Artemis (DCLRE1C) Acts as a Target to Enhance Radiotherapy Response in Triple-Negative Breast Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Reagents
2.2. Genome-Wide CRISPR/Cas9 Knockout Screening
2.3. CRISPR/Cas9 Artemis-Specific Knockout
2.4. The Cancer Genome Atlas (TCGA) Data Analysis
2.5. Colony-Forming Assay
2.6. In Vivo Studies
2.7. Immunoblotting
2.8. Cell Cycle Analysis
2.9. Galactosidase Staining
2.10. RNA-Seq Analysis
2.11. Pathway Analysis and Gene Set Enrichment Analysis (GSEA)
2.12. Reverse Transcription Quantitative PCR (RT-qPCR)
2.13. Statistical Analysis
3. Results
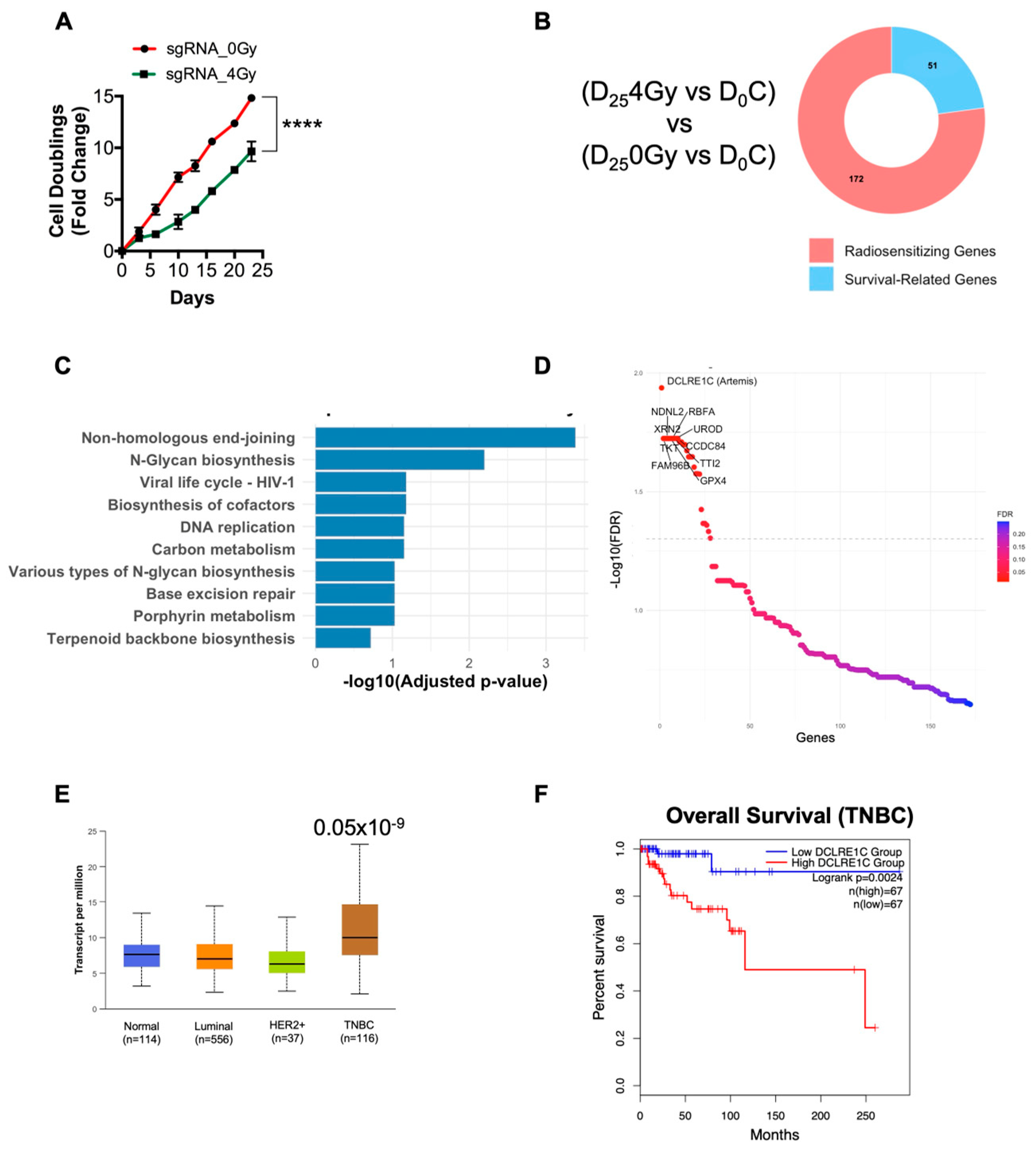
3.1. Genome-Wide CRISPR Knockout Screen Identifies ARTEMIS as the Top Radiosensitizing Target
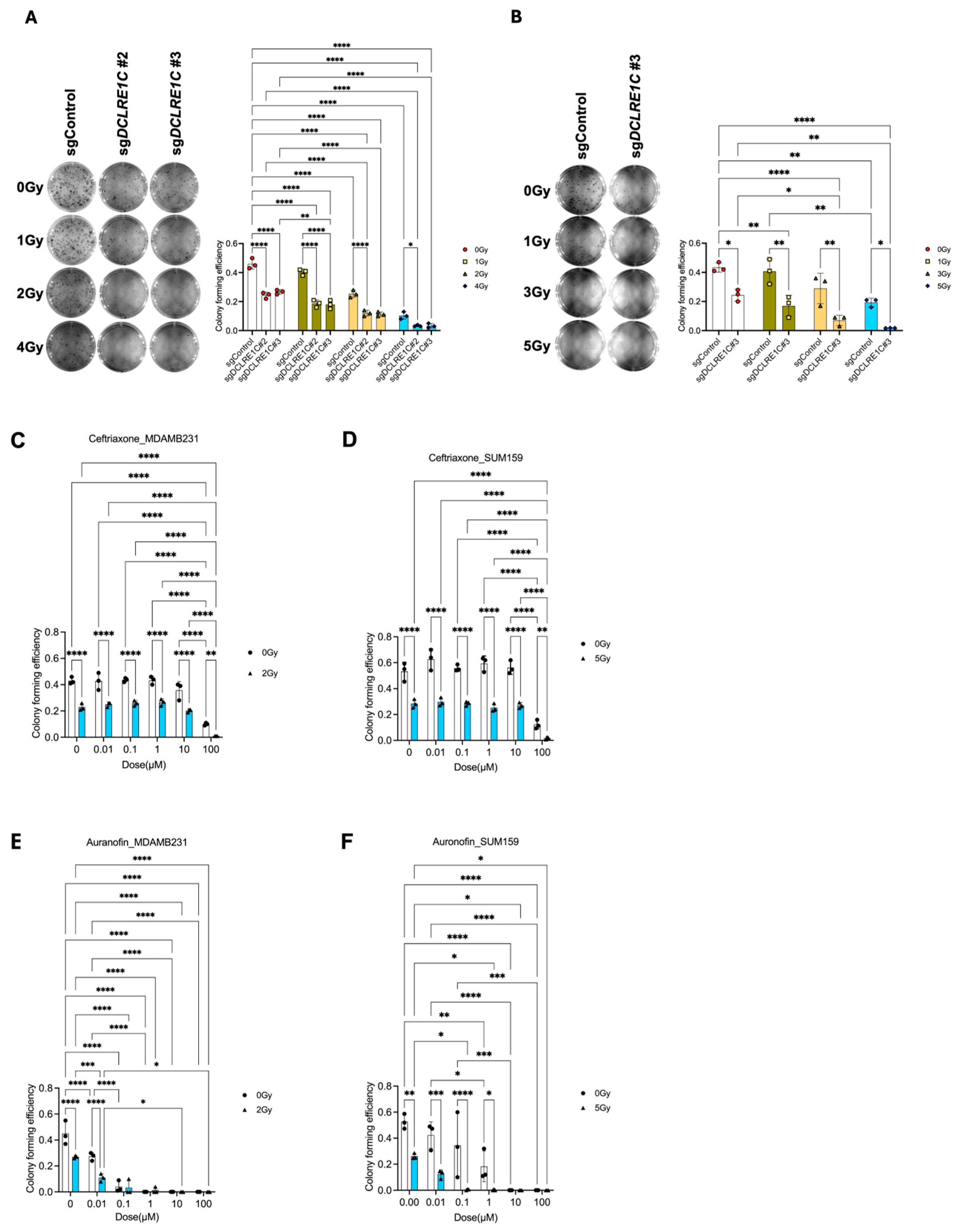
3.2. Loss of Function of Artemis Significantly Decreases Proliferation and Enhances RT Response in TNBC Cells In Vitro
3.3. Putative Pharmacological Inhibitors of Artemis Exert Antiproliferative and Radiosensitization Effects in TNBC
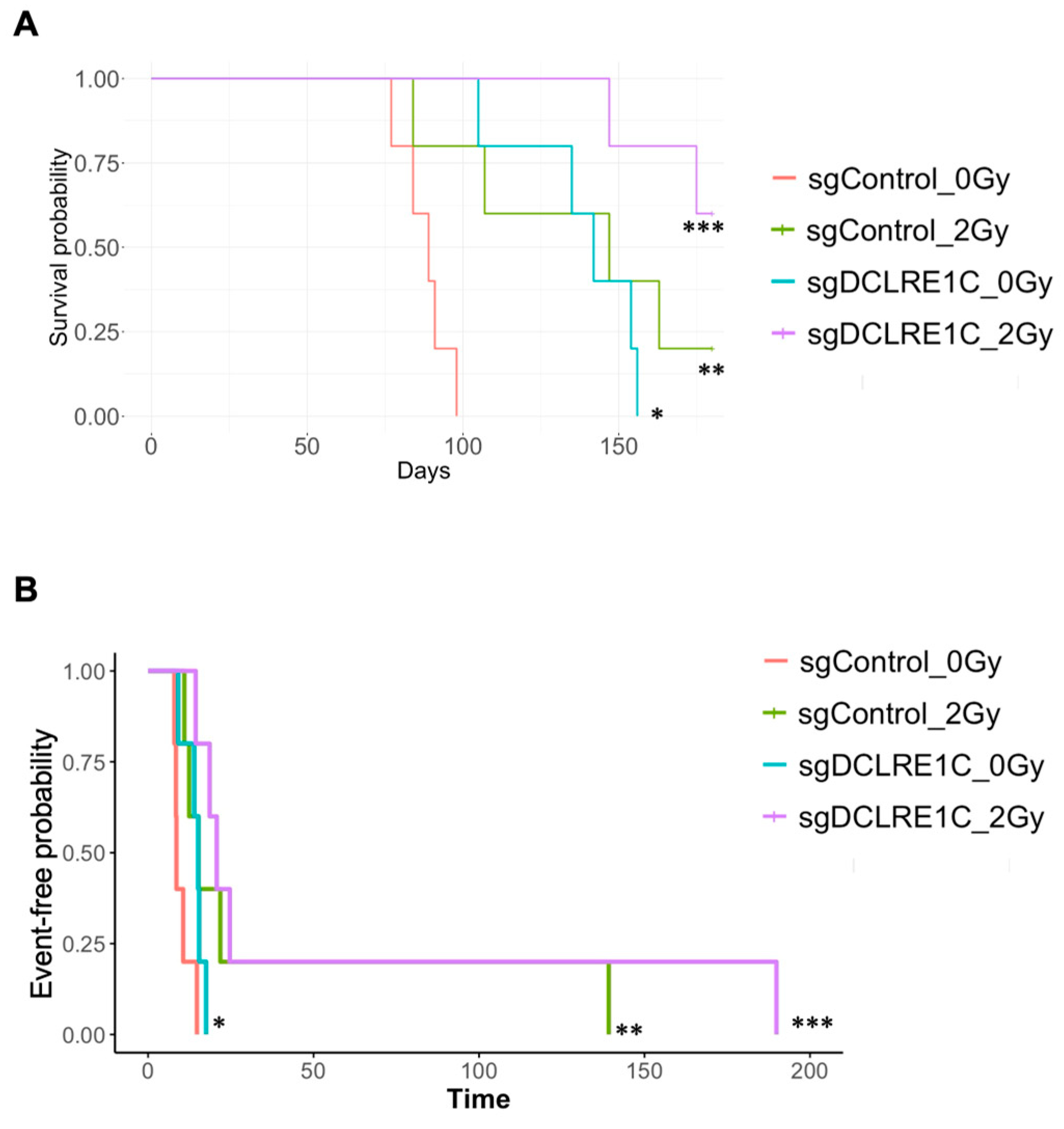
3.4. Loss of Function of Artemis Significantly Impairs Growth and Enhances Radiotherapy Response of TNBC Tumors In Vivo
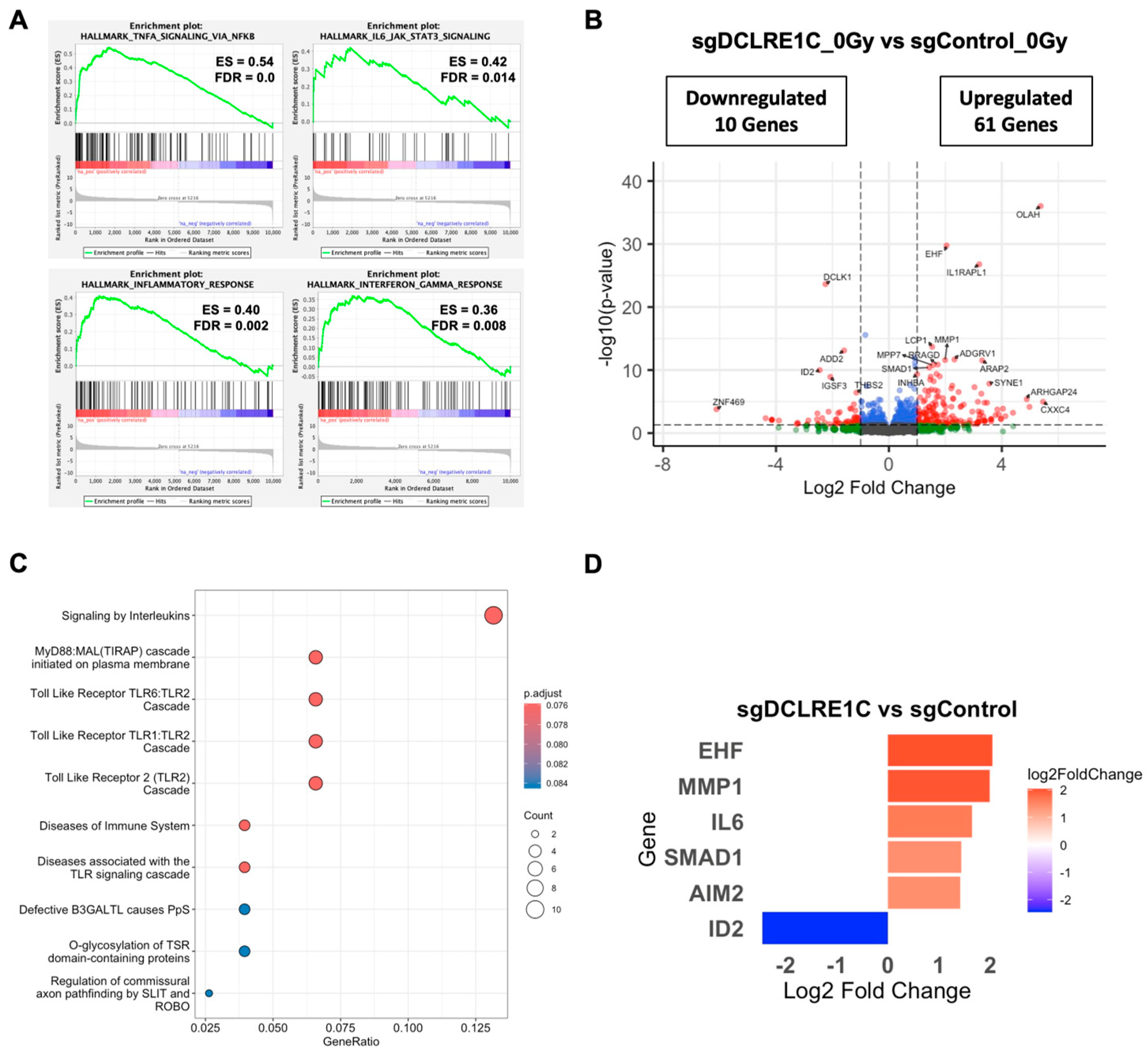
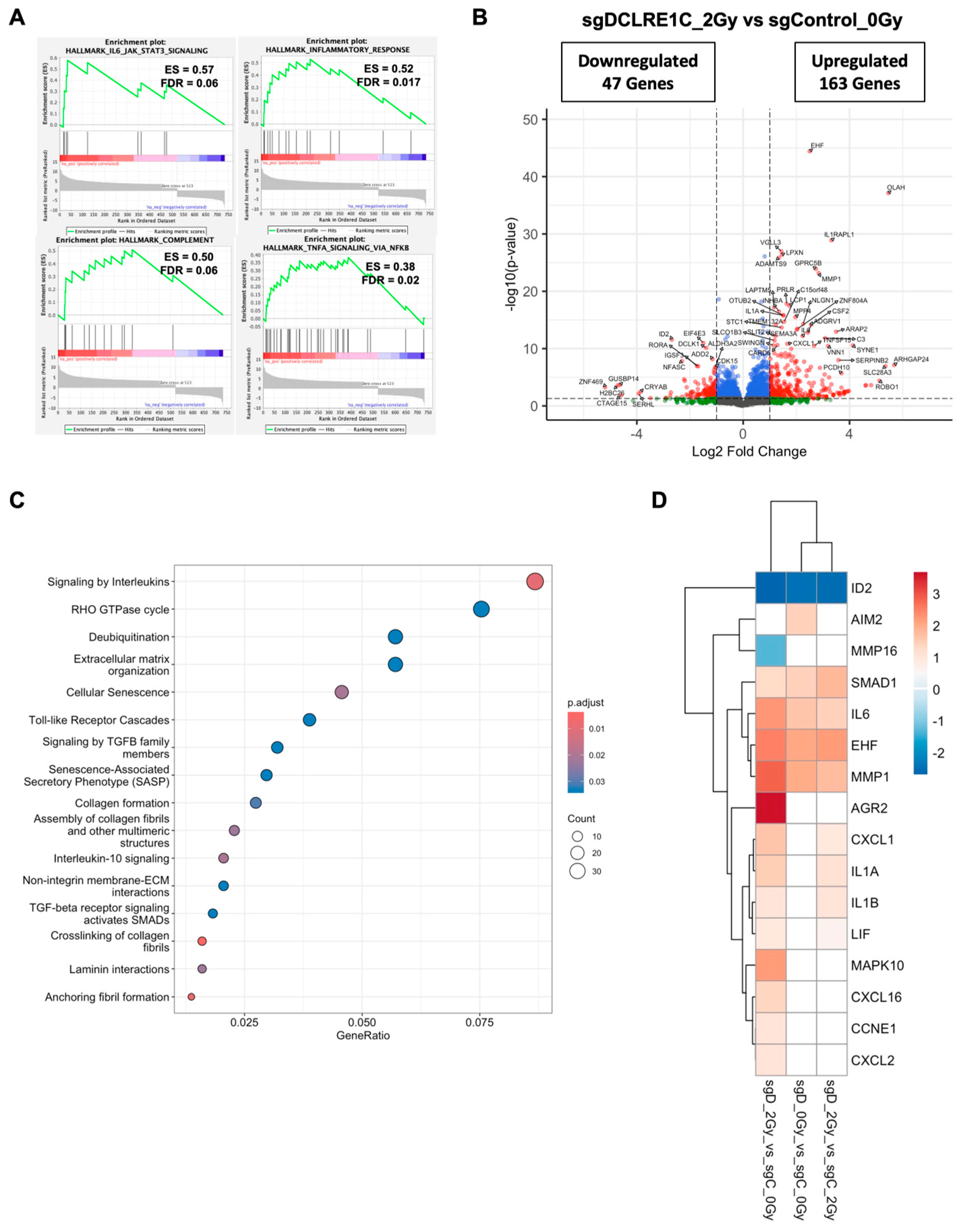
3.5. Loss of Artemis Expression Activates Anti-Cancer Phenotypes in TNBC
3.6. Artemis Deficiency Further Enhances RT Effects in TNBC via Induction of G2/M Arrest and Senescence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, R.; Ordóñez-Morán, P.; Allegrucci, C. Challenges for Triple Negative Breast Cancer Treatment: Defeating Heterogeneity and Cancer Stemness. Cancers 2022, 14, 4280. [Google Scholar] [CrossRef]
- Xiong, N.; Wu, H.; Yu, Z. Advancements and challenges in triple-negative breast cancer: A comprehensive review of therapeutic and diagnostic strategies. Front. Oncol. 2024, 14, 1405491. [Google Scholar] [CrossRef] [PubMed]
- Darby, S.; McGale, P.; Correa, C.; Taylor, C.; Arriagada, R.; Clarke, M.; Cutter, D.; Davies, C.; Ewertz, M.; Godwin, J.; et al. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: Meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet 2011, 378, 1707–1716. [Google Scholar] [CrossRef]
- McGale, P.; Taylor, C.; Correa, C.; Cutter, D.; Duane, F.; Ewertz, M.; Gray, R.; Mannu, G.; Peto, R.; Whelan, T.; et al. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: Meta-analysis of individual patient data for 8135 women in 22 randomised trials. Lancet 2014, 383, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Gradishar, W.J.; Moran, M.S.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Anderson, B.; Burstein, H.J.; Chew, H.; Dang, C.; et al. Breast Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 691–722. [Google Scholar] [CrossRef]
