Lead Exposure and Bladder Cancer: Molecular Insights from TCGA RNA-Seq and Toxicogenomic Integration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria and Cohort Definition
2.2. Data Source
2.3. Preprocessing and Quality Control
2.4. Differential Gene Expression Analysis
2.5. Visualization and Candidate Gene Selection
2.6. Survival Analysis
2.7. Multivariable Survival Modeling
2.8. Sensitivity (Stage Parameterization)
2.9. Toxicogenomic Integration (Lead)
2.10. Pathway Over-Representation Analysis (ORA)
2.11. Lead-Response Gene Set Score and Survival Modeling
2.12. Operationalization of the Lead-Response Composite Score
2.13. Subtype Context (Exploratory Plan)
2.14. Statistical Software and Multiplicity Control
3. Results
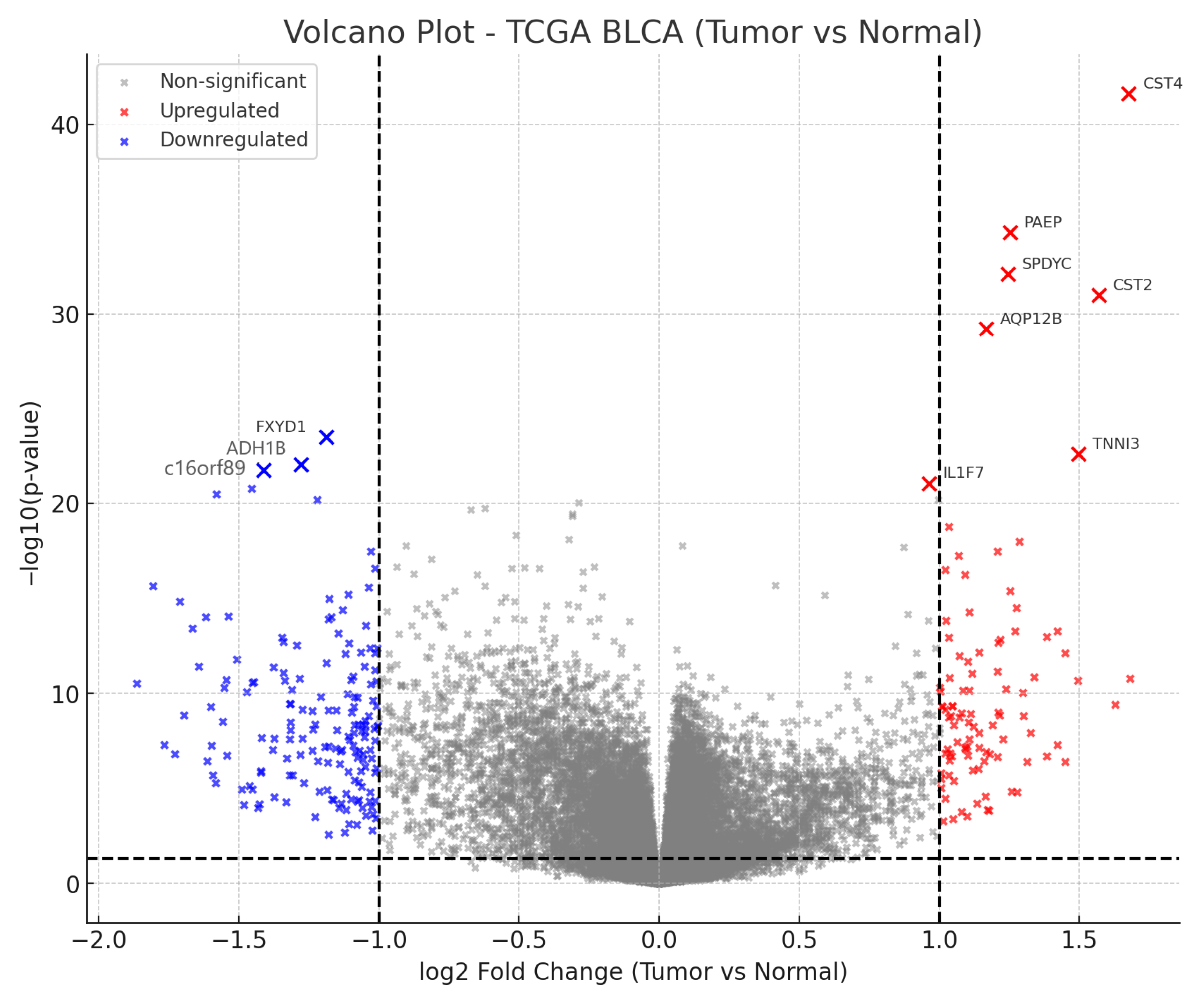
3.1. Differential Gene Expression in BLCA
3.2. Top Ten Most Significant Genes
3.3. Biological Interpretation
3.4. Survival Findings
3.5. Multivariable Survival Analysis
3.6. Sensitivity Analysis (Stage Parameterization)
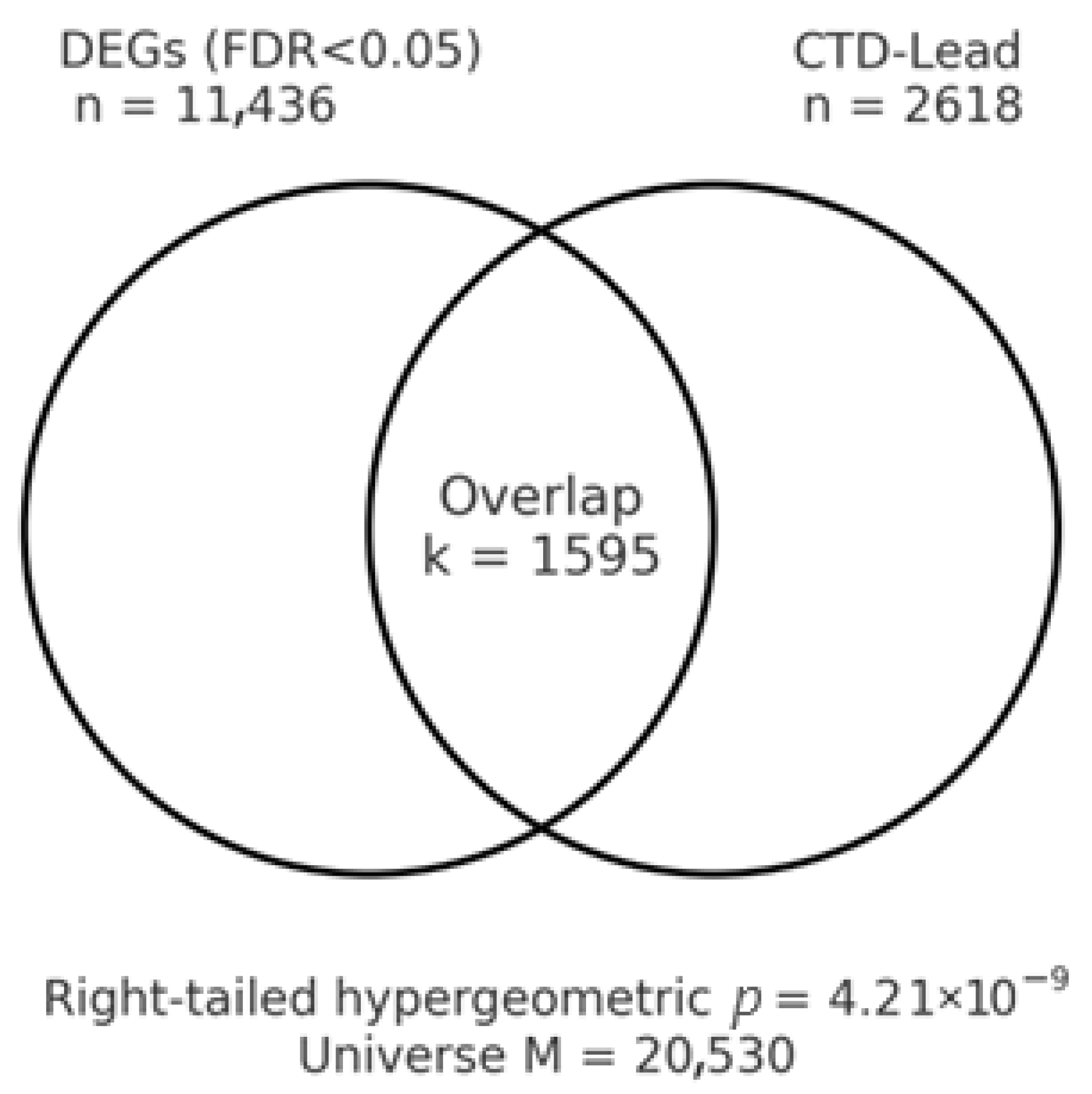
3.7. Toxicogenomic Overlap with Lead
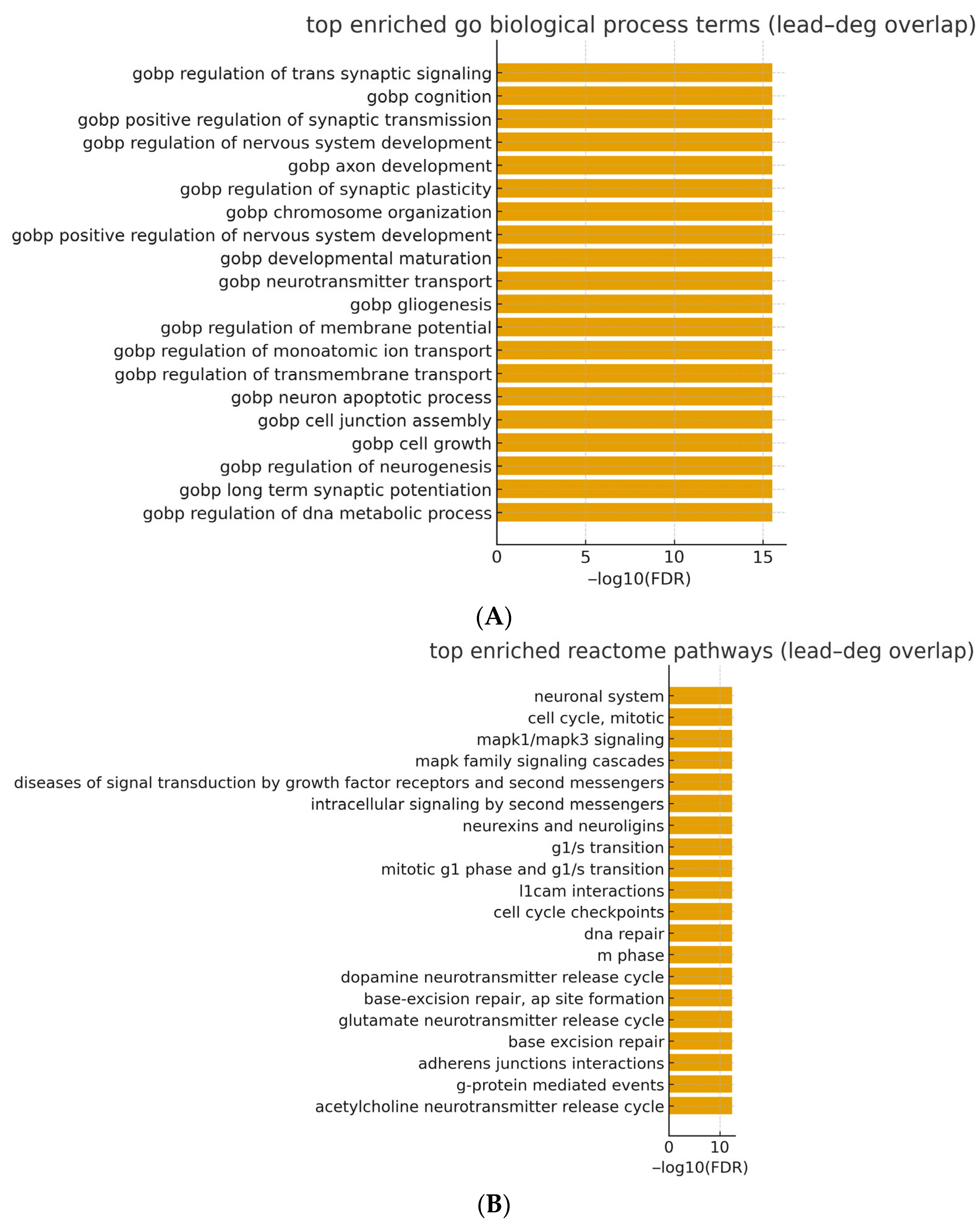
3.8. Pathway Over-Representation Results (Lead–DEG Overlap)
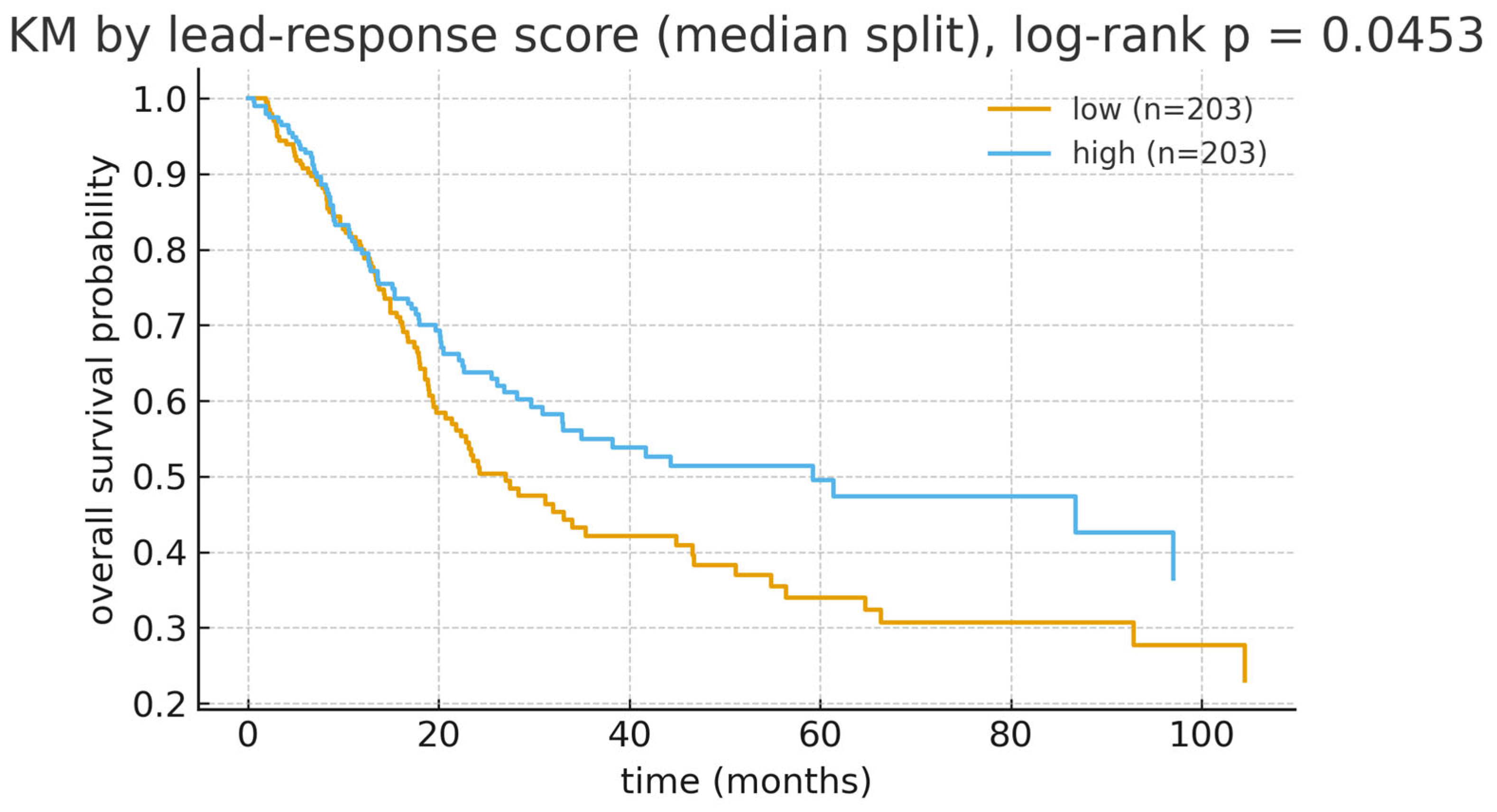
3.9. Lead-Response Gene Set Score: Survival Results
3.10. Sensitivity and Subgroup Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zhang, Y.; Rumgay, H.; Li, M.; Yu, H.; Pan, H.; Ni, J. The global landscape of bladder cancer incidence and mortality in 2020 and projections to 2040. J. Glob. Health 2023, 13, 04109. [Google Scholar] [CrossRef]
- Jankovic, S.; Radosavljevic, V. Risk factors for bladder cancer. Tumori 2007, 93, 4–12. [Google Scholar] [CrossRef]
- Jubber, I.; Ong, S.; Bukavina, L.; Black, P.C.; Comperat, E.; Kamat, A.M.; Kiemeney, L.; Lawrentschuk, N.; Lerner, S.P.; Meeks, J.J.; et al. Epidemiology of Bladder Cancer in 2023: A Systematic Review of Risk Factors. Eur. Urol. 2023, 84, 176–190. [Google Scholar] [CrossRef]
- Cumberbatch, M.G.K.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T.; et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef]
- Naujokas, M.F.; Anderson, B.; Ahsan, H.; Aposhian, H.V.; Graziano, J.H.; Thompson, C.; Suk, W.A. The broad scope of health effects from chronic arsenic exposure: Update on a worldwide public health problem. Environ. Health Perspect. 2013, 121, 295–302. [Google Scholar] [CrossRef]
- Piyathilake, C. Dietary factors associated with bladder cancer. Investig. Clin. Urol. 2016, 57 (Suppl. S1), S14–S25. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Arsenic, metals, fibres, and dusts. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100 Pt C, 11–465. [Google Scholar]
- Issanov, A.; Adewusi, B.; Dummer, T.J.B.; Saint-Jacques, N. Arsenic in Drinking Water and Urinary Tract Cancers: A Systematic Review Update. Water 2023, 15, 2185. [Google Scholar] [CrossRef]
- Crippa, A.; Larsson, S.C.; Discacciati, A.; Wolk, A.; Orsini, N. Red and processed meat consumption and risk of bladder cancer: A dose-response meta-analysis of epidemiological studies. Eur. J. Nutr. 2018, 57, 689–701. [Google Scholar] [CrossRef]
