Impact of Somatic Gene Mutations on Prognosis Prediction in De Novo AML: Unraveling Insights from a Systematic Review and Meta-Analysis
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment
2.5. Statistical Analysis
3. Results
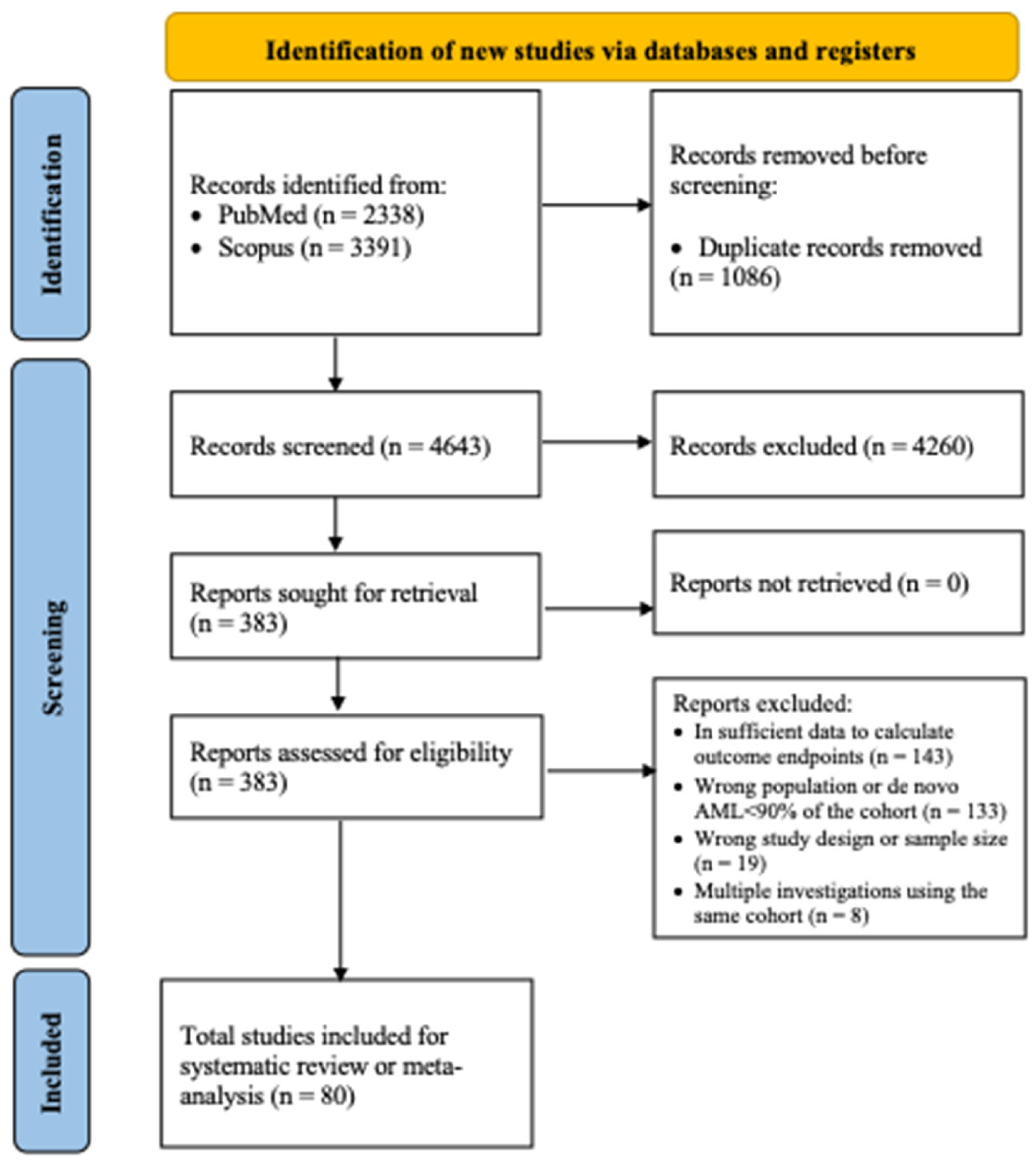
3.1. Search Results
3.2. Studies Characteristics
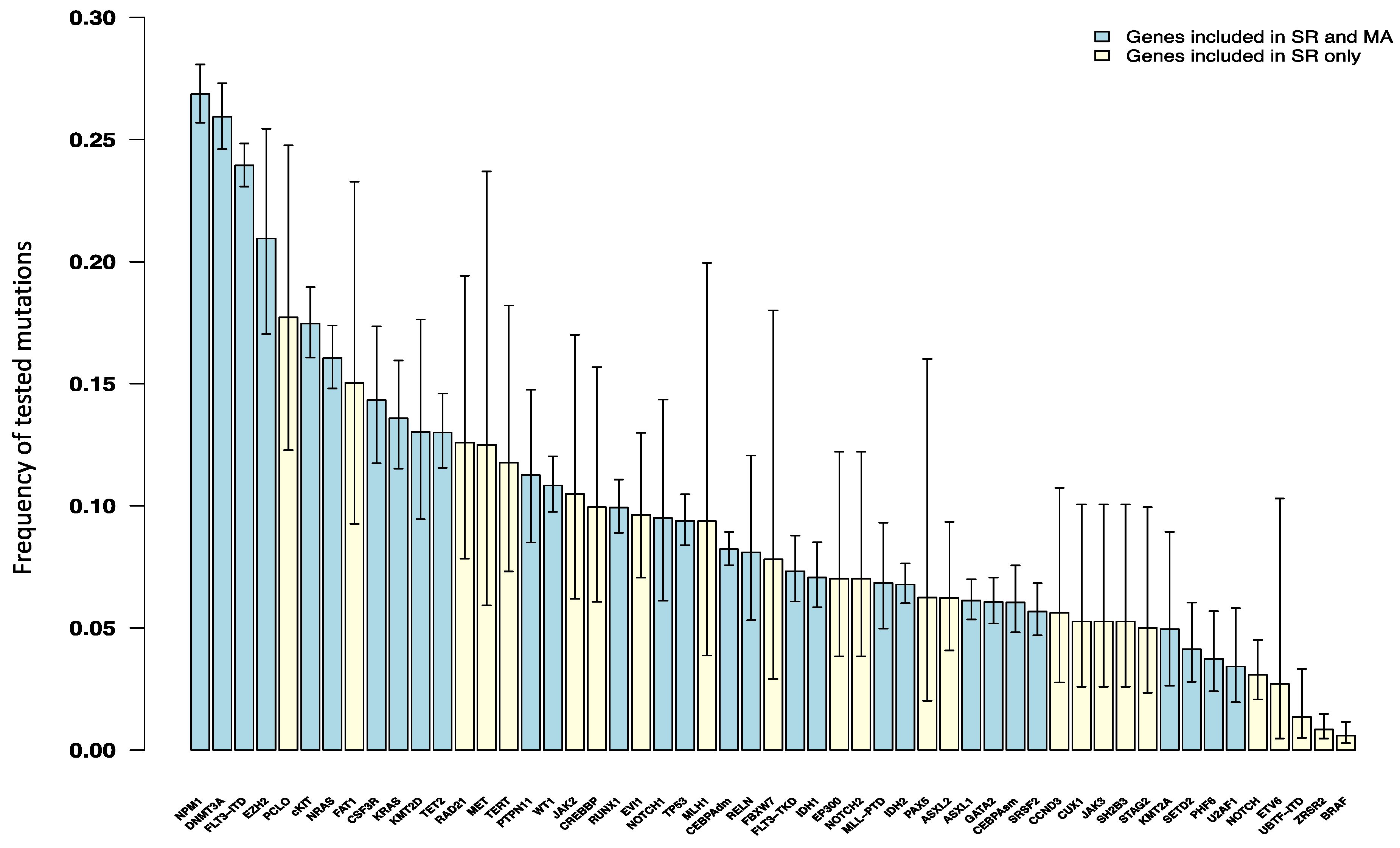
3.3. Frequency of Somatic Genetic Alterations in De Novo AML Patients
3.4. Molecular Determinants of Overall Survival and Relapse-Free Survival
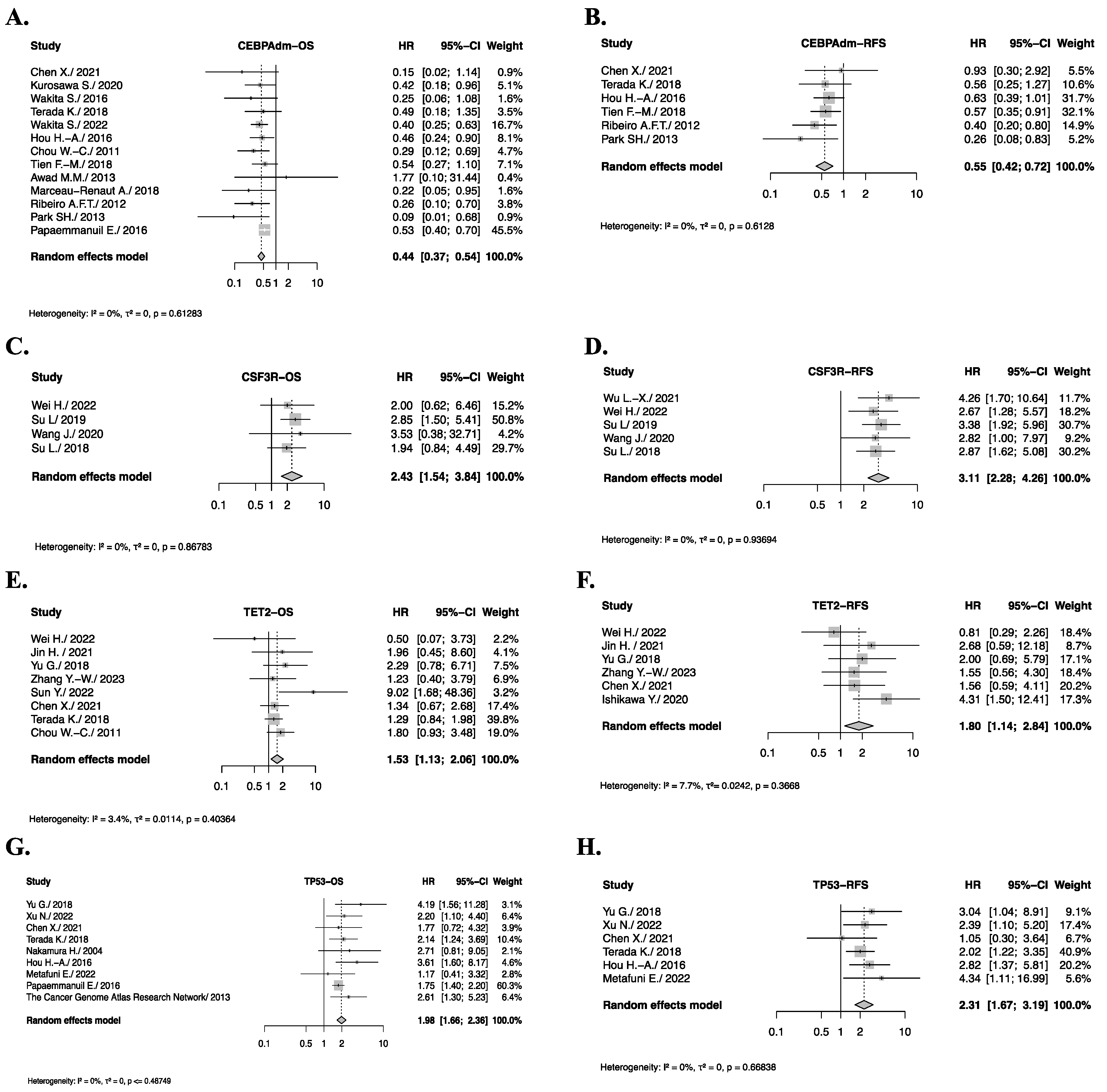
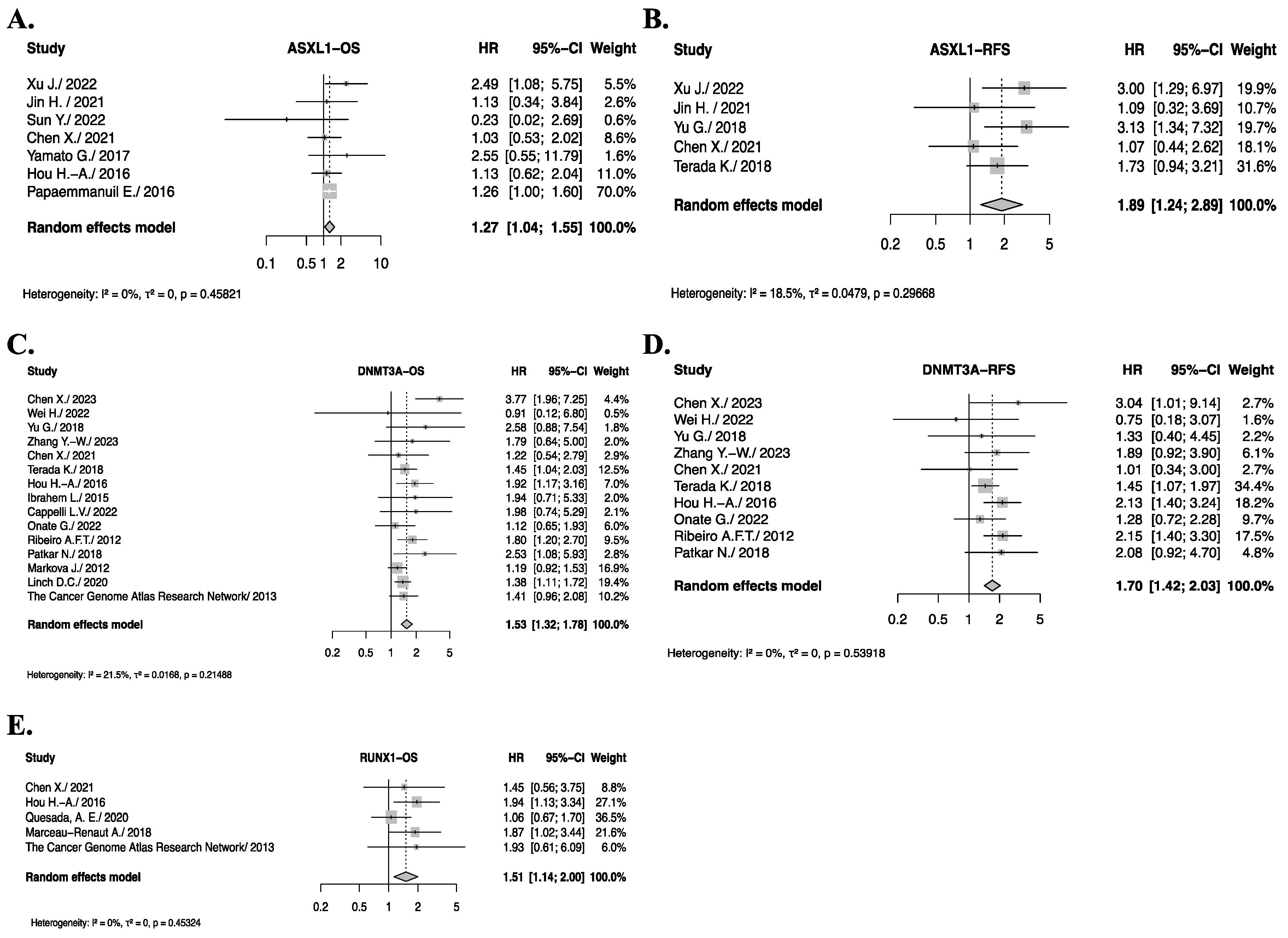
3.5. Pooled Analysis of Somatic Gene Mutations with Significant Impact on OS or RFS
3.6. Genes with Significant Impact on OS or RFS After Sensitivity Analysis
3.7. Genes with Significant Impact on OS or RFS After Sub-Group Analysis
3.8. Genes with Non-Significant Impact on OS or RFS of De Novo AML Patients
3.9. Publication Bias
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute Myeloid Leukemia: Current Progress and Future Directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, B.C.; Chan, S.M.; Daver, N.G.; Jonas, B.A.; Pollyea, D.A. Optimizing Survival Outcomes with Post-remission Therapy in Acute Myeloid Leukemia. Am. J. Hematol. 2019, 94, 803–811. [Google Scholar] [CrossRef]
- Sanders, M.A.; Valk, P.J.M. The Evolving Molecular Genetic Landscape in Acute Myeloid Leukaemia. Curr. Opin. Hematol. 2013, 20, 79–85. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating Morphologic, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- PROSPERO. Available online: https://www.crd.york.ac.uk/PROSPERO/ (accessed on 25 February 2024).
- PRISMA. Available online: http://www.prisma-statement.org/?AspxAutoDetectCookieSupport=1 (accessed on 25 February 2024).
- Rayyan—Intelligent Systematic Review-Rayyan. Available online: https://www.rayyan.ai/ (accessed on 25 February 2024).
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical Methods for Incorporating Summary Time-to-Event Data into Meta-Analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef]
- Gierisch, J.M.; Beadles, C.; Shapiro, A.; McDuffie, J.R.; Cunningham, N.; Bradford, D.; Strauss, J.; Callahan, M.; Chen, M.; Hemminger, A.; et al. Newcastle-ottawa scale coding manual for cohort studies. In Health Disparities in Quality Indicators of Healthcare Among Adults with Mental Illness [Internet]; Department of Veterans Affairs (US): Washington, DC, USA, 2014. [Google Scholar]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ Br. Med. J. 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Begg, C.B.; Berlin, J.A. Publication Bias: A Problem in Interpreting Medical Data. J. R. Stat. Soc. Ser. A Stat. Soc. 1988, 151, 419–445. [Google Scholar] [CrossRef]
- Matsuo, H.; Yoshida, K.; Nakatani, K.; Harata, Y.; Higashitani, M.; Ito, Y.; Kamikubo, Y.; Shiozawa, Y.; Shiraishi, Y.; Chiba, K.; et al. Fusion Partner–Specific Mutation Profiles and KRAS Mutations as Adverse Prognostic Factors in MLL-Rearranged AML. Blood Adv. 2020, 4, 4623–4631. [Google Scholar] [CrossRef]
- Christen, F.; Hoyer, K.; Yoshida, K.; Hou, H.A.; Waldhueter, N.; Heuser, M.; Hills, R.K.; Chan, W.; Hablesreiter, R.; Blau, O.; et al. Genomic Landscape and Clonal Evolution of Acute Myeloid Leukemia with t(8;21): An International Study on 331 Patients. Blood 2019, 133, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Zhou, L.; Li, Y.; Yang, E.; Liu, Y.; Lv, N.; Fu, L.; Ding, Y.; Wang, N.; Fang, N.; et al. Profiling of Somatic Mutations and Fusion Genes in Acute Myeloid Leukemia Patients with FLT3-ITD or FLT3-TKD Mutation at Diagnosis Reveals Distinct Evolutionary Patterns. Exp. Hematol. Oncol. 2021, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Aref, S.; Rizk, R.; El Agdar, M.; Fakhry, W.; El Zafrany, M.; Sabry, M. NOTCH-1 Gene Mutations Influence Survival in Acute Myeloid Leukemia Patients. Asian Pac. J. Cancer Prev. 2020, 21, 1987–1992. [Google Scholar] [CrossRef]
- Wakita, S.; Yamaguchi, H.; Ueki, T.; Usuki, K.; Kurosawa, S.; Kobayashi, Y.; Kawata, E.; Tajika, K.; Gomi, S.; Koizumi, M.; et al. Complex Molecular Genetic Abnormalities Involving Three or More Genetic Mutations Are Important Prognostic Factors for Acute Myeloid Leukemia. Leukemia 2016, 30, 545–554. [Google Scholar] [CrossRef]
- Park, S.H.; Chi, H.-S.; Cho, Y.-U.; Jang, S.; Park, C.-J. CEBPA Single Mutation Can Be a Possible Favorable Prognostic Indicator in NPM1 and FLT3-ITD Wild-Type Acute Myeloid Leukemia Patients with Intermediate Cytogenetic Risk. Leuk Res. 2013, 37, 1488–1494. [Google Scholar] [CrossRef]
- Sengsayadeth, S.M.; Jagasia, M.; Engelhardt, B.G.; Kassim, A.; Strickland, S.A.; Goodman, S.; Lucid, C.; Vnencak-Jones, C.L.; Greer, J.P.; Savani, B.N. Allo-SCT for High-Risk AML-CR1 in the Molecular Era: Impact of FLT3/ITD Outweighs the Conventional Markers. Bone Marrow Transplant. 2012, 47, 1535–1537. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamaguchi, H.; Kuboyama, M.; Najima, Y.; Usuki, K.; Ueki, T.; Oh, I.; Mori, S.; Kawata, E.; Uoshima, N.; et al. Significance of FLT3-Tyrosine Kinase Domain Mutation as a Prognostic Factor for Acute Myeloid Leukemia. Int. J. Hematol. 2019, 110, 566–574. [Google Scholar] [CrossRef]
- Han, H.; Yao, Y.; Wang, H.; Zhou, M.; Zhang, Z.; Xu, X.; Qi, J.; Liu, Y.; Wu, D.; Han, Y. Landscape and Clinical Impact of NOTCH Mutations in Newly Diagnosed Acute Myeloid Leukemia. Cancer 2023, 129, 245–254. [Google Scholar] [CrossRef]
- Mason, E.F.; Hasserjian, R.P.; Aggarwal, N.; Seegmiller, A.C.; Pozdnyakova, O. Blast Phenotype and Comutations in Acute Myeloid Leukemia with Mutated NPM1 Influence Disease Biology and Outcome. Blood Adv. 2019, 3, 3322–3332. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.-J.; Liu, X.-D.; Zhong, L.-Y.; Li, K.-B.; Sun, Q.-X.; Xu, X.; Wei, T.; Li, Q.-S.; Zhu, Z.-G. Comparison of Gene Mutation Spectra in Younger and Older Chinese Acute Myeloid Leukemia Patients and Its Prognostic Value. Gene 2021, 770, 145344. [Google Scholar] [CrossRef] [PubMed]