- Zhou, Z.R.; Wang, X.Y.; Yu, X.L.; Mei, X.; Chen, X.X.; Hu, Q.C.; Yang, Z.Z.; Guo, X.M. Building radiation-resistant model in triple-negative breast cancer to screen radioresistance-related molecular markers. Ann. Transl. Med. 2020, 8, 108. [Google Scholar] [CrossRef]
- Chen, X.; Yu, X.; Chen, J.; Zhang, Z.; Tuan, J.; Shao, Z.; Guo, X.; Feng, Y. Analysis in early stage triple-negative breast cancer treated with mastectomy without adjuvant radiotherapy: Patterns of failure and prognostic factors. Cancer 2013, 119, 2366–2374. [Google Scholar] [CrossRef]
- Chen, X.; Yu, X.; Chen, J.; Yang, Z.; Shao, Z.; Zhang, Z.; Guo, X.; Feng, Y. Radiotherapy can improve the disease-free survival rate in triple-negative breast cancer patients with T1-T2 disease and one to three positive lymph nodes after mastectomy. Oncologist 2013, 18, 141–147. [Google Scholar] [CrossRef]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef]
- Wu, Y.; Song, Y.; Wang, R.; Wang, T. Molecular mechanisms of tumor resistance to radiotherapy. Mol. Cancer 2023, 22, 96. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Chen, Y.; Liang, N.; Xie, J.; Deng, G.; Chen, F.; Wang, X.; Liu, F.; Li, Y.; Zhang, J. Targeting Epithelial-to-Mesenchymal Transition in Radioresistance: Crosslinked Mechanisms and Strategies. Front. Oncol. 2022, 12, 775238. [Google Scholar] [CrossRef]
- Suwa, T.; Kobayashi, M.; Nam, J.M.; Harada, H. Tumor microenvironment and radioresistance. Exp. Mol. Med. 2021, 53, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Carlos-Reyes, A.; Muñiz-Lino, M.A.; Romero-Garcia, S.; López-Camarillo, C.; Hernández-de la Cruz, O.N. Biological Adaptations of Tumor Cells to Radiation Therapy. Front. Oncol. 2021, 11, 718636. [Google Scholar] [CrossRef]
- Ouellette, M.M.; Zhou, S.; Yan, Y. Cell Signaling Pathways That Promote Radioresistance of Cancer Cells. Diagnostics 2022, 12, 656. [Google Scholar] [CrossRef]
- Bhat, V.; Pellizzari, S.; Allan, A.L.; Wong, E.; Lock, M.; Brackstone, M.; Lohmann, A.E.; Cescon, D.W.; Parsyan, A. Radiotherapy and radiosensitization in breast cancer: Molecular targets and clinical applications. Crit. Rev. Oncol. Hematol. 2022, 169, 103566. [Google Scholar] [CrossRef]
- Tufail, M. DNA repair pathways in breast cancer: From mechanisms to clinical applications. Breast Cancer Res. Treat. 2023, 200, 305–321. [Google Scholar] [CrossRef]
- Wiegmans, A.P.; Ward, A.; Ivanova, E.; Duijf, P.H.G.; Adams, M.N.; Najib, I.M.; Van Oosterhout, R.; Sadowski, M.C.; Kelly, G.; Morrical, S.W.; et al. Genome instability and pressure on non-homologous end joining drives chemotherapy resistance via a DNA repair crisis switch in triple negative breast cancer. NAR Cancer 2021, 3, zcab022. [Google Scholar] [CrossRef]
- Dylgjeri, E.; Knudsen, K.E. DNA-PKcs: A Targetable Protumorigenic Protein Kinase. Cancer Res. 2022, 82, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Dragojevic, S.; Ji, J.; Singh, P.K.; Connors, M.A.; Mutter, R.W.; Lester, S.C.; Talele, S.M.; Zhang, W.; Carlson, B.L.; Remmes, N.B.; et al. Preclinical Risk Evaluation of Normal Tissue Injury with Novel Radiosensitizers. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, e54–e62. [Google Scholar] [CrossRef]
