390/cancers17203314
He R, Chen H, Zou W, Gu M, Zhao X, Jia N, Liu W. Transformer-Based Deep Learning for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Cancers. 2025; 17(20):3314. https://doi.org/10.3390/cancers17203314
Chicago/Turabian StyleHe, Ruilin, Huilin Chen, Wenjie Zou, Mengting Gu, Xingyu Zhao, Ningyang Jia, and Wanmin Liu. 2025. "Transformer-Based Deep Learning for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma" Cancers 17, no. 20: 3314. https://doi.org/10.3390/cancers17203314
APA StyleHe, R., Chen, H., Zou, W., Gu, M., Zhao, X., Jia, N., & Liu, W. (2025). Transformer-Based Deep Learning for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Cancers, 17(20), 3314. https://doi.org/10.3390/cancers17203314