- Wakita, S.; Sakaguchi, M.; Oh, I.; Kako, S.; Toya, T.; Najima, Y.; Doki, N.; Kanda, J.; Kuroda, J.; Mori, S.; et al. Prognostic Impact of CEBPA BZIP Domain Mutation in Acute Myeloid Leukemia. Blood Adv. 2022, 6, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-X.; Jiang, H.; Chang, Y.-J.; Zhou, Y.-L.; Wang, J.; Wang, Z.-L.; Cao, L.-M.; Li, J.-L.; Sun, Q.-Y.; Cao, S.-B.; et al. Risk Stratification of Cytogenetically Normal Acute Myeloid Leukemia With Biallelic CEBPA Mutations Based on a Multi-Gene Panel and Nomogram Model. Front. Oncol. 2021, 11, 706935. [Google Scholar] [CrossRef]
- Cappelli, L.V.; Meggendorfer, M.; Baer, C.; Nadarajah, N.; Hutter, S.; Jeromin, S.; Dicker, F.; Kern, W.; Haferlach, T.; Haferlach, C.; et al. Indeterminate and Oncogenic Potential: CHIP vs CHOP Mutations in AML with NPM1 Alteration. Leukemia 2022, 36, 394–402. [Google Scholar] [CrossRef]
- Duan, W.; Liu, X.; Zhao, X.; Jia, J.; Wang, J.; Gong, L.; Jiang, Q.; Zhao, T.; Wang, Y.; Zhang, X.; et al. Both the Subtypes of KIT Mutation and Minimal Residual Disease Are Associated with Prognosis in Core Binding Factor Acute Myeloid Leukemia: A Retrospective Clinical Cohort Study in Single Center. Ann. Hematol. 2021, 100, 1203–1212. [Google Scholar] [CrossRef]
- Tarlock, K.; Alonzo, T.A.; Wang, Y.C.; Gerbing, R.B.; Ries, R.; Loken, M.R.; Pardo, L.; Hylkema, T.; Joaquin, J.; Sarukkai, L.; et al. Functional Properties of KIT Mutations Are Associated with Differential Clinical Outcomes and Response to Targeted Therapeutics in CBF Acute Myeloid Leukemia. Clin. Cancer Res. 2019, 25, 5038–5048. [Google Scholar] [CrossRef]
- Kurosawa, S.; Yamaguchi, H.; Yamaguchi, T.; Fukunaga, K.; Yui, S.; Kanamori, H.; Usuki, K.; Uoshima, N.; Yanada, M.; Takeuchi, J.; et al. The Prognostic Impact of FLT3-ITD, NPM1 and CEBPa in Cytogenetically Intermediate-Risk AML after First Relapse. Int. J. Hematol. 2020, 112, 200–209. [Google Scholar] [CrossRef]
- Terada, K.; Yamaguchi, H.; Ueki, T.; Usuki, K.; Kobayashi, Y.; Tajika, K.; Gomi, S.; Kurosawa, S.; Miyadera, K.; Tokura, T.; et al. Full-Length Mutation Search of the TP53 Gene in Acute Myeloid Leukemia Has Increased Significance as a Prognostic Factor. Ann. Hematol. 2018, 97, 51–61. [Google Scholar] [CrossRef]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Mrózek, K.; Chen, H.; Kittles, R.A.; Vukosavljevic, T.; Perrotti, D.; Vardiman, J.W.; Carroll, A.J.; et al. Adverse Prognostic Significance of KIT Mutations in Adult Acute Myeloid Leukemia with Inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006, 24, 3904–3911. [Google Scholar] [CrossRef]
- Patkar, N.; Kodgule, R.; Kakirde, C.; Raval, G.; Bhanshe, P.; Joshi, S.; Chaudhary, S.; Badrinath, Y.; Ghoghale, S.; Kadechkar, S.; et al. Clinical Impact of Measurable Residual Disease Monitoring by Ultradeep next Generation Sequencing in NPM1 Mutated Acute Myeloid Leukemia. Oncotarget 2018, 9, 36616–36624. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, Y.; Huang, Y.; Tan, J.; Chen, Y.; Yang, J.; Dou, H.; Zou, L.; Yu, J.; Bao, L. WT1 Mutations and Single Nucleotide Polymorphism Rs16754 Analysis of Patients with Pediatric Acute Myeloid Leukemia in a Chinese Population. Leuk. Lymphoma 2012, 53, 2195–2204. [Google Scholar] [CrossRef]
- Boissel, N.; Nibourel, O.; Renneville, A.; Gardin, C.; Reman, O.; Contentin, N.; Bordessoule, D.; Pautas, C.; de Revel, T.; Quesnel, B.; et al. Prognostic Impact of Isocitrate Dehydrogenase Enzyme Isoforms 1 and 2 Mutations in Acute Myeloid Leukemia: A Study by the Acute Leukemia French Association Group. J. Clin. Oncol. 2010, 28, 3717–3723. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Taki, T.; Tabuchi, K.; Taketani, T.; Hanada, R.; Tawa, A.; Tsuchida, M.; Horibe, K.; Tsukimoto, I.; Hayashi, Y. Tandem Duplications of MLL and FLT3 Are Correlated with Poor Prognoses in Pediatric Acute Myeloid Leukemia: A Study of the Japanese Childhood AML Cooperative Study Group. Pediatr. Blood Cancer 2008, 50, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Shouval, R.; Labopin, M.; Bomze, D.; Baerlocher, G.M.; Capria, S.; Blaise, D.; Hänel, M.; Forcade, E.; Huynh, A.; Saccardi, R.; et al. Risk Stratification Using FLT3 and NPM1 in Acute Myeloid Leukemia Patients Autografted in First Complete Remission. Bone Marrow Transplant. 2020, 55, 2244–2253. [Google Scholar] [CrossRef]
- Xu, J.; Hao, Z.; Chen, X.; Hong, M.; Muyey, D.M.; Chen, X.; Wang, H. The Characteristics and Clinical Prognosis Analysis of ASXL1 Mutations in Chinese Adult Patients with Primary Cytogenetically Normal Acute Myeloid Leukemia by Next-Generation Sequencing. Leuk. Lymphoma 2022, 63, 2321–2329. [Google Scholar] [CrossRef]
- Chen, X.; Tian, C.; Hao, Z.; Pan, L.; Hong, M.; Wei, W.; Muyey, D.M.; Wang, H.; Chen, X. The Impact of DNMT3A Variant Allele Frequency and Two Different Comutations on Patients with de Novo Cytogenetically Normal Acute Myeloid Leukemia. Cancer Med. 2023, 12, 10340–10350. [Google Scholar] [CrossRef]
- Kaburagi, T.; Shiba, N.; Yamato, G.; Yoshida, K.; Tabuchi, K.; Ohki, K.; Ishikita, E.; Hara, Y.; Shiraishi, Y.; Kawasaki, H.; et al. UBTF-Internal Tandem Duplication as a Novel Poor Prognostic Factor in Pediatric Acute Myeloid Leukemia. Genes Chromosomes Cancer 2023, 62, 202–209. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, H.; Qiao, C.; Zhao, S.; Liu, L.; Wang, Y.; Jin, H.; Qian, S.; Wu, Y. Next-Generation Sequencing Reveals Gene Mutations Landscape and Clonal Evolution in Patients with Acute Myeloid Leukemia. Hematology 2021, 26, 111–122. [Google Scholar] [CrossRef]