- Mansilla-Soto, J.; Cortes, P. VDJ recombination: Artemis and its in vivo role in hairpin opening. J. Exp. Med. 2003, 197, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.; He, Y.; Lees-Miller, S.P. How to fix DNA breaks: New insights into the mechanism of non-homologous end joining. Biochem. Soc. Trans. 2023, 51, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pluth, J.M.; Cooper, P.K.; Cowan, M.J.; Chen, D.J.; Yannone, S.M. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair 2005, 4, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Parsyan, A.; Bhat, V.; Athwal, H.; Goebel, E.A.; Allan, A.L. Artemis and its role in cancer. Transl. Oncol. 2025, 51, 102165. [Google Scholar] [CrossRef]
- Shahi, R.B.; De Brakeleer, S.; Caljon, B.; Pauwels, I.; Bonduelle, M.; Joris, S.; Fontaine, C.; Vanhoeij, M.; Van Dooren, S.; Teugels, E.; et al. Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer 2019, 19, 313. [Google Scholar] [CrossRef]
- Yosaatmadja, Y.; Baddock, H.T.; Newman, J.A.; Bielinski, M.; Gavard, A.E.; Mukhopadhyay, S.M.M.; Dannerfjord, A.A.; Schofield, C.J.; McHugh, P.J.; Gileadi, O. Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition. Nucleic Acids Res. 2021, 49, 9310–9326. [Google Scholar] [CrossRef]
- Thu, K.L.; Silvester, J.; Elliott, M.J.; Ba-Alawi, W.; Duncan, M.H.; Elia, A.C.; Mer, A.S.; Smirnov, P.; Safikhani, Z.; Haibe-Kains, B.; et al. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E1570–E1577. [Google Scholar] [CrossRef]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Li, M.; Yu, J.S.L.; Tilgner, K.; Ong, S.H.; Koike-Yusa, H.; Yusa, K. Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution. Cell Rep. 2018, 24, 489–502. [Google Scholar] [CrossRef]
- Dai, M.; Yan, G.; Wang, N.; Daliah, G.; Edick, A.M.; Poulet, S.; Boudreault, J.; Ali, S.; Burgos, S.A.; Lebrun, J.J. In vivo genome-wide CRISPR screen reveals breast cancer vulnerabilities and synergistic mTOR/Hippo targeted combination therapy. Nat. Commun. 2021, 12, 3055. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Parsyan, A.; Cruickshank, J.; Hodgson, K.; Wakeham, D.; Pellizzari, S.; Bhat, V.; Cescon, D.W. Anticancer effects of radiation therapy combined with Polo-Like Kinase 4 (PLK4) inhibitor CFI-400945 in triple negative breast cancer. Breast 2021, 58, 6–9. [Google Scholar] [CrossRef]