- Yu, J.; Li, H.; Liu, Z.; Wang, T.; Zhou, F.; Ma, S.; Chen, B.; Chen, W. Meat Intake and the Risk of Bladder Cancer: A Systematic Review and Meta-Analysis of Observational Studies. Nutr. Cancer 2023, 75, 825–845. [Google Scholar] [CrossRef] [PubMed]
- Bayne, C.E.; Farah, D.; Herbst, K.W.; Hsieh, M.H. Role of urinary tract infection in bladder cancer: A systematic review and meta-analysis. World J. Urol. 2018, 36, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Al-Shammari, A.; Al-Abkal, J. Chronic urinary tract infection and bladder carcinoma risk: A meta-analysis of case-control and cohort studies. World J. Urol. 2018, 36, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Finocchiaro, A.; Tylecki, A.; Vigano, S.; Bertini, A.; Ficarra, V.; Di Trapani, E.; Salonia, A.; Briganti, A.; Montorsi, F.; Lughezzani, G.; et al. Socioeconomic disparities and bladder cancer stage at diagnosis: A statewide cohort analysis. JNCI Cancer Spectr. 2025, 9, pkaf054. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Sciaky, D.; Barkalow, F.; Strong, M.; Wyatt, B.; Wiegers, J.; McMorran, R.; Abrar, S.; Mattingly, C.J. Comparative Toxicogenomics Database’s 20th anniversary: Update 2025. Nucleic Acids Res. 2025, 53, D1328–D1334. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e525. [Google Scholar] [CrossRef]
- Rebouissou, S.; Bernard-Pierrot, I.; de Reynies, A.; Lepage, M.L.; Krucker, C.; Chapeaublanc, E.; Herault, A.; Kamoun, A.; Caillault, A.; Letouze, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra291. [Google Scholar] [CrossRef]
- Jiang, L.J.; Guo, S.B.; Huang, Z.Y.; Li, X.L.; Jin, X.H.; Huang, W.J.; Tian, X.P. PHB promotes bladder cancer cell epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway. Pathol. Res. Pract. 2023, 247, 154536. [Google Scholar] [CrossRef]
- Jiang, L.J.; Guo, S.B.; Zhou, Z.H.; Li, Z.Y.; Zhou, F.J.; Yu, C.P.; Li, M.; Huang, W.J.; Liu, Z.W.; Tian, X.P. Snai2-mediated upregulation of NADSYN1 promotes bladder cancer progression by interacting with PHB. Clin. Transl. Med. 2024, 14, e1555. [Google Scholar] [CrossRef]
- Bates, A.S.; Ayers, J.; Kostakopoulos, N.; Lumsden, T.; Schoots, I.G.; Willemse, P.M.; Yuan, Y.; van den Bergh, R.C.N.; Grummet, J.P.; van der Poel, H.G.; et al. A Systematic Review of Focal Ablative Therapy for Clinically Localised Prostate Cancer in Comparison with Standard Management Options: Limitations of the Available Evidence and Recommendations for Clinical Practice and Further Research. Eur. Urol. Oncol. 2021, 4, 405–423. [Google Scholar] [CrossRef]
- Sjodahl, G.; Lauss, M.; Lovgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Ferno, M.; Ringner, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Dyrskjot, L.; Hansel, D.E.; Efstathiou, J.A.; Knowles, M.A.; Galsky, M.D.; Teoh, J.; Theodorescu, D. Bladder cancer. Nat. Rev. Dis. Primers 2023, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Kumagai, R.; Tanaka, T.; Nakano, T.; Fujie, T.; Fujiwara, Y.; Yamamoto, C.; Kaji, T. Lead suppresses perlecan expression via EGFR-ERK1/2-COX-2-PGI(2) pathway in cultured bovine vascular endothelial cells. J. Toxicol. Sci. 2023, 48, 655–663. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Inorganic and organic lead compounds. IARC Monogr. Eval. Carcinog. Risks Hum. 2006, 87, 1–471. [Google Scholar]