- Jin, H.; Zhu, Y.; Hong, M.; Wu, Y.; Qiu, H.; Wang, R.; Jin, H.; Sun, Q.; Fu, J.; Li, J.; et al. Co-Occurrence of KIT and NRAS Mutations Defines an Adverse Prognostic Core-Binding Factor Acute Myeloid Leukemia. Leuk. Lymphoma 2021, 62, 2428–2437. [Google Scholar] [CrossRef]
- Abousamra, N.; Elghannam, D.M.; Shahin, D.A.; Goda, E.F.; Azzam, H.; Azmy, E.; El-Din, M.S. Prognostic Implication of N-RAS Gene Mutations in Egyptian Adult Acute Myeloid Leukemia. Egypt. J. Immunol. 2009, 16, 9–15. [Google Scholar] [CrossRef]
- Aly, R.; El-Sharnoby, M.R.; Hagag, A.A. Prognostic Significance of NRAS Gene Mutations in Children with Acute Myelogenous Leukemia. Mediterr. J. Hematol. Infect. Dis. 2011, 3, e2011055. [Google Scholar] [CrossRef] [PubMed]
- Mechaal, A.; Menif, S.; Abbes, S.; Safra, I. EZH2, New Diagnosis and Prognosis Marker in Acute Myeloid Leukemia Patients. Adv. Med. Sci. 2019, 64, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Dou, H.; Wang, X.; Huang, Y.; Lu, L.; Bin, J.; Su, Y.; Zou, L.; Yu, J.; Bao, L. KIT Mutations Correlate with Adverse Survival in Children with Core-Binding Factor Acute Myeloid Leukemia. Leuk. Lymphoma 2018, 59, 829–836. [Google Scholar] [CrossRef]
- Zhang, Y.-W.; Su, L.; Tan, Y.-H.; Lin, H.; Liu, X.-L.; Liu, Q.-J.; Sun, J.-N.; Zhang, M.; Du, Y.-Z.; Song, F.; et al. Measurable Residual Disease Detected by Flow Cytometry Independently Predicts Prognoses of NPM1-Mutated Acute Myeloid Leukemia. Ann. Hematol. 2023, 102, 337–347. [Google Scholar] [CrossRef]
- Aref, S.; El-Ghonemy, M.S.; Abouzeid, T.E.; El-Sabbagh, A.M.; El-Baiomy, M.A. Telomerase Reverse Transcriptase (TERT) A1062T Mutation as a Prognostic Factor in Egyptian Patients with Acute Myeloid Leukemia (AML). Med. Oncol. 2014, 31, 158. [Google Scholar] [CrossRef]
- Yang, J.; Yao, D.-M.; Ma, J.-C.; Yang, L.; Guo, H.; Wen, X.-M.; Xiao, G.-F.; Qian, Z.; Lin, J.; Qian, J. The Prognostic Implication of SRSF2 Mutations in Chinese Patients with Acute Myeloid Leukemia. Tumor Biol. 2016, 37, 10107–10114. [Google Scholar] [CrossRef]
- Su, L.; Gao, S.; Tan, Y.; Lin, H.; Liu, X.; Liu, S.; Yang, Y.; Sun, J.; Li, W. CSF3R Mutations Were Associated with an Unfavorable Prognosis in Patients with Acute Myeloid Leukemia with CEBPA Double Mutations. Ann. Hematol. 2019, 98, 1641–1646. [Google Scholar] [CrossRef]
- Riera, L.; Marmont, F.; Toppino, D.; Frairia, C.; Sismondi, F.; Audisio, E.; Di Bello, C.; D’Ardia, S.; Di Celle, P.F.; Messa, E.; et al. Core Binding Factor Acute Myeloid Leukaemia and C-KIT Mutations. Oncol. Rep. 2013, 29, 1867–1872. [Google Scholar] [CrossRef]
- Awad, M.M.; Aladle, D.A.; Abousamra, N.K.; Elghannam, D.M.; Fawzy, I.M. CEBPA Gene Mutations in Egyptian Acute Myeloid Leukemia Patients: Impact on Prognosis. Hematology 2013, 18, 61–68. [Google Scholar] [CrossRef]
- Virijevic, M.; Karan-Djurasevic, T.; Marjanovic, I.; Tosic, N.; Mitrovic, M.; Djunic, I.; Colovic, N.; Vidovic, A.; Suvajdzic-Vukovic, N.; Tomin, D.; et al. Somatic Mutations of Isocitrate Dehydrogenases 1 and 2 Are Prognostic and Follow-up Markers in Patients with Acute Myeloid Leukaemia with Normal Karyotype. Radiol. Oncol. 2016, 50, 385–393. [Google Scholar] [CrossRef]
- Moualla, Y.; Moassass, F.; Al-Halabi, B.; Al-Achkar, W.; Georgeos, M.; Yazigi, H.; Khamis, A. Evaluating the Clinical Significance of FLT3 Mutation Status in Syrian Newly Diagnosed Acute Myeloid Leukemia Patients with Normal Karyotype. Heliyon 2022, 8, e11858. [Google Scholar] [CrossRef] [PubMed]
- Zidan, M.A.A.; Kamal Shaaban, H.M.; Elghannam, D.M. Prognostic Impact of Wilms tumor Gene Mutations in Egyptian Patients with Acute Myeloid Leukemia with Normal Karyotype. Hematology 2014, 19, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Al-Arbeed, I.F.; Wafa, A.; Moassass, F.; Al-Halabi, B.; Al-Achkar, W.; Abou-Khamis, I. Frequency of FLT3 Internal Tandem Duplications in Adult Syrian Patients with Acute Myeloid Leukemia and Normal Karyotype. Asian Pac. J. Cancer Prev. 2021, 22, 3245–3251. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, F.; Huo, L.; Cai, W.; Wang, Q.; Wen, L.; Yan, L.; Shen, H.; Xu, X.; Chen, S. Clinical Characteristics and Prognostic Analysis of Acute Myeloid Leukemia Patients with PTPN11 Mutations. Hematology 2022, 27, 1184–1190. [Google Scholar] [CrossRef]
- Hollink, I.H.I.M.; Van Den Heuvel-Eibrink, M.M.; Zimmermann, M.; Balgobind, B.V.; Arentsen-Peters, S.T.C.J.M.; Alders, M.; Willasch, A.; Kaspers, G.J.L.; Trka, J.; Baruchel, A.; et al. Clinical Relevance of Wilms Tumor 1 Gene Mutations in Childhood Acute Myeloid Leukemia. Blood 2009, 113, 5951–5960. [Google Scholar] [CrossRef]
- Suzuki, T. Clinical Characteristics and Prognostic Implications of NPM1 Mutations in Acute Myeloid Leukemia. Blood 2005, 106, 2854–2861. [Google Scholar] [CrossRef]
- Nakamura, H.; Inokuchi, K.; Yamaguchi, H.; Dan, K. Abnormalities of P51, P53, FLT3 and N-Ras Genes and Their Prognostic Value in Relapsed Acute Myeloid Leukemia. J. Nippon. Med. Sch. 2004, 71, 270–278. [Google Scholar] [CrossRef]
- Ribeiro, A.F.T.; Pratcorona, M.; Erpelinck-Verschueren, C.; Rockova, V.; Sanders, M.; Abbas, S.; Figueroa, M.E.; Zeilemaker, A.; Melnick, A.; Löwenberg, B.; et al. Mutant DNMT3A: A Marker of Poor Prognosis in Acute Myeloid Leukemia. Blood 2012, 119, 5824–5831. [Google Scholar] [CrossRef]
- Wei, H.; Zhou, C.; Liu, B.; Lin, D.; Li, Y.; Wei, S.; Gong, B.; Zhang, G.; Liu, K.; Gong, X.; et al. The Prognostic Factors in Acute Myeloid Leukaemia with Double-Mutated CCAAT/Enhancer-Binding Protein Alpha (CEBPAdm). Br. J. Haematol. 2022, 197, 442–451. [Google Scholar] [CrossRef]