- Brix, N.; Samaga, D.; Belka, C.; Zitzelsberger, H.; Lauber, K. Analysis of clonogenic growth in vitro. Nat. Protoc. 2021, 16, 4963–4991. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Pellizzari, S.; Bhat, V.; Athwal, H.; Cescon, D.W.; Allan, A.L.; Parsyan, A. PLK4 as a potential target to enhance radiosensitivity in triple-negative breast cancer. Radiat. Oncol. 2024, 19, 24. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Redwood, A.B.; Zhang, X.; Seth, S.B.; Ge, Z.; Bindeman, W.E.; Zhou, X.; Sinha, V.C.; Heffernan, T.P.; Piwnica-Worms, H. The cytosolic iron-sulfur cluster assembly (CIA) pathway is required for replication stress tolerance of cancer cells to Chk1 and ATR inhibitors. NPJ Breast Cancer 2021, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Ito, E.; Yue, S.; Moriyama, E.H.; Hui, A.B.; Kim, I.; Shi, W.; Alajez, N.M.; Bhogal, N.; Li, G.; Datti, A.; et al. Uroporphyrinogen decarboxylase is a radiosensitizing target for head and neck cancer. Sci. Transl. Med. 2011, 3, 67ra67. [Google Scholar] [CrossRef] [PubMed]
- Hurov, K.E.; Cotta-Ramusino, C.; Elledge, S.J. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010, 24, 1939–1950. [Google Scholar] [CrossRef]
- Geng, L.; Zhu, M.; Luo, D.; Chen, H.; Li, B.; Lao, Y.; An, H.; Wu, Y.; Li, Y.; Xia, A.; et al. TKT-PARP1 axis induces radioresistance by promoting DNA double-strand break repair in hepatocellular carcinoma. Oncogene 2024, 43, 682–692. [Google Scholar] [CrossRef]
- van der Crabben, S.N.; Hennus, M.P.; McGregor, G.A.; Ritter, D.I.; Nagamani, S.C.; Wells, O.S.; Harakalova, M.; Chinn, I.K.; Alt, A.; Vondrova, L.; et al. Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J. Clin. Investig. 2016, 126, 2881–2892. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Lei, G.; Zhang, Y.; Koppula, P.; Liu, X.; Zhang, J.; Lin, S.H.; Ajani, J.A.; Xiao, Q.; Liao, Z.; Wang, H.; et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020, 30, 146–162. [Google Scholar] [CrossRef] [PubMed]
- de Wolf, B.; Oghabian, A.; Akinyi, M.V.; Hanks, S.; Tromer, E.C.; van Hooff, J.J.E.; van Voorthuijsen, L.; van Rooijen, L.E.; Verbeeren, J.; Uijttewaal, E.C.H.; et al. Chromosomal instability by mutations in the novel minor spliceosome component CENATAC. EMBO J. 2021, 40, e106536. [Google Scholar] [CrossRef]
- Li, S.; Chang, H.H.; Niewolik, D.; Hedrick, M.P.; Pinkerton, A.B.; Hassig, C.A.; Schwarz, K.; Lieber, M.R. Evidence that the DNA endonuclease ARTEMIS also has intrinsic 5′-exonuclease activity. J. Biol. Chem. 2014, 289, 7825–7834. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, G.; Lieber, M.R.; Williams, D.R. Structural analysis of the basal state of the Artemis:DNA-PKcs complex. Nucleic Acids Res. 2022, 50, 7697–7720. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.; Scuoppo, C.; Wang, X.; Fang, X.; Balgley, B.; Bolden, J.E.; Premsrirut, P.; Luo, W.; Chicas, A.; Lee, C.S.; et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011, 25, 2125–2136. [Google Scholar] [CrossRef]
- Kandhaya-Pillai, R.; Yang, X.; Tchkonia, T.; Martin, G.M.; Kirkland, J.L.; Oshima, J. TNF-α/IFN-γ synergy amplifies senescence-associated inflammation and SARS-CoV-2 receptor expression via hyper-activated JAK/STAT1. Aging Cell 2022, 21, e13646. [Google Scholar] [CrossRef]