- Barbosa, F., Jr.; Tanus-Santos, J.E.; Gerlach, R.F.; Parsons, P.J. A critical review of biomarkers used for monitoring human exposure to lead: Advantages, limitations, and future needs. Environ. Health Perspect. 2005, 113, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Lanphear, B.P.; Rauch, S.; Auinger, P.; Allen, R.W.; Hornung, R.W. Low-level lead exposure and mortality in US adults: A population-based cohort study. Lancet Public Health 2018, 3, e177–e184. [Google Scholar] [CrossRef]
- Bergdahl, I.A.; Skerfving, S. Biomonitoring of lead exposure-alternatives to blood. J. Toxicol. Environ. Health Part A 2008, 71, 1235–1243. [Google Scholar] [CrossRef]
- Wright, R.O.; Schwartz, J.; Wright, R.J.; Bollati, V.; Tarantini, L.; Park, S.K.; Hu, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. 2010, 118, 790–795. [Google Scholar] [CrossRef]
- Elkin, E.R.; Higgins, C.; Aung, M.T.; Bakulski, K.M. Metals Exposures and DNA Methylation: Current Evidence and Future Directions. Curr. Environ. Health Rep. 2022, 9, 673–696. [Google Scholar] [CrossRef]
- Heiss, J.A.; Tellez-Rojo, M.M.; Estrada-Gutierrez, G.; Schnaas, L.; Amarasiriwardena, C.; Baccarelli, A.A.; Wright, R.O.; Just, A.C. Prenatal lead exposure and cord blood DNA methylation in PROGRESS: An epigenome-wide association study. Environ. Epigenet. 2020, 6, dvaa014. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hivert, M.F.; Cardenas, A.; Zhong, J.; Rifas-Shiman, S.L.; Agha, G.; Colicino, E.; Just, A.C.; Amarasiriwardena, C.; Lin, X.; et al. Exposure to Low Levels of Lead in Utero and Umbilical Cord Blood DNA Methylation in Project Viva: An Epigenome-Wide Association Study. Environ. Health Perspect. 2017, 125, 087019. [Google Scholar] [CrossRef] [PubMed]
- Flora, G.; Gupta, D.; Tiwari, A. Toxicity of lead: A review with recent updates. Interdiscip. Toxicol. 2012, 5, 47–58. [Google Scholar] [CrossRef]
- Ijomone, O.M.; Iroegbu, J.D.; Aschner, M.; Bornhorst, J. Impact of environmental toxicants on p38- and ERK-MAPK signaling pathways in the central nervous system. Neurotoxicology 2021, 86, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.; Cheong, Y.K.; Kim, N.H.; Chung, H.T.; Kang, D.G.; Pae, H.O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J. Signal Transduct. 2011, 2011, 792639. [Google Scholar] [CrossRef]
- Zhang, Z.; Hernandez, K.; Savage, J.; Li, S.; Miller, D.; Agrawal, S.; Ortuno, F.; Staudt, L.M.; Heath, A.; Grossman, R.L. Uniform genomic data analysis in the NCI Genomic Data Commons. Nat. Commun. 2021, 12, 1226. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Huang, M.; Li, H.; Chen, J.; Li, L.; Zhan, Y.; Du, Y.; Bian, J.; Chen, M.; Lai, D. Blood lead levels and bladder cancer among US participants: NHANES 1999–2018. BMC Public Health 2025, 25, 416. [Google Scholar] [CrossRef]
- Golabek, T.; Darewicz, B.; Borawska, M.; Markiewicz, R.; Socha, K.; Kudelski, J. Lead concentration in the bladder tissue and blood of patients with bladder cancer. Scand. J. Urol. Nephrol. 2009, 43, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, J.; Zhang, B.; Liu, Y.; Lu, T.; Shi, Y.; Shan, G.; Dong, L. Urinary Lead Concentration Is an Independent Predictor of Cancer Mortality in the U.S. General Population. Front. Oncol. 2018, 8, 242. [Google Scholar] [CrossRef]