- Cairoli, R.; Beghini, A.; Turrini, M.; Bertani, G.; Nadali, G.; Rodeghiero, F.; Castagnola, C.; Lazzaroni, F.; Nichelatti, M.; Ferrara, F.; et al. Old and New Prognostic Factors in Acute Myeloid Leukemia with Deranged Core-Binding Factor Beta. Am. J. Hematol. 2013, 88, 594–600. [Google Scholar] [CrossRef]
- Linch, D.C.; Hills, R.K.; Burnett, A.K.; Russell, N.; Gale, R.E. Analysis of the Clinical Impact of NPM1 Mutant Allele Burden in a Large Cohort of Younger Adult Patients with Acute Myeloid Leukaemia. Br. J. Haematol. 2020, 188, 852–859. [Google Scholar] [CrossRef]
- Hou, H.-A.; Liu, C.-Y.; Kuo, Y.-Y.; Chou, W.-C.; Tsai, C.-H.; Lin, L.-I.; Tseng, M.-H.; Chiang, Y.-C.; Liu, M.-C.; Liu, C.-W.; et al. Splicing Factor Mutations Predict Poor Prognosis in Patients with de Novo Acute Myeloid Leukemia. Oncotarget 2016, 7, 9084–9101. [Google Scholar] [CrossRef]
- Chou, W.C.; Chou, S.C.; Liu, C.W.C.Y.; Chen, C.Y.; Hou, H.A.; Kuo, Y.Y.Y.Y.; Lee, M.C.; Ko, B.S.; Tang, J.L.; Yao, M.; et al. TET2 Mutation Is an Unfavorable Prognostic Factor in Acute Myeloid Leukemia Patients with Intermediate-Risk Cytogenetics. Blood 2011, 118, 3803–3810. [Google Scholar] [CrossRef]
- Pollard, J.A.; Alonzo, T.A.; Gerbing, R.B.; Ho, P.A.; Zeng, R.; Ravindranath, Y.; Dahl, G.; Lacayo, N.J.; Becton, D.; Chang, M.; et al. Prevalence and Prognostic Significance of KIT Mutations in Pediatric Patients with Core Binding Factor AML Enrolled on Serial Pediatric Cooperative Trials for de Novo AML. Blood 2010, 115, 2372–2379. [Google Scholar] [CrossRef]
- Ibrahem, L.; Mahfouz, R.; Elhelw, L.; Abdsalam, E.M.; Soliman, R. Prognostic Significance of DNMT3A Mutations in Patients with Acute Myeloid Leukemia. Blood Cells Mol. Dis. 2015, 54, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hua, H.; Wang, Z.; Wang, B.; Cao, L.; Qin, W.; Wu, P.; Cai, X.; Chao, H.; Lu, X. Frequency and Clinical Impact of WT1 Mutations in the Context of CEBPA-Mutated Acute Myeloid Leukemia. Hematology 2022, 27, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Lai, Y.Y.; Chen, W.M.; Jiang, H.; Wang, Y.; Wang, X.; Zhao, X.S.; Huang, X.J.; Jiang, Q.; Qin, Y.Z.; et al. Independent Prognostic Significance of TP53 Mutations in Adult Acute Myeloid Leukaemia with Complex Karyotype. Int. J. Lab. Hematol. 2022, 44, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Lugthart, S.; Kavelaars, F.G.; Schelen, A.; Koenders, J.E.; Zeilemaker, A.; Van Putten, W.J.L.; Rijneveld, A.W.; Löwenberg, B.; Valk, P.J.M.; et al. Acquired Mutations in the Genes Encoding IDH1 and IDH2 Both Are Recurrent Aberrations in Acute Myeloid Leukemia: Prevalence and Prognostic Value. Blood 2010, 116, 2122–2126. [Google Scholar] [CrossRef]
- Yamato, G.; Shiba, N.; Yoshida, K.; Shiraishi, Y.; Hara, Y.; Ohki, K.; Okubo, J.; Okuno, H.; Chiba, K.; Tanaka, H.; et al. ASXL2 Mutations Are Frequently Found in Pediatric AML Patients with t(8;21)/ RUNX1-RUNX1T1 and Associated with a Better Prognosis. Genes Chromosomes Cancer 2017, 56, 382–393. [Google Scholar] [CrossRef]
- Yuen, K.; Liu, Y.; Zhou, Y.; Wang, Y.; Zhou, D.; Fang, J.; Xu, L. Mutational Landscape and Clinical Outcome of Pediatric Acute Myeloid Leukemia with 11q23/KMT2A Rearrangements. Cancer Med. 2023, 12, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, R.Q.; Wu, Y.; Jia, J.S.; Gong, L.; Liu, X.H.; Lu, S.Y.; Wang, Y.; Yan, C.H.; Liu, K.Y.; et al. Detection of Measurable Residual Disease May Better Predict Outcomes than Mutations Based on Next-Generation Sequencing in Acute Myeloid Leukaemia with Biallelic Mutations of CEBPA. Br. J. Haematol. 2020, 190, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Toogeh, G.; Ramzi, M.; Faranoush, M.; Amirizadeh, N.; Haghpanah, S.; Moghadam, M.; Cohan, N. Prevalence and Prognostic Impact of Wilms’ Tumor 1 (WT1) Gene, Including SNP Rs16754 in Cytogenetically Normal Acute Myeloblastic Leukemia (CN-AML): An Iranian Experience. Clin. Lymphoma Myeloma Leuk. 2016, 16, e21–e26. [Google Scholar] [CrossRef] [PubMed]
- Bachas, C.; Schuurhuis, G.J.; Reinhardt, D.; Creutzig, U.; Kwidama, Z.J.; Zwaan, C.M.; van den Heuvel-Eibrink, M.M.; De Bont, E.S.J.M.; Elitzur, S.; Rizzari, C.; et al. Clinical Relevance of Molecular Aberrations in Paediatric Acute Myeloid Leukaemia at First Relapse. Br. J. Haematol. 2014, 166, 902–910. [Google Scholar] [CrossRef]
- Marková, J.; Michková, P.; Burčková, K.; Březinová, J.; Michalová, K.; Dohnalová, A.; Maaloufová, J.S.; Soukup, P.; Vítek, A.; Cetkovský, P.; et al. Prognostic Impact of DNMT3A Mutations in Patients with Intermediate Cytogenetic Risk Profile Acute Myeloid Leukemia. Eur. J. Haematol. 2012, 88, 128–135. [Google Scholar] [CrossRef]
- Quesada, A.E.; Montalban-Bravo, G.; Luthra, R.; Patel, K.P.; Sasaki, K.; Bueso-Ramos, C.E.; Khoury, J.D.; Routbort, M.J.; Bassett, R.; Hidalgo-Lopez, J.E.; et al. Clinico-Pathologic Characteristics and Outcomes of the World Health Organization (WHO) Provisional Entity de Novo Acute Myeloid Leukemia with Mutated RUNX1. Mod. Pathol. 2020, 33, 1678–1689. [Google Scholar] [CrossRef]
- Su, L.; Tan, Y.; Lin, H.; Liu, X.; Yu, L.; Yang, Y.; Liu, S.; Bai, O.; Yang, Y.; Jin, F.; et al. Mutational Spectrum of Acute Myeloid Leukemia Patients with Double CEBPA Mutations Based on Next-Generation Sequencing and Its Prognostic Significance. Oncotarget 2018, 9, 24970–24979. [Google Scholar] [CrossRef]
- Tien, F.-M.; Hou, H.-A.; Tsai, C.-H.; Tang, J.-L.; Chiu, Y.-C.; Chen, C.-Y.; Kuo, Y.-Y.; Tseng, M.-H.; Peng, Y.-L.; Liu, M.-C.; et al. GATA2 Zinc Finger 1 Mutations Are Associated with Distinct Clinico-Biological Features and Outcomes Different from GATA2 Zinc Finger 2 Mutations in Adult Acute Myeloid Leukemia. Blood Cancer J. 2018, 8, 87. [Google Scholar] [CrossRef]