- Bae, E.J.; Choi, M.; Kim, J.T.; Kim, D.K.; Jung, M.K.; Kim, C.; Kim, T.K.; Lee, J.S.; Jung, B.C.; Shin, S.J.; et al. TNF-α promotes α-synuclein propagation through stimulation of senescence-associated lysosomal exocytosis. Exp. Mol. Med. 2022, 54, 788–800. [Google Scholar] [CrossRef]
- Han, D.; Gong, H.; Wei, Y.; Xu, Y.; Zhou, X.; Wang, Z.; Feng, F. Hesperidin inhibits lung fibroblast senescence via IL-6/STAT3 signaling pathway to suppress pulmonary fibrosis. Phytomedicine 2023, 112, 154680. [Google Scholar] [CrossRef]
- Prattichizzo, F.; Giuliani, A.; Recchioni, R.; Bonafè, M.; Marcheselli, F.; De Carolis, S.; Campanati, A.; Giuliodori, K.; Rippo, M.R.; Brugè, F.; et al. Anti-TNF-α treatment modulates SASP and SASP-related microRNAs in endothelial cells and in circulating angiogenic cells. Oncotarget 2016, 7, 11945–11958. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.; Lee, J.H.; Hwangbo, C. Toll-like receptor 4 (TLR4): New insight immune and aging. Immun. Ageing 2023, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Ponomareva, L.; Veeranki, S.; Panchanathan, R.; Dickerson, E.; Choubey, D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol. Cancer Res. 2011, 9, 589–602. [Google Scholar] [CrossRef]
- Lim, S.; Lim, J.; Lee, A.; Kim, K.I.; Lim, J.S. Anticancer Effect of E26 Transformation-Specific Homologous Factor through the Induction of Senescence and the Inhibition of Epithelial-Mesenchymal Transition in Triple-Negative Breast Cancer Cells. Cancers 2023, 15, 5270. [Google Scholar] [CrossRef]
- Vernot, J.P. Senescence-Associated Pro-inflammatory Cytokines and Tumor Cell Plasticity. Front. Mol. Biosci. 2020, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Levi, N.; Papismadov, N.; Solomonov, I.; Sagi, I.; Krizhanovsky, V. The ECM path of senescence in aging: Components and modifiers. FEBS J. 2020, 287, 2636–2646. [Google Scholar] [CrossRef]
- Xu, J.Z.; Zhou, Y.M.; Zhang, L.L.; Chen, X.J.; Yang, Y.Y.; Zhang, D.; Zhu, K.C.; Kong, X.K.; Sun, L.H.; Tao, B.; et al. BMP9 reduces age-related bone loss in mice by inhibiting osteoblast senescence through Smad1-Stat1-P21 axis. Cell Death Discov. 2022, 8, 254. [Google Scholar] [CrossRef] [PubMed]
- Zebedee, Z.; Hara, E. Id proteins in cell cycle control and cellular senescence. Oncogene 2001, 20, 8317–8325. [Google Scholar] [CrossRef] [PubMed]
- White, T.L.; Deshpande, N.; Kumar, V.; Gauthier, A.G.; Jurkunas, U.V. Cell cycle re-entry and arrest in G2/M phase induces senescence and fibrosis in Fuchs Endothelial Corneal Dystrophy. Free Radic. Biol. Med. 2021, 164, 34–43. [Google Scholar] [CrossRef]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 645593. [Google Scholar] [CrossRef]
- Gire, V.; Dulic, V. Senescence from G2 arrest, revisited. Cell Cycle 2015, 14, 297–304. [Google Scholar] [CrossRef]