- Kirel, B.; Aksit, M.A.; Bulut, H. Blood lead levels of maternal-cord pairs, children and adults who live in a central urban area in Turkey. Turk. J. Pediatr. 2005, 47, 125–131. [Google Scholar] [PubMed]
- Wang, L.; Wang, C.; Liu, T.; Xuan, H.; Li, X.; Shi, X.; Dai, F.; Chen, J.; Li, D.; Xu, T. Association of low-level lead exposure with all-cause and cardiovascular disease mortality in US adults with hypertension: Evidence from the National Health and Nutrition Examination Survey 2003–2010. Arch. Public Health 2023, 81, 146. [Google Scholar] [CrossRef]
- Laouali, N.; Benmarhnia, T.; Lanphear, B.P.; Oulhote, Y. Associations with Blood Lead and Urinary Cadmium Concentrations in Relation to Mortality in the US Population: A Causal Survival Analysis with G-Computation. Toxics 2023, 11, 133. [Google Scholar] [CrossRef]
- Biswas, A.; Sahoo, S.; Riedlinger, G.M.; Ghodoussipour, S.; Jolly, M.K.; De, S. Transcriptional state dynamics lead to heterogeneity and adaptive tumor evolution in urothelial bladder carcinoma. Commun. Biol. 2023, 6, 1292. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Duan, L.; Cui, D.; Deng, K.; Dong, Z.; Wei, S. Whole transcriptome sequencing identifies a competitive endogenous RNA network that regulates the immunity of bladder cancer. Heliyon 2024, 10, e29344. [Google Scholar] [CrossRef]
- Yoshihara, K.; Ito, K.; Kimura, T.; Yamamoto, Y.; Urabe, F. Single-cell RNA sequencing and spatial transcriptome analysis in bladder cancer: Current status and future perspectives. Bladder Cancer 2025, 11, 23523735251322017. [Google Scholar] [CrossRef]
- Metryka, E.; Chibowska, K.; Gutowska, I.; Falkowska, A.; Kupnicka, P.; Barczak, K.; Chlubek, D.; Baranowska-Bosiacka, I. Lead (Pb) Exposure Enhances Expression of Factors Associated with Inflammation. Int. J. Mol. Sci. 2018, 19, 1813. [Google Scholar] [CrossRef]
- Wei, J.; Du, K.; Cai, Q.; Ma, L.; Jiao, Z.; Tan, J.; Xu, Z.; Li, J.; Luo, W.; Chen, J.; et al. Lead induces COX-2 expression in glial cells in a NFAT-dependent, AP-1/NFkappaB-independent manner. Toxicology 2014, 325, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.W.; Choi, B.Y.; Son, S.Y.; Lee, J.H.; Min, J.Y.; Min, K.B. Lead and cadmium exposure was associated with faster epigenetic aging in a representative sample of adults aged 50 and older in the United States. Chemosphere 2025, 374, 144194. [Google Scholar] [CrossRef]
- Lieberman-Cribbin, W.; Domingo-Relloso, A.; Navas-Acien, A.; Cole, S.; Haack, K.; Umans, J.; Tellez-Plaza, M.; Colicino, E.; Baccarelli, A.A.; Gao, X.; et al. Epigenetic Biomarkers of Lead Exposure and Cardiovascular Disease: Prospective Evidence in the Strong Heart Study. J. Am. Heart Assoc. 2022, 11, e026934. [Google Scholar] [CrossRef]
- Fang, Y.W.; Chen, C.W.; Su, T.C.; Wang, C.; Lin, C.Y. Investigating the associations of blood lead and cadmium with smoking-related DNA methylation and mortality among U.S. adults. Ecotoxicol. Environ. Saf. 2025, 299, 118360. [Google Scholar] [CrossRef]
- Senut, M.C.; Cingolani, P.; Sen, A.; Kruger, A.; Shaik, A.; Hirsch, H.; Suhr, S.T.; Ruden, D. Epigenetics of early-life lead exposure and effects on brain development. Epigenomics 2012, 4, 665–674. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef]