- Canaani, J.; Labopin, M.; Huang, X.J.; Arcese, W.; Ciceri, F.; Blaise, D.; Irrera, G.; Corral, L.L.; Bruno, B.; Santarone, S.; et al. T-Cell Replete Haploidentical Stem Cell Transplantation Attenuates the Prognostic Impact of FLT3-ITD in Acute Myeloid Leukemia: A Report from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation. Am. J. Hematol. 2018, 93, 736–744. [Google Scholar] [CrossRef]
- Koczkodaj, D.; Zmorzyński, S.; Grygalewicz, B.; Pieńkowska-Grela, B.; Styk, W.; Popek-Marciniec, S.; Filip, A.A. WT1 Gene Mutations, Rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients. J. Clin. Med. 2022, 11, 1873. [Google Scholar] [CrossRef] [PubMed]
- Zare-Abdollahi, D.; Safari, S.; Movafagh, A.; Riazi-Isfahani, S.; Ghadyani, M.; Feyzollah, H.-G.; Nasrollahi, M.F.; Omrani, M.D. A Mutational and Expressional Analysis of DNMT3A in Acute Myeloid Leukemia Cytogenetic Subgroups. Hematology 2015, 20, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Pratcorona, M.; Brunet, S.; Nomdedéu, J.; Ribera, J.M.; Tormo, M.; Duarte, R.; Escoda, L.; Guàrdia, R.; de Llano, M.P.Q.; Salamero, O.; et al. Favorable Outcome of Patients with Acute Myeloid Leukemia Harboring a Low-Allelic Burden FLT3-ITD Mutation and Concomitant NPM1 Mutation: Relevance to Post-Remission Therapy. Blood 2013, 121, 2734–2738. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Kawashima, N.; Atsuta, Y.; Sugiura, I.; Sawa, M.; Dobashi, N.; Yokoyama, H.; Doki, N.; Tomita, A.; Kiguchi, T.; et al. Prospective Evaluation of Prognostic Impact of KIT Mutations on Acute Myeloid Leukemia with RUNX1-RUNX1T1 and CBFB-MYH11. Blood Adv. 2020, 4, 66–75. [Google Scholar] [CrossRef]
- Yui, S.; Kurosawa, S.; Yamaguchi, H.; Kanamori, H.; Ueki, T.; Uoshima, N.; Mizuno, I.; Shono, K.; Usuki, K.; Chiba, S.; et al. D816 Mutation of the KIT Gene in Core Binding Factor Acute Myeloid Leukemia Is Associated with Poorer Prognosis than Other KIT Gene Mutations. Ann. Hematol. 2017, 96, 1641–1652. [Google Scholar] [CrossRef]
- Yu, G.; Yin, C.; Wu, F.; Jiang, L.; Zheng, Z.; Xu, D.; Zhou, J.; Jiang, X.; Liu, Q.; Meng, F. Gene Mutation Profile and Risk Stratification in AML1-ETO-Positive Acute Myeloid Leukemia Based on next-Generation Sequencing. Oncol. Rep. 2019, 42, 2333–2344. [Google Scholar] [CrossRef]
- Marceau-Renaut, A.; Duployez, N.; Ducourneau, B.; Labopin, M.; Petit, A.; Rousseau, A.; Geffroy, S.; Bucci, M.; Cuccuini, W.; Fenneteau, O.; et al. Molecular Profiling Defines Distinct Prognostic Subgroups in Childhood AML: A Report from the French ELAM02 Study Group. Hemasphere 2018, 2, e31. [Google Scholar] [CrossRef]
- Oñate, G.; Bataller, A.; Garrido, A.; Hoyos, M.; Arnan, M.; Vives, S.; Coll, R.; Tormo, M.; Sampol, A.; Escoda, L.; et al. Prognostic Impact of DNMT3A Mutation in Acute Myeloid Leukemia with Mutated NPM1. Blood Adv. 2022, 6, 882–890. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamaguchi, H.; Najima, Y.; Usuki, K.; Ueki, T.; Oh, I.; Mori, S.; Kawata, E.; Uoshima, N.; Kobayashi, Y.; et al. Prognostic Impact of Low Allelic Ratio FLT3-ITD and NPM1 Mutation in Acute Myeloid Leukemia. Blood Adv. 2018, 2, 2744–2754. [Google Scholar] [CrossRef]
- Metafuni, E.; Amato, V.; Giammarco, S.; Bellesi, S.; Rossi, M.; Minnella, G.; Frioni, F.; Limongiello, M.A.; Pagano, L.; Bacigalupo, A.; et al. Pre-Transplant Gene Profiling Characterization by next-Generation DNA Sequencing Might Predict Relapse Occurrence after Hematopoietic Stem Cell Transplantation in Patients Affected by AML. Front. Oncol. 2022, 12, 939819. [Google Scholar] [CrossRef] [PubMed]
- Duncavage, E.J.; Bagg, A.; Hasserjian, R.P.; DiNardo, C.D.; Godley, L.A.; Iacobucci, I.; Jaiswal, S.; Malcovati, L.; Vannucchi, A.M.; Patel, K.P.; et al. Genomic Profiling for Clinical Decision Making in Myeloid Neoplasms and Acute Leukemia. Blood 2022, 140, 2228–2247. [Google Scholar] [CrossRef] [PubMed]
- Tien, F.M.; Yao, C.Y.; Tsai, X.C.H.; Lo, M.Y.; Chen, C.Y.; Lee, W.H.; Lin, C.C.; Kuo, Y.Y.; Peng, Y.L.; Tseng, M.H.; et al. Dysregulated Immune and Metabolic Pathways Are Associated with Poor Survival in Adult Acute Myeloid Leukemia with CEBPA BZIP In-Frame Mutations. Blood Cancer J. 2024, 14, 15. [Google Scholar] [CrossRef]
- Blau, O.; Berenstein, R.; Sindram, A.; Blau, I.W. Molecular Analysis of Different FLT3-ITD Mutations in Acute Myeloid Leukemia. Leuk. Lymphoma 2013, 54, 145–152. [Google Scholar] [CrossRef]
- Falini, B.; Brunetti, L.; Sportoletti, P.; Paola Martelli, M. NPM1-Mutated Acute Myeloid Leukemia: From Bench to Bedside. Blood 2020, 136, 1707–1721. [Google Scholar] [CrossRef]
- Thol, F.; Damm, F.; Lüdeking, A.; Winschel, C.; Wagner, K.; Morgan, M.; Yun, H.; Göhring, G.; Schlegelberger, B.; Hoelzer, D.; et al. Incidence and Prognostic Influence of DNMT3A Mutations in Acute Myeloid Leukemia. J. Clin. Oncol. 2011, 29, 2889–2896. [Google Scholar] [CrossRef]
- Schnittger, S.; Kohl, T.M.; Haferlach, T.; Kern, W.; Hiddemann, W.; Spiekermann, K.; Schoch, C. KIT-D816 Mutations in AML1-ETO-Positive AML Are Associated with Impaired Event-Free and Overall Survival. Blood 2006, 107, 1791–1799. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Altman, J.K.; Assi, R.; Bixby, D.; Fathi, A.T.; Foran, J.M.; Gojo, I.; Hall, A.C.; Jonas, B.A.; Kishtagari, A.; et al. Acute Myeloid Leukemia, Version 3.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 503–513. [Google Scholar] [CrossRef]
- Bacher, U.; Haferlach, T.; Schoch, C.; Kern, W.; Schnittger, S. Implications of NRAS Mutations in AML: A Study of 2502 Patients. Blood 2006, 107, 3847–3853. [Google Scholar] [CrossRef]
- Bezerra, M.F.; Lima, A.S.; Piqué-Borràs, M.R.; Silveira, D.R.; Coelho-Silva, J.L.; Pereira-Martins, D.A.; Weinhäuser, I.; Franca-Neto, P.L.; Quek, L.; Corby, A.; et al. Co-Occurrence of DNMT3A, NPM1, FLT3 Mutations Identifies a Subset of Acute Myeloid Leukemia with Adverse Prognosis. Blood 2020, 135, 870–875. [Google Scholar] [CrossRef]