- He, L.; Lv, Y.; Song, Y.; Zhang, B. The prognosis comparison of different molecular subtypes of breast tumors after radiotherapy and the intrinsic reasons for their distinct radiosensitivity. Cancer Manag. Res. 2019, 11, 5765–5775. [Google Scholar] [CrossRef]
- Kawale, A.S.; Akopiants, K.; Valerie, K.; Ruis, B.; Hendrickson, E.A.; Huang, S.N.; Pommier, Y.; Povirk, L.F. TDP1 suppresses mis-joining of radiomimetic DNA double-strand breaks and cooperates with Artemis to promote optimal nonhomologous end joining. Nucleic Acids Res. 2018, 46, 8926–8939. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Huang, A.; Gao, H.; Sun, Y.; Jiang, T.; Shi, L.; Wu, X.; Dong, Q.; Sun, X. Silencing Artemis Enhances Colorectal Cancer Cell Sensitivity to DNA-Damaging Agents. Oncol. Res. 2018, 27, 29–38. [Google Scholar] [CrossRef]
- Liu, H.; Sun, X.; Zhang, S.; Ge, W.; Zhu, Y.; Zhang, J.; Zheng, S. The dominant negative mutant Artemis enhances tumor cell radiosensitivity. Radiother. Oncol. 2011, 101, 66–72. [Google Scholar] [CrossRef]
- Calugaru, V.; Nauraye, C.; Cordelières, F.P.; Biard, D.; De Marzi, L.; Hall, J.; Favaudon, V.; Mégnin-Chanet, F. Involvement of the Artemis protein in the relative biological efficiency observed with the 76-MeV proton beam used at the Institut Curie Proton Therapy Center in Orsay. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, 36–43. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, X.; Li, P.; Zhu, Y.; Sun, Y.; Hu, J.; Liu, H.; Sun, X. Developing a Peptide That Inhibits DNA Repair by Blocking the Binding of Artemis and DNA Ligase IV to Enhance Tumor Radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, 515–527. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- Volk, T.; Pannicke, U.; Reisli, I.; Bulashevska, A.; Ritter, J.; Björkman, A.; Schäffer, A.A.; Fliegauf, M.; Sayar, E.H.; Salzer, U.; et al. DCLRE1C (ARTEMIS) mutations causing phenotypes ranging from atypical severe combined immunodeficiency to mere antibody deficiency. Hum. Mol. Genet. 2015, 24, 7361–7372. [Google Scholar] [CrossRef]
- Woodbine, L.; Grigoriadou, S.; Goodarzi, A.A.; Riballo, E.; Tape, C.; Oliver, A.W.; van Zelm, M.C.; Buckland, M.S.; Davies, E.G.; Pearl, L.H.; et al. An Artemis polymorphic variant reduces Artemis activity and confers cellular radiosensitivity. DNA Repair 2010, 9, 1003–1010. [Google Scholar] [CrossRef]
- Moscariello, M.; Wieloch, R.; Kurosawa, A.; Li, F.; Adachi, N.; Mladenov, E.; Iliakis, G. Role for Artemis nuclease in the repair of radiation-induced DNA double strand breaks by alternative end joining. DNA Repair 2015, 31, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, L.; Gao, H.; Kuang, X.; Xiao, H.; Yang, C.; Cheng, Y.; Zhang, L.; Guo, X.; Zhong, Y.; et al. APE1 promotes non-homologous end joining by initiating DNA double-strand break formation and decreasing ubiquitination of artemis following oxidative genotoxic stress. J. Transl. Med. 2023, 21, 183. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, R.; Shan, J.; Xie, X.; Wang, C.; Hu, P.; Sun, X. Artemis as Predictive Biomarker of Responsiveness to Preoperative Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer. Curr. Oncol. 2024, 31, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Farkas, S.A.; Vymetalkova, V.; Vodickova, L.; Vodicka, P.; Nilsson, T.K. DNA methylation changes in genes frequently mutated in sporadic colorectal cancer and in the DNA repair and Wnt/β-catenin signaling pathway genes. Epigenomics 2014, 6, 179–191. [Google Scholar] [CrossRef]
- Liu, S.; Liu, W.; Ding, Z.; Yang, X.; Jiang, Y.; Wu, Y.; Liu, Y.; Wu, J. Identification and validation of a novel tumor driver gene signature for diagnosis and prognosis of head and neck squamous cell carcinoma. Front. Mol. Biosci. 2022, 9, 912620. [Google Scholar] [CrossRef]
- Ogana, H.A.; Hurwitz, S.; Hsieh, C.L.; Geng, H.; Müschen, M.; Bhojwani, D.; Wolf, M.A.; Larocque, J.; Lieber, M.R.; Kim, Y.M. Artemis inhibition as a therapeutic strategy for acute lymphoblastic leukemia. Front. Cell Dev. Biol. 2023, 11, 1134121. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.; Geng, L.; Wang, H.; Legerski, R.J. Artemis is a negative regulator of p53 in response to oxidative stress. Oncogene 2009, 28, 2196–2204. [Google Scholar] [CrossRef]
- Richter, C.; Marquardt, S.; Li, F.; Spitschak, A.; Murr, N.; Edelhäuser, B.A.H.; Iliakis, G.; Pützer, B.M.; Logotheti, S. Rewiring E2F1 with classical NHEJ via APLF suppression promotes bladder cancer invasiveness. J. Exp. Clin. Cancer Res. 2019, 38, 292. [Google Scholar] [CrossRef]
- Labay, E.; Mauceri, H.J.; Efimova, E.V.; Flor, A.C.; Sutton, H.G.; Kron, S.J.; Weichselbaum, R.R. Repurposing cephalosporin antibiotics as pro-senescent radiosensitizers. Oncotarget 2016, 7, 33919–33933. [Google Scholar] [CrossRef]
- Nag, D.; Bhanja, P.; Riha, R.; Sanchez-Guerrero, G.; Kimler, B.F.; Tsue, T.T.; Lominska, C.; Saha, S. Auranofin Protects Intestine against Radiation Injury by Modulating p53/p21 Pathway and Radiosensitizes Human Colon Tumor. Clin. Cancer Res. 2019, 25, 4791–4807. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, H.; Li, S.; Zhu, F.; Kim, D.J.; Xie, H.; Li, Y.; Nadas, J.; Oi, N.; Zykova, T.A.; et al. Ceftriaxone, an FDA-approved cephalosporin antibiotic, suppresses lung cancer growth by targeting Aurora B. Carcinogenesis 2012, 33, 2548–2557. [Google Scholar] [CrossRef] [PubMed]
- Chittavanich, P.; Saengwimol, D.; Roytrakul, S.; Rojanaporn, D.; Chaitankar, V.; Srimongkol, A.; Anurathapan, U.; Hongeng, S.; Kaewkhaw, R. Ceftriaxone exerts antitumor effects in MYCN-driven retinoblastoma and neuroblastoma by targeting DDX3X for translation repression. Mol. Oncol. 2024, 18, 918–938. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Park, S.H.; Park, W.H. Anti-Cancer Effects of Auranofin in Human Lung Cancer Cells by Increasing Intracellular ROS Levels and Depleting GSH Levels. Molecules 2022, 27, 5207. [Google Scholar] [CrossRef]