- Yang, B.; Li, G.; Huang, J.; Zhang, X.; Wang, R.; Zhang, J. Notch signaling pathway-based classification of bladder cancer in relation to tumor immune infiltration. Cent. Eur. J. Immunol. 2023, 48, 274–289. [Google Scholar] [CrossRef]
- Bekele, R.T.; Samant, A.S.; Nassar, A.H.; So, J.; Garcia, E.P.; Curran, C.R.; Hwang, J.H.; Mayhew, D.L.; Nag, A.; Thorner, A.R.; et al. RAF1 amplification drives a subset of bladder tumors and confers sensitivity to MAPK-directed therapeutics. J. Clin. Investig. 2021, 131, e147849. [Google Scholar] [CrossRef]
- Mei, Z.; Liu, G.; Zhao, B.; He, Z.; Gu, S. Emerging roles of epigenetics in lead-induced neurotoxicity. Environ. Int. 2023, 181, 108253. [Google Scholar] [CrossRef] [PubMed]
- Maraver, A.; Fernandez-Marcos, P.J.; Cash, T.P.; Mendez-Pertuz, M.; Duenas, M.; Maietta, P.; Martinelli, P.; Munoz-Martin, M.; Martinez-Fernandez, M.; Canamero, M.; et al. NOTCH pathway inactivation promotes bladder cancer progression. J. Clin. Investig. 2015, 125, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Ferrandino, F.; Grazioli, P.; Bellavia, D.; Campese, A.F.; Screpanti, I.; Felli, M.P. Notch and NF-kappaB: Coach and Players of Regulatory T-Cell Response in Cancer. Front. Immunol. 2018, 9, 2165. [Google Scholar] [CrossRef] [PubMed]
- Friesen, M.C.; Costello, S.; Eisen, E.A. Quantitative exposure to metalworking fluids and bladder cancer incidence in a cohort of autoworkers. Am. J. Epidemiol. 2009, 169, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, U.; Kumari, N.; Vasudeva, P.; Mohanty, N.K.; Saxena, S. Overexpression of COX2 indicates poor survival in urothelial bladder cancer. Ann. Diagn. Pathol. 2018, 34, 50–55. [Google Scholar] [CrossRef] [PubMed]



| Characteristic | Value |
|---|---|
| N (tumor cohort) | 412 |
| Age, years (median [IQR]) | 69 [60–76] |
| Sex—Male | 304 (73.8%) |
| Sex—Female | 108 (26.2%) |
| Pathological stage—I | 2 (0.5%) |
| Pathological stage—II | 131 (31.8%) |
| Pathological stage—III | 141 (34.2%) |
| Pathological stage—IV | 136 (33.0%) |
| Pathological stage—Missing | 2 (0.5%) |
| OS (N/events; median months [IQR]) | 411/180; 17.6 [10.8–31.6] |
| PFI (N/events; median months [IQR]) | 412/177; 14.1 [7.1–26.2] |
| DFI (N/events; median months [IQR]) | 189/32; 19.0 [12.2–41.7] |
| DSS (N/events; median months [IQR]) | 411/123; 17.6 [10.8–31.6] |
| Gene | log2FC | p-Value | FDR |
|---|---|---|---|
| CST4 | 1.676252 | 2.45 × 10−42 | 4.19 × 10−38 |
| PAEP | 1.252275 | 5.14 × 10−35 | 4.4 × 10−31 |
| SPDYC | 1.244989 | 8.1 × 10−33 | 4.62 × 10−29 |
| CST2 | 1.568127 | 1.03 × 10−31 | 4.42 × 10−28 |
| AQP12B | 1.16774 | 6.07 × 10−30 | 2.08 × 10−26 |
| FXYD1 | −1.18892 | 3.29 × 10−24 | 9.38 × 10−21 |
| TNNI3 | 1.497404 | 2.64 × 10−23 | 6.47 × 10−20 |
| ADH1B | −1.27773 | 9.12 × 10−23 | 1.95 × 10−19 |
| C16orf89 | −1.41286 | 1.88 × 10−22 | 3.57 × 10−19 |
| IL1F7 | 0.962661 | 9.21 × 10−22 | 1.58 × 10−18 |
| Gene | N_high | N_low | Logrank_p_OS | Cox_HR_ per_SD_OS | Cox_95%CI_low | Cox_95%CI_high | Cox_p_OS | PFI_N | PFI_logrank_p | DFI_N | DFI_logrank_p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AQP12B | 203 | 203 | 0.004 | 0.771 | 0.644 | 0.923 | 0.004 | 407 | 0.023 | 187 | 0.121 |