| Study ID | Country of Origin | Sample Size | M | F | U | Age in Years (Range) | Follow up in Months | Type of Molecular Test | Tested Somatic Mutations | Accompanied Somatic Mutations | NOS Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abbas S./2010 | The Netherlands | 895 | 429 | 466 | 0 | NA (15–77) | 33.2 | Direct sequencing | IDH1 U, IDH2 U | FLT3-ITD, FLT3-TKD, NPM1, NRAS, KRAS | 6 |
| Al-Arbeed I.F./2021 | Syria | 44 | 23 | 21 | 0 | 35.3 ± 12.4 | 14 | RFLP-PCR | FLT3-ITD K | NA | 5 |
| Aly R.M./2011 | Egypt | 39 | 21 | 18 | 0 | 7.4 (5.6–13) | 32 ± 2.24 | PCR-SSCP | NRAS K | NA | 7 |
| Aref S./2014 | Egypt | 153 | 75 | 78 | 0 | NA (17–65) | 48 | Direct sequencing | TERT M | NA | 6 |
| Aref S./2020 | Egypt | 50 | 26 | 24 | 0 | NA (24–59) | 24 | Direct sequencing | NOTCH1 M | NA | 8 |
| Awad M.M./2013 | Egypt | 55 | 25 | 30 | 0 | 45.65 ± 16 | NA | PCR-SSCP | CEBPAsm K, CEBPAdm K | NA | 6 |
| Bachas C./2014 | Germany and The Netherlands | 198 | 122 | 76 | 0 | 10.2 (0.4–19.5) | 40.6 (3.9–159.3) | HRM analysis | FLT3-ITD U, FLT3-TKD U, KRAS U, WT1 U, cKIT U, NRAS U, NPM1 U | NA | 7 |
| Boissel N./2010 | France | 205 | 90 | 115 | 0 | 48 (17–70) | NA | Direct sequencing | IDH2 M | FLT3-ITD, CEBPAsm, IDH1, WT1, NPM1 | 7 |
| Cairoli R./2013 | Italy | 58 | 40 | 18 | 0 | 42 (15–60) | 50 | Direct sequencing | cKIT M | NA | 8 |
| Canaani J./2018 | Israel | 293 | 172 | 121 | 0 | NA (18–73.8) | NA | NA | FLT3-ITD M | NA | 8 |
| Cappelli L.V./2022 | Germany | 150 | 73 | 77 | 0 | 57 (19–82) | 39.6 (2.4–104.4) | WGS or NGS panel (63 genes) | FLT3-ITD U, DNMT3A U | NA | 7 |
| Chen X./2012 | China | 127 | 67 | 60 | 0 | NA (0.3–15) | NA | Direct sequencing | WT1 U | FLT3-ITD, NPM1, CEBPA, cKIT | 5 |
| Chen X./2018 | China | 50 | 31 | 19 | 0 | 7 | 32 (1–90) | Direct sequencing | cKIT M | WT1, CEBPA | 8 |
| Chen X./2021 | China | 204 | 103 | 101 | 0 | 54.4 (20–86) | NA | NGS panel (22 genes) | NPM1 M, CEBPAdm M, TET2 M, ASXL1 M, cKIT M, IDH1 M, IDH2 M, DNMT3A M, RUNX1 M, TP53 M, PHF6 M, NRAS M | FLT3-ITD, DNMT3A | 8 |
| Chen X./2023 | China | 171 | 90 | 81 | 0 | 53 (19–86) | 47 | NGS panel (34 genes) | DNMT3A M | NPM1, BCOR, FLT3-ITD, CEBPAsm, NRAS, TET2 | 9 |
| Chou W.-C./2011 | Taiwan | 486 | 274 | 212 | 0 | 51.5 (15–90) | NA | Direct sequencing | TET2 M, CEBPAdm M, NPM1 M | cKIT, KRAS, NRAS, FLT3-ITD, MLL-PTD, ASXL1, FLT3-TKD, WT1, RUNX1 | 8 |
| Christen F./2019 | Multicenter | 331 | 188 | 143 | 0 | 41.7 (15–84) | 80.88 (3–253.2) | NGS panel (66 genes) | RAD21 M, JAK2 M | ASXL1, ASXL2, CBL, DHX15, EZH2, FLT3-ITD, cKIT, KRAS, NRAS, TET2 | 7 |
| Duan W./2021 | China | 215 | 124 | 91 | 0 | 39 (15–70) | 26 (7–12) | TaqMan based RT-PCR | cKIT M | FLT3-ITD, CEBPAsm, NPM1, EV11, MLL-PTD | 8 |
| Elghannam D.M./2009 | Egypt | 150 | 110 | 40 | 0 | 55 (19–74) | NA | PCR-SSCP | NRAS M | NA | 7 |
| Guan W./2021 | China | 207 | 115 | 92 | 0 | 45.4 (14–76) | NA | NGS panel | FLT3-ITD K, FLT3-TKD K | NPM1, DNMT3A, RUNX1, KIT, PTPN11. TET2, CEBPAdm, ASXL1, TP53 | 9 |
| Han H./2023 | China | 878 | 466 | 412 | 0 | 44 (8–78) | 45.2 | 2 NGS panel (51 or 172 genes panels) | NOTCH M | CBL, CSMD1, FLT3-TKD, JAK, PTPN11, STAG2, ZRSR2, TET2, TP53, WT1, CSF3R, FLT3-ITD, IDH2, NPM1, SETD2, CEBPAdm, EZH2, IDH1, MPL, RUNX1, CEBPAsm, DNMT3A, GATA2, cKIT, RAS | 8 |
| Hollink I. H. I./2009 | Multicenter | 232 | 133 | 99 | 0 | 9.6 | 52 | Direct sequencing | FLT3-ITD M, WT1 M | cKIT, NPM1, MLL-PTD | 7 |
| Hou H.-A./2016 | Taiwan | 500 | 285 | 215 | 0 | 51 (15–90) | 55 (1–160) | Direct sequencing | CEBPAdm M, RUNX1 M, WT1 M, ASXL1 M, IDH2 M, DNMT3A M, TP53 M | cKIT, JAK2, NPM1, MLL-PTD, SF3B1, U2AF1, SRSF2, FLT-ITD, FLT3-TKD, NRAS, KRAS, PTPN11 | 9 |
| Ibrahem L./2015 | Egypt | 120 | 66 | 54 | 0 | 47 (33–60) | 30 | Cycle sequencing | DNMT3A M | NA | 8 |
| Ishikawa Y./2020 | Japan | 199 | 125 | 74 | 0 | 41 (16–64) | 52.2 (11.9–81.8) | NGS panel | cKIT M, TET2 M, NRAS M | FLT3-ITD, FLT3-TKD, KRAS, JAK2, PTPN11, ASXL1, BCORL1, EZH2, KDM6A, SMC, SMC1A, RAD21, RUNX1, WT1, CSRF3R, ASXL2, DNMT3A, ETV6 | 7 |
| Jin H./2021 | China | 62 | 33 | 29 | 0 | 49.5 (19–83) | 21.5 | NGS Targeted deep sequencing | cKIT U, NRAS M, ASXL1 U, FLT3-ITD U, TET2 U | DMNT3A | 5 |
| Kaburagi T./2023 | Japan | 369 | 194 | 175 | 0 | 7 (0–17) | NA | NGS panel (343 genes) or Direct sequencing | UBTF-ITD K | FLT3-ITD, WT1 | 5 |
| Koczkodaj D./2022 | Poland | 90 | 42 | 48 | 0 | 62.63 (18–85) | NA | Direct sequencing | FLT3-ITD M, WT1 M, NPM1 M | NA | 7 |
| Kurosawa S./2020 | Japan | 235 | 141 | 94 | 0 | 51 (18–65) | NA | NGS (Ion torrent) + Direct sequencing | FLT3-ITD M, NPM1 U, CEBPAdm M | NA | 8 |
| Linch D.C./2020 | UK | 876 | 368 | 508 | 0 | NA (16–59) | 108 (4–260) | Capillary electrophoresis | FLT3-ITD U, DNMT3A U | NA | 7 |
| Marceau-Renaut A./2018 | France | 385 | 210 | 175 | 0 | 8.6 (0–18) | 59 | NGS panel (36 genes) | NPM1 M, FLT3-ITD K, CEBPAdm M, WT1 M, RUNX1 M, PHF6 M | FLT3-TKD, cKIT, NRAS, KRAS, CBL, TET2, PTPN11, ASXL1, SMC1A, SF3B1, JAK2, EZH2, SMC3, ZRSR2, MPL, BCOR, RAD21, U2AF1, SETBP1, BCORL1, STAG2, DNMT3A, GATA2, IDH1, IDH2, ETV6, TP53, NPM1, WT1, GATA1, GATA2 | 7 |
| Markova J./2012 | Czech Republic | 226 | 107 | 119 | 0 | 54.9 (18.2–81.7) | 1.6 (0–202) | Direct sequencing | DNMT3A K | NA | 7 |
| Mason EF./2019 | US | 239 | 110 | 129 | 0 | 64.8 (14–89) | 14.2 (0.1–88.4) | NGS panel (95 genes) | FLT3-ITD M | NPM1, SRSF2, IDH2, TET2, IDH1, DNMT3A, RAS, WT1 | 8 |
| Matsuo H./2020 | Japan | 160 | 72 | 88 | 0 | 3.9 (0.0–18.2) | NA | NGS panel (338 genes) | FLT3-ITD M, KRAS M, NRAS M, PTPN11 M, SETD2 M, STAG2 M, CCND3 M, U2AF1 M | NA | 8 |
| Mechaal A./2019 | Tunisia | 211 | 110 | 101 | 0 | 35 (2–80) | NA | Direct sequencing | EZH2 K | IDH2, NPM1, FLT3-ITD, DNMT3A | 5 |