- Hou, G.X.; Liu, P.P.; Zhang, S.; Yang, M.; Liao, J.; Yang, J.; Hu, Y.; Jiang, W.Q.; Wen, S.; Huang, P. Elimination of stem-like cancer cell side-population by auranofin through modulation of ROS and glycolysis. Cell Death Dis. 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Hales, C.N.; Ozanne, S.E. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007, 35, 7417–7428. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Montero, P.; Londoño-Vallejo, A.; Vernot, J.P. Senescence-associated IL-6 and IL-8 cytokines induce a self- and cross-reinforced senescence/inflammatory milieu strengthening tumorigenic capabilities in the MCF-7 breast cancer cell line. Cell Commun. Signal 2017, 15, 17. [Google Scholar] [CrossRef]
- Kojima, H.; Inoue, T.; Kunimoto, H.; Nakajima, K. IL-6-STAT3 signaling and premature senescence. JAK-STAT 2013, 2, e25763. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Asaithamby, A.; Hu, B.; Chen, D.J. Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8293–8298. [Google Scholar] [CrossRef]
- Ulus-Senguloglu, G.; Arlett, C.F.; Plowman, P.N.; Parnell, J.; Oatel, N.; Bourton, E.C.; Parris, C.N. Elevated Expression of Artemis in Human Fibroblast Cells Is Associated with Cellular Radiosensitivity and Increased Apoptosis. Br. J. Cancer 2012, 107, 1506–1513. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, V.; Thu, K.L.; Goncalves Santiago, A.d.F.; Bonvissuto, A.C.; Ghasemi, F.; Goodale, D.; Roes, M.V.; Passos, D.T.; Dick, F.A.; Cescon, D.W.; et al. Artemis (DCLRE1C) Acts as a Target to Enhance Radiotherapy Response in Triple-Negative Breast Cancer. Cancers 2025, 17, 3279. https://doi.org/10.3390/cancers17203279
Bhat V, Thu KL, Goncalves Santiago AdF, Bonvissuto AC, Ghasemi F, Goodale D, Roes MV, Passos DT, Dick FA, Cescon DW, et al. Artemis (DCLRE1C) Acts as a Target to Enhance Radiotherapy Response in Triple-Negative Breast Cancer. Cancers. 2025; 17(20):3279. https://doi.org/10.3390/cancers17203279
Chicago/Turabian StyleBhat, Vasudeva, Kelsie L. Thu, Anayra de Fatima Goncalves Santiago, Anna C. Bonvissuto, Farhad Ghasemi, David Goodale, Michael V. Roes, Daniel T. Passos, Frederick A. Dick, David W. Cescon, and et al. 2025. "Artemis (DCLRE1C) Acts as a Target to Enhance Radiotherapy Response in Triple-Negative Breast Cancer" Cancers 17, no. 20: 3279. https://doi.org/10.3390/cancers17203279
APA StyleBhat, V., Thu, K. L., Goncalves Santiago, A. d. F., Bonvissuto, A. C., Ghasemi, F., Goodale, D., Roes, M. V., Passos, D. T., Dick, F. A., Cescon, D. W., Allan, A. L., & Parsyan, A. (2025). Artemis (DCLRE1C) Acts as a Target to Enhance Radiotherapy Response in Triple-Negative Breast Cancer. Cancers, 17(20), 3279. https://doi.org/10.3390/cancers17203279