| C16orf89 | 203 | 203 | 0.015 | 1.116 | 0.971 | 1.282 | 0.121 | 407 | 0.134 | 187 | 0.693 |
| CST4 | 203 | 203 | 0.134 | 0.865 | 0.735 | 1.019 | 0.083 | 407 | 0.136 | 187 | 0.764 |
| FXYD1 | 203 | 203 | 0.142 | 1.126 | 0.977 | 1.297 | 0.099 | 407 | 0.354 | 187 | 0.806 |
| SPDYC | 203 | 203 | 0.190 | 1.115 | 0.979 | 1.270 | 0.099 | 407 | 0.397 | 187 | 0.300 |
| TNNI3 | 203 | 203 | 0.190 | 1.038 | 0.895 | 1.203 | 0.618 | 407 | 0.198 | 187 | 0.430 |
| CST2 | 203 | 203 | 0.286 | 1.127 | 0.976 | 1.301 | 0.100 | 407 | 0.856 | 187 | 0.290 |
| PAEP | 203 | 203 | 0.395 | 1.078 | 0.932 | 1.247 | 0.310 | 407 | 0.168 | 187 | 0.593 |
| ADH1B | 203 | 203 | 0.451 | 1.070 | 0.928 | 1.235 | 0.348 | 407 | 0.212 | 187 | 0.982 |
| IL1F7 | 203 | 203 | 0.814 | 0.925 | 0.782 | 1.094 | 0.364 | 407 | 0.835 | 187 | 0.566 |
| Gene | Endpoint | N | HR_gene_perSD | CI_low | CI_high | p_Wald |
|---|---|---|---|---|---|---|
| CST4 | DFI | 186 | 0.762 | 0.506 | 1.147 | 0.193 |
| AQP12B | DFI | 186 | 0.750 | 0.486 | 1.157 | 0.194 |
| IL1F7 | DFI | 186 | 0.804 | 0.530 | 1.217 | 0.303 |
| FXYD1 | DFI | 186 | 0.844 | 0.581 | 1.225 | 0.374 |
| C16orf89 | DFI | 186 | 0.851 | 0.576 | 1.258 | 0.421 |
| TNNI3 | DFI | 186 | 0.877 | 0.596 | 1.290 | 0.506 |
| PAEP | DFI | 186 | 1.079 | 0.763 | 1.525 | 0.665 |
| CST2 | DFI | 186 | 0.936 | 0.637 | 1.373 | 0.735 |
| SPDYC | DFI | 186 | 1.050 | 0.750 | 1.469 | 0.775 |
| ADH1B | DFI | 186 | 0.999 | 0.692 | 1.443 | 0.998 |
| AQP12B | DSS | 391 | 0.710 | 0.560 | 0.899 | 0.004 |
| CST4 | DSS | 391 | 0.799 | 0.645 | 0.990 | 0.040 |
| IL1F7 | DSS | 391 | 0.855 | 0.684 | 1.070 | 0.172 |
| PAEP | DSS | 391 | 1.082 | 0.906 | 1.292 | 0.380 |
| SPDYC | DSS | 391 | 1.071 | 0.908 | 1.263 | 0.411 |
| ADH1B | DSS | 391 | 0.961 | 0.804 | 1.148 | 0.662 |
| TNNI3 | DSS | 391 | 0.965 | 0.806 | 1.154 | 0.697 |
| C16orf89 | DSS | 391 | 1.028 | 0.862 | 1.225 | 0.756 |
| FXYD1 | DSS | 391 | 0.981 | 0.818 | 1.176 | 0.836 |
| CST2 | DSS | 391 | 0.981 | 0.817 | 1.177 | 0.839 |
| AQP12B | OS | 404 | 0.761 | 0.633 | 0.915 | 0.003 |
| CST4 | OS | 404 | 0.841 | 0.708 | 1.000 | 0.050 |
| IL1F7 | OS | 404 | 0.874 | 0.731 | 1.044 | 0.139 |
| SPDYC | OS | 404 | 1.089 | 0.952 | 1.245 | 0.211 |
| C16orf89 | OS | 404 | 1.082 | 0.939 | 1.247 | 0.273 |
| PAEP | OS | 404 | 1.075 | 0.926 | 1.248 | 0.336 |
| CST2 | OS | 404 | 1.023 | 0.879 | 1.190 | 0.765 |
| FXYD1 | OS | 404 | 1.020 | 0.879 | 1.184 | 0.790 |
| TNNI3 | OS | 404 | 0.980 | 0.846 | 1.136 | 0.795 |
| ADH1B | OS | 404 | 1.001 | 0.865 | 1.159 | 0.983 |
| CST4 | PFI | 405 | 0.809 | 0.682 | 0.960 | 0.015 |
| PAEP | PFI | 405 | 1.140 | 0.988 | 1.316 | 0.070 |
| AQP12B | PFI | 405 | 0.853 | 0.716 | 1.017 | 0.077 |
| IL1F7 | PFI | 405 | 0.870 | 0.726 | 1.042 | 0.130 |
| CST2 | PFI | 405 | 0.945 | 0.810 | 1.104 | 0.481 |
| FXYD1 | PFI | 405 | 0.947 | 0.816 | 1.101 | 0.484 |
| C16orf89 | PFI | 405 | 0.954 | 0.821 | 1.109 | 0.546 |
| TNNI3 | PFI | 405 | 1.030 | 0.888 | 1.195 | 0.690 |
| SPDYC | PFI | 405 | 1.028 | 0.890 | 1.187 | 0.700 |
| ADH1B | PFI | 405 | 0.992 | 0.855 | 1.150 | 0.917 |
| Gene | Direction | log2FC | FDR |
|---|---|---|---|
| SNORA72 | Up | 0.459 | 3.39 × 10−20 |
| TNNI3 | Up | 2.45 | 4.52 × 10−20 |