| Metafuni E./2022 | Italy | 96 | 57 | 39 | 0 | 56 (17–73) | NA | NGS panel (26 gene) | TP53 M, NRAS M, WT1 M, FLT3-ITD M | ASXL1, cKIT, DNMT3A, EZH2, TET2, SRSF2, RUNX1, KRAS, IDH1, IDH2, U2AF1 | 6 |
| Moualla Y./2022 | Syria | 100 | 51 | 49 | 0 | NA | NA | Direct sequencing | FLT3-ITD M | FLT3-TKD | 6 |
| Nakamura H./2004 | Japan | 24 | 12 | 12 | 0 | 54 (34–78) | 9 (6–81) | Direct sequencing | NRAS K, TP53 K, FLT3-ITD K | NA | 9 |
| Onate G./2022 | Spain | 164 | 58 | 106 | 0 | NA (18–72) | 30 | Direct sequencing | DNMT3A K | NA | 8 |
| Papaemmanuil E./2016 | UK | 1540 | 823 | 717 | 0 | 54 (18–84) | 70.8 (1–179) | NGS panel (111 genes) | FLT3-ITD M, GATA2 M, TP53 M, BRAF M, SRSF2 M, NPM1 M, CEBPAdm M, ASXL1 M, ZRSR2 M, RUNX1 M, IDH2 M | DNMT3A, FLT3-TKD, STAG2, RAD21 | 8 |
| Park SH./2013 | South Korea | 157 | 91 | 66 | 0 | 50.65 ± 17.2 | NA | Direct sequencing | FLT3-ITD M, CEBPAsm M, CEBPAdm M, NPM1 M | DNMT3A, IDH1, IDH2 | 8 |
| Paschka et al. 2006 | US | 110 | 59 | 51 | 0 | NA | 64 | Direct sequencing and DHLPC | cKIT M | NA | 9 |
| Patkar N./2018 | India | 83 | 46 | 37 | 0 | 36.7 (18–62) | 23.5 | NGS Targeted deep sequencing | FLT3-ITD U, DNMT3A U | NPM1 | 7 |
| Pollard J.A./2010 | US | 203 | 106 | 97 | 0 | NA (0.6–19.6) | 66.8 (1.9–104.5) | Direct sequencing | cKIT K | FLT3-ITD, WT1 | 8 |
| Pratcorona M./2013 | Spain | 303 | 173 | 130 | 0 | 47 (17–60) | NA | Direct sequencing | FLT3-ITD M, NPM1 M | NA | 8 |
| Quesada, A. E./2020 | US | 140 | 79 | 61 | 0 | NA (20–87) | 21.4 | Direct sequencing | RUNX1 U, NPM1 U | FLT3-ITD, NRAS, IDH2, SRSF2, EZH2, DNMT3A, CEBPA, TET2, ASXL1 | 9 |
| Ribeiro A.F.T./2012 | The Netherlands | 415 | 210 | 205 | 0 | 41 (15–60) | 115.7 (7.2–224.1) | HPLC | DNMT3A M, FLT3-ITD M, NPM1 M, CEBPAdm M, NRAS M, IDH1 M, IDH2 M, EV11 M, WT1 M, cKIT M | NA | 8 |
| Riera L./2013 | Italy | 23 | 11 | 12 | 0 | 42.7 (19–64) | 88 | Direct sequencing | cKIT K | NA | 6 |
| Sakaguchi M./2018 | Japan | 147 | 66 | 77 | 4 | 56 (18–90) | 11.5 | Fragment analysis | NPM1 U | CEBPAsm, CEBPAdm, FLT3-ITD | 8 |
| Sakaguchi M./2019 | Japan | 674 | 395 | 276 | 3 | 57 (15–94) | NA | RFLP-PCR | FLT3-ITD K, FLT3-TKD K | NPM1, CEBPAsm, CEBPAdm | 7 |
| Sengsayadeth S.M./2012 | US | 75 | 37 | 38 | 0 | 49 (20–68) | NA | NA | FLT3-ITD K | NA | 8 |
| Shimada A./2008 | Japan | 158 | 89 | 69 | 0 | 6 (0–15) | NA | Direct sequencing | FLT3-ITD M, MLL-PTD M | NA | 7 |
| Shouval R./2020 | France | 405 | 200 | 205 | 0 | 52.5 (42.9–60) | 66 (43.2–93.6) | NA | FLT3-ITD K, NPM1 K | NA | 8 |
| Su L./2018 | China | 81 | 45 | 36 | 0 | 44 (9–67) | 8 (2–66) | NGS panel (112 genes) | CSF3R K, WT1 K, GATA2 K | NRAS, TET2, CEBPAdm | 6 |
| Su L/2019 | China | 101 | 53 | 48 | 0 | 43 (9–79) | 18.5 (3–78) | Direct sequencing | CSF3R K | NA | 8 |
| Sun Y./2022 | China | 74 | 35 | 39 | 0 | 43 (6–68) | NA | NGS panel | ETV6 M, TET2 M, ASXL1 M | PTPN11, DNMT3A, NPM1, CEBPA, FLT3-ITD, EZH2, NRAS, cKIT | 8 |
| Suzuki T./2005 | Japan | 190 | NA | NA | 190 | 50 (15–85) | NA | Direct sequencing | NPM1 K | FLT3-ITD, TP53, NRAS | 7 |
| Tarlock K./2019 | US | 205 | 103 | 102 | 0 | 11.5 (0.33–22.76) | 64. 9 (0–96.9) | Targeted exome capture sequencing | cKIT U | FLT3-ITD | 7 |
| Terada K./2018 | Japan | 412 | 235 | 174 | 3 | 55.1 (15–91) | NA | NGS Ion PGM™ and Direct sequencing | TP53 M, FLT3-ITD M, TET2 M, DNMT3A M, NRAS M, cKIT M, CEBPAdm M, MLL-PTD M, ASXL1 M | NOTCH1, NCOR2, IDH2, WT1, CEBPAsm, IDH1, PTPN11, GATA2, BCOR, NPM1, BCORL1, FLT3-TKD | 8 |
| The Cancer Genome Atlas Research Network/2013 | US | 200 | 108 | 92 | 0 | 55.0 ± 16.1 | NA | WGS or WES | TP53 M, DNMT3A M, FLT3-ITD M, RUNX1 M | NPM1, TET2, CEBPAdm, WT1, PTPN11, KIT | 8 |
| Tien F.-M./2018 | Taiwan | 693 | 395 | 298 | 0 | 55 (15–94) | 78.6 (0.1–236) | Ion torrent NGS and Direct sequencing | GATA2 M, CEBPAdm M | IDH1, IDH2, NPM1, TET2, CEBPAsm, DNMT3A, KRAS, WT1, PTPN11, NRAS, ETV6, RUNX1, MLL-PTD, TP53, cKIT, ASXL1, FLT3-ITD, FLT3-TKD | 7 |
| Toogeh G./2016 | Iran | 88 | 55 | 33 | 0 | 42 ± 12 | 24 | Direct sequencing | WT1 K | NA | 8 |
| Virijevic M./2016 | Serbia | 110 | 62 | 48 | 0 | 53.5 (19–78) | NA | Direct sequencing | FLT3-ITD M | NA | 8 |
| Wakita S./2016 | Japan | 271 | 157 | 114 | 0 | 54. (17–86) | NA | NGS (Ion torrent) | NPM1 M, FLT3-ITD M, CEBPAdm M, NRAS M | CEBPAsm, IDH1, IDH2, FLT3-TKD, KMT2A, KRAS, TET2, DNMT3A, ASXL1, KMT2A, RUNX1, cKIT, TP53, PTPN11, GATA2, WT1, STAG2, SMC1A, SMC3, DAXX, BCOR, BCORL1, NF1, DDX41, PHF6 | 9 |
| Wakita S./2022 | Japan | 1028 | 580 | 448 | 0 | 54.3 (16–70) | NA | Direct sequencing | CEBPAdm K, CEBPAsm K | FLT3-ITD, NPM1 | 8 |
| Wang J./2020 | China | 124 | 73 | 51 | 0 | 37.5 (16–69) | 33.5 (4–69) | NGS panel (87 genes) | NRAS K, WT1 K, GATA2 K, CSF3R K | FLT3-ITD | 7 |
| Wang T./2022 | China | 220 | 114 | 106 | 0 | 39 (18–88) | 30.5 (0.5–60.6) | NGS panel (112 genes) and Direct sequencing | WT1 K | CEBPAdm, RUNX1, IDH1, JAK2, CSF3R, ZRSR2, SMC3, SRSF2, SF3B1, RAD21, BCOR, BCORL, cKIT, TP53, ASXL1, FAT1, EZH2, GATA2, IDH2, SH2B3, RELN, NRAS, SETBP1, DNMT3A, PTPN11, NOTCH11, KRAS, CEBPAsm, ETV6, TET2, NOTCH2, FLT3-ITD | 7 |
| Wei H./2022 | China | 171 | 100 | 71 | 0 | 38 (14–59) | 39 (0.3–106) | Direct sequencing and NGS panel (69 genes) | CSF3R M, WT1 M, CUX1 M, GATA2 M, NRAS M, FLT3-ITD M, JAK3 M, TET2 M, CREBBP M, cKIT M, NOTCH1 M, KMT2D M, DNMT3A M, EZH2 M, EP300 M, NOTCH2 M, RELN M, SH2B3 M | NA | 8 |
| Wu L.-X./2021 | China | 158 | 55 | 103 | 0 | 41 (17–74) | NA | NGS panel (236 genes) | NRAS U, PCLO U, KMT2A M, CSF3R M | GATA2, WT1, TET2, FLT3-ITD, DNMT3A, BAZ2A, NPM1, AHNAK2 | 8 |
| Xu J./2022 | China | 156 | 82 | 74 | 0 | NA | NA | NGS panel (34 genes) | ASXL1 M | TET2, CBL, TP53, SH2B3, CEBPAsm, DNMT3A, FLT3-ITD, NPM1, JAK2, CSF3R, cKIT, U2AF1, GATA2, PHF6, SRSF2, ETV6, MPL | 7 |
| Xu N./2022 | China | 84 | 46 | 38 | 0 | 54 (18–69) | 10 (1–102) | Bidirectional sequencing on an ABI 3730 sequencer | TP53 M | FLT3-ITD, NPM1, CEBPAdm | 5 |
| Yamato G./2017 | Japan | 369 | 179 | 190 | 0 | 8.4 (0–17.9) | 36 | NGS Targeted deep sequencing | ASXL1 K, ASXL2 K | FLT3-ITD, NRAS, WT1, KMT2A, BCOR, BCORL1, STAG2, CSF3R, SMC3, CEBPA | 8 |
| Yang J./2016 | China | 249 | 145 | 104 | 0 | NA (18–93) | NA | Direct sequencing | SRSF2 M, U2AF1 M | cKIT, IDH1, IDH2, NPM1, DNMT3A, FLT3-ITD, CEBPA, SF3B1 | 8 |
| Yu G./2018 | China | 64 | 39 | 25 | 0 | 27.5 (2–65) | 23.5 (4–85) | NGS panel (67 genes) | cKIT M, ASXL1 M, MET U, MLH1 U, TET2 U, FBXW7 U, TP53 U, DNMT3A U, KMT2A U, PAX5 U | NRAS, APC, RUNX1, NPM1, KRAS, SH2B3, HRAS, SMAD4, DNMT3L | 6 |
| Yuen K.-Y./2023 | China | 493 | 255 | 238 | 0 | 8.7 | 55 | NGS panel (177 genes) | KRAS M, FLT3-ITD M, NRAS M, SETD2 M | FLT3-TKD, WT1, cKIT | 9 |
| Yui S/2017 | Japan | 136 | 91 | 45 | 0 | 45 (15–80) | NA | Direct sequencing | cKIT M | NA | 8 |
| Zare-Abdollahi D./2015 | Iran | 96 | 53 | 43 | 0 | 42 (18–60) | 33 | Direct sequencing | DNMT3A M | NA | 7 |
| Zhang Y.-W./2023 | China | 266 | 114 | 152 | 0 | 52.5 (12–78) | 26.0 (4.0–101.3) | NGS panel (112 genes) | DNMT3A M, TET2 M, FLT3-TKD M, PTPN11 M, FLT3-ITD M | NPM1, IDH, RAS | 7 |
| Zhong WJ/2021 | China | 113 | 66 | 47 | 0 | 56 (18–89) | 15 (1–54) | NGS panel (141 genes) | SRSF2 K, KRAS K, KMT2D K, FAT1 K, RELN K | FLT3-ITD, NPM1, SETBP1, NOTCH11, ASXL1, NRAS, DNMT3A, CUX1, TET2, JAK2, RUNX1, ATM, TP53, cKIT, CREBBP, CARD11, SH2B3, BRAF, DIS3, DDX41, BCORL1, DNMT3B, FGFR3, ARID1A | 8 |
| Zidan M./2014 | Egypt | 216 | 109 | 107 | 0 | 44.16 ± 15.7 | NA | PCR-SSCP | WT1 M, NPM1 M | NA | 9 |
| COMPARISON VARIABLES | FLT3-ITD | cKIT | WT1 | NPM1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K | HR (95%CI) | I2 (%), Ph | p* | K | HR (95%CI) | I2 (%), Ph | p* | K | HR (95%CI) | I2 (%), Ph | p* | K | HR (95%CI) | I2 (%), Ph | p* | |
| TOTAL | 28 | 1.70 (1.45–1.99) | 48%, 0.003 | <0.0001 | 15 | 1.65 (1.13–2.41) | 72%, <0.0001 | 0.002 | 13 | 1.65 (1.14–2.38) | 72%, 0.008 | 0.01 | 16 | 0.67 (0.51–0.88) | 69%, <0.001 | 0.01 |
| REGION | ||||||||||||||||
| EUROPEAN | 10 | 1.67 (1.47–1.89) | 25% | 0.8 | 4 | 0.93 (0.46–1.89) | 60% | 0.05 | 5 | 1.45 (1.10–1.91) | 17% | 0.44 | 7 | 0.59 (0.38–0.93) | 58.5% | 0.23 |
| NON-EUROPEAN | 18 | 1.73 (1.36–2.21) | 56% | 11 | 2.04 (1.46–2.87) | 59% | 8 | 1.89 (1.04–3.44) | 81% | 9 | 0.83 (0.61–1.13) | 61% | ||||
| AGE GROUP | ||||||||||||||||
| PEDIATRIC | 5 | 1.55 (0.92–2.60) | 61% | 0.6 | 3 | 1.58 (0.50–4.98) | 79% | 0.1 | 4 | 1.73 (1.27–2.36) | 0% | 0.97 | 2 | 0.33 (0.06–1.95) | 78% | 0.57 |
| ADULT | 15 | 1.66 (1.26–2.17) | 59% | 5 | 2.74 (1.84–4.08) | 0% | 5 | 1.69 (0.63–4.54) | 87% | 11 | 0.76 (0.60–0.96) | 56% | ||||
| MIXED | 8 | 1.87 (1.68–2.10) | 0% | 7 | 1.4 (0.86–2.28) | 65% | 4 | 1.84 (1.26–2.68) | 24% | 3 | 0.60 (0.28–1.30) | 86% | ||||
| DATA TYPE | ||||||||||||||||
| MULTIVARIATE | 16 | 1.68 (1.40–2.01) | 47% | 0.9 | 11 | 1.85 (1.24–2.76) | 66% | 0.47 | 8 | 1.25 (0.82–1.91) | 75% | 0.01 | 10 | 0.61 (0.42–0.87) | 46% | 0.12 |
| OTHERS * | 12 | 1.71 (1.28–2.30) | 48% | 4 | 1.36 (0.65–2.86) | 64% | 5 | 2.64 (1.79–3.88) | 0% | 6 | 0.9 (0.64–1.25) | 79% | ||||
| COMPARISON VARIABLES | FLT3-ITD | cKIT | ||||||
|---|---|---|---|---|---|---|---|---|
| K | HR (95%CI) | I2 (%), Ph | p* | K | HR (95%CI) | I2 (%), Ph | p* | |
| TOTAL | 20 | 1.62 (1.36–1.92) | 40%, 0.03 | <0.0001 | 11 | 1.42 (0.98–2.07) | 69%, <0.0001 | 0.06 |
| REGION | ||||||||
| EUROPEAN | 8 | 1.62 (1.32–1.98) | 4% | 1 | 2 | 0.85 (0.40–1.78) | 29% | 0.15 |
| NON-EUROPEAN | 12 | 1.61 (1.26–2.05) | 53% | 9 | 1.59 (1.06–2.38) | 71% | ||
| AGE GROUP | ||||||||
| PEDIATRIC | 4 | 1.64 (0.88–3.05) | 71% | 0.6 | 3 | 0.77 (0.46–1.30) | 63% | 0 |
| ADULT | 10 | 1.54 (1.16–2.04) | 39% | 3 | 2.65 (1.69–4.16) | 0% | ||
| MIXED | 6 | 1.80 (1.54–2.11) | 0% | 5 | 1.81 (1.22–2.69) | 19% | ||
| DATA TYPE | ||||||||
| MULTIVARIATE | 13 | 1.73 (1.42–2.10) | 35% | 0.3 | 7 | 2.02 (1.41–2.90) | 29% | 0.03 |
| OTHERS * | 7 | 1.43 (1.05–1.96) | 49% | 4 | 0.94 (0.52–1.69) | 68% | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elfatih, A.; Ahmed, N.; Srour, L.; Mohammed, I.; Villiers, W.; Al-Barazenji, T.; Mbarek, H.; El Akiki, S.; Jithesh, P.V.; Muneer, M.; et al. Impact of Somatic Gene Mutations on Prognosis Prediction in De Novo AML: Unraveling Insights from a Systematic Review and Meta-Analysis. Cancers 2025, 17, 3189. https://doi.org/10.3390/cancers17193189
Elfatih A, Ahmed N, Srour L, Mohammed I, Villiers W, Al-Barazenji T, Mbarek H, El Akiki S, Jithesh PV, Muneer M, et al. Impact of Somatic Gene Mutations on Prognosis Prediction in De Novo AML: Unraveling Insights from a Systematic Review and Meta-Analysis. Cancers. 2025; 17(19):3189. https://doi.org/10.3390/cancers17193189
Chicago/Turabian StyleElfatih, Amal, Nisar Ahmed, Luma Srour, Idris Mohammed, William Villiers, Tara Al-Barazenji, Hamdi Mbarek, Susanna El Akiki, Puthen Veettil Jithesh, Mohammed Muneer, and et al. 2025. "Impact of Somatic Gene Mutations on Prognosis Prediction in De Novo AML: Unraveling Insights from a Systematic Review and Meta-Analysis" Cancers 17, no. 19: 3189. https://doi.org/10.3390/cancers17193189
APA StyleElfatih, A., Ahmed, N., Srour, L., Mohammed, I., Villiers, W., Al-Barazenji, T., Mbarek, H., El Akiki, S., Jithesh, P. V., Muneer, M., Fareed, S., & Mifsud, B. (2025). Impact of Somatic Gene Mutations on Prognosis Prediction in De Novo AML: Unraveling Insights from a Systematic Review and Meta-Analysis. Cancers, 17(19), 3189. https://doi.org/10.3390/cancers17193189