| SERPINA10 | Up | 0.45 | 4.41 × 10−17 |
| SMPD4 | Up | 0.671 | 8.35 × 10−16 |
| AHSG | Up | 0.823 | 2.31 × 10−15 |
| ORM2 | Up | 0.658 | 8.56 × 10−15 |
| KNG1 | Up | 0.59 | 3.32 × 10−13 |
| ILF3 | Up | 0.579 | 5.54 × 10−11 |
| XPO5 | Up | 0.764 | 3.05 × 10−10 |
| FOXK2 | Up | 0.575 | 3.15 × 10−10 |
| APCS | Up | 0.383 | 8.81 × 10−10 |
| GC | Up | 0.756 | 9.97 × 10−10 |
| ALB | Up | 1.576 | 1.02 × 10−09 |
| CCT6A | Up | 0.537 | 7.59 × 10−09 |
| ACP1 | Up | 0.65 | 9.97 × 10−09 |
| FXYD1 | Down | −5.271 | 6.75 × 10−21 |
| ADH1B | Down | −7.567 | 1.34 × 10−19 |
| SOX10 | Down | −4.782 | 1.87 × 10−18 |
| CADM3 | Down | −5.809 | 6.06 × 10−18 |
| BCL2 | Down | −2.033 | 4.44 × 10−16 |
| SYBU | Down | −2.374 | 8.61 × 10−15 |
| F10 | Down | −4.712 | 9.37 × 10−15 |
| PHYHIP | Down | −3.931 | 1.09 × 10−13 |
| RCBTB2 | Down | −1.306 | 1.96 × 10−13 |
| NRXN1 | Down | −5.583 | 3.50 × 10−13 |
| GFRA1 | Down | −4.808 | 9.58 × 10−13 |
| PTH1R | Down | −3.298 | 9.58 × 10−13 |
| SORCS1 | Down | −5.391 | 1.53 × 10−12 |
| RASGEF1C | Down | −2.593 | 1.72 × 10−12 |
| MATN2 | Down | −3.231 | 1.98 × 10−12 |
| Collection | Term | k/n | K | p | FDR |
|---|---|---|---|---|---|
| GO BP | Gobp regulation of trans synaptic signaling | 0.0627 | 483 | 6.17 × 10−20 | 2.95 × 10−16 |
| GO BP | Gobp cognition | 0.0420 | 308 | 7.36 × 10−15 | 2.95 × 10−16 |
| GO BP | Gobp positive regulation of synaptic transmission | 0.0282 | 158 | 7.62 × 10−15 | 2.95 × 10−16 |
| GO BP | Gobp regulation of nervous system development | 0.0514 | 424 | 8.75 × 10−15 | 2.95 × 10−16 |
| GO BP | Gobp axon development | 0.0571 | 499 | 1.19 × 10−14 | 2.95 × 10−16 |
| GO BP | Gobp regulation of synaptic plasticity | 0.0320 | 204 | 4.03 × 10−14 | 2.95 × 10−16 |
| GO BP | Gobp chromosome organization | 0.0527 | 475 | 7.28 × 10−13 | 2.95 × 10−16 |
| GO BP | Gobp positive regulation of nervous system development | 0.0357 | 264 | 1.06 × 10−12 | 2.95 × 10−16 |
| GO BP | Gobp developmental maturation | 0.0408 | 332 | 3.06 × 10−12 | 2.95 × 10−16 |
| GO BP | Gobp neurotransmitter transport | 0.0301 | 208 | 5.27 × 10−12 | 2.95 × 10−16 |
| GO BP | Gobp gliogenesis | 0.0395 | 327 | 1.37 × 10−11 | 2.95 × 10−16 |
| GO BP | Gobp regulation of membrane potential | 0.0483 | 444 | 1.80 × 10−11 | 2.95 × 10−16 |
| GO BP | Gobp regulation of monoatomic ion transport | 0.0451 | 403 | 1.99 × 10−11 | 2.95 × 10−16 |
| GO BP | Gobp regulation of transmembrane transport | 0.0433 | 388 | 6.49 × 10−11 | 2.95 × 10−16 |
| GO BP | Gobp neuron apoptotic process | 0.0357 | 297 | 1.50 × 10−10 | 2.95 × 10−16 |
| GO BP | Gobp cell junction assembly | 0.0502 | 492 | 1.92 × 10−10 | 2.95 × 10−16 |
| GO BP | Gobp cell growth | 0.0470 | 453 | 3.07 × 10−10 | 2.95 × 10−16 |
| GO BP | Gobp regulation of neurogenesis | 0.0389 | 349 | 6.31 × 10−10 | 2.95 × 10−16 |
| GO BP | Gobp long term synaptic potentiation | 0.0176 | 97 | 6.43 × 10−10 | 2.95 × 10−16 |
| GO BP | Gobp regulation of DNA metabolic process | 0.0445 | 427 | 7.56 × 10−10 | 2.95 × 10−16 |
| Reactome | Neuronal system | 0.0520 | 407 | 2.44 × 10−16 | 3.77 × 10−13 |
| Reactome | Cell cycle, mitotic | 0.0470 | 452 | 2.77 × 10−10 | 3.77 × 10−13 |
| Reactome | MAPK1/MAPK3 signaling | 0.0307 | 264 | 9.07 × 10−09 | 3.77 × 10−13 |
| Reactome | Mapk family signaling cascades | 0.0332 | 299 | 1.18 × 10−08 | 3.77 × 10−13 |
| Reactome | Diseases of signal transduction by growth factor receptors and second messengers | 0.0433 | 438 | 1.29 × 10−08 | 3.77 × 10−13 |
| Reactome | Intracellular signaling by second messengers | 0.0326 | 294 | 1.75 × 10−08 | 3.77 × 10−13 |
| Reactome | Neurexins and neuroligins | 0.0119 | 57 | 2.70 × 10−08 | 3.77 × 10−13 |
| Reactome | G1/s transition | 0.0176 | 114 | 3.29 × 10−08 | 3.77 × 10−13 |
| Reactome | Mitotic g1 phase and g1/s transition | 0.0188 | 132 | 7.12 × 10−08 | 3.77 × 10−13 |
| Reactome | L1cam interactions | 0.0169 | 116 | 1.89 × 10−07 | 3.77 × 10−13 |
| Reactome | Cell cycle checkpoints | 0.0257 | 231 | 5.02 × 10−07 | 3.77 × 10−13 |
| Reactome | DNA repair | 0.0282 | 267 | 6.47 × 10−07 | 3.77 × 10−13 |
| Reactome | M phase | 0.0313 | 315 | 1.01 × 10−06 | 3.77 × 10−13 |
| Reactome | Dopamine neurotransmitter release cycle | 0.0063 | 22 | 2.11 × 10−06 | 3.77 × 10−13 |
| Reactome | Base-excision repair, ap site formation | 0.0056 | 18 | 2.57 × 10−06 | 3.77 × 10−13 |
| Reactome | Glutamate neurotransmitter release cycle | 0.0063 | 23 | 3.48 × 10−06 | 3.77 × 10−13 |
| Reactome | Base excision repair | 0.0088 | 45 | 4.62 × 10−06 | 3.77 × 10−13 |
| Reactome | Adherens junctions interactions | 0.0194 | 168 | 5.24 × 10−06 | 3.77 × 10−13 |
| Reactome | G-protein mediated events | 0.0094 | 53 | 7.86 × 10−06 | 3.77 × 10−13 |
| Reactome | Acetylcholine neurotransmitter release cycle | 0.0050 | 16 | 9.53 × 10−06 | 3.77 × 10−13 |
| Endpoint | Model | HR_per_SD | CI_low | CI_high | p | N |
|---|---|---|---|---|---|---|
| OS | Univariable | 0.902 | 0.788 | 1.03 | 0.138 | 406 |
| OS | Multivariable (age, sex, stage-ordinal) | 0.925 | 0.812 | 1.05 | 0.244 | 404 |
| PFI | Univariable | 0.956 | 0.832 | 1.1 | 0.533 | 407 |
| PFI | Multivariable (age, sex, stage-ordinal) | 0.99 | 0.865 | 1.13 | 0.888 | 405 |
| DFI | Univariable | 1.02 | 0.707 | 1.48 | 0.907 | 187 |
| DFI | Multivariable (age, sex, stage-ordinal) | 1.1 | 0.747 | 1.63 | 0.625 | 186 |
| DSS | Univariable | 0.901 | 0.765 | 1.06 | 0.214 | 393 |
| DSS | Multivariable (age, sex, stage-ordinal) | 0.941 | 0.803 | 1.1 | 0.452 | 391 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öztan, G.; İşsever, H.; İşsever, T.; Şahin, L. Lead Exposure and Bladder Cancer: Molecular Insights from TCGA RNA-Seq and Toxicogenomic Integration. Cancers 2025, 17, 3291. https://doi.org/10.3390/cancers17203291
Öztan G, İşsever H, İşsever T, Şahin L. Lead Exposure and Bladder Cancer: Molecular Insights from TCGA RNA-Seq and Toxicogenomic Integration. Cancers. 2025; 17(20):3291. https://doi.org/10.3390/cancers17203291
Chicago/Turabian StyleÖztan, Gözde, Halim İşsever, Tuğçe İşsever, and Levent Şahin. 2025. "Lead Exposure and Bladder Cancer: Molecular Insights from TCGA RNA-Seq and Toxicogenomic Integration" Cancers 17, no. 20: 3291. https://doi.org/10.3390/cancers17203291
APA StyleÖztan, G., İşsever, H., İşsever, T., & Şahin, L. (2025). Lead Exposure and Bladder Cancer: Molecular Insights from TCGA RNA-Seq and Toxicogenomic Integration. Cancers, 17(20), 3291. https://doi.org/10.3390/cancers17203291






