Impact of Epigenome-Wide Methylation and Breast Cancer Recurrence in Women Tested Negative for BRCA Genes: The Breast Methylation Risk (BREMERI) Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
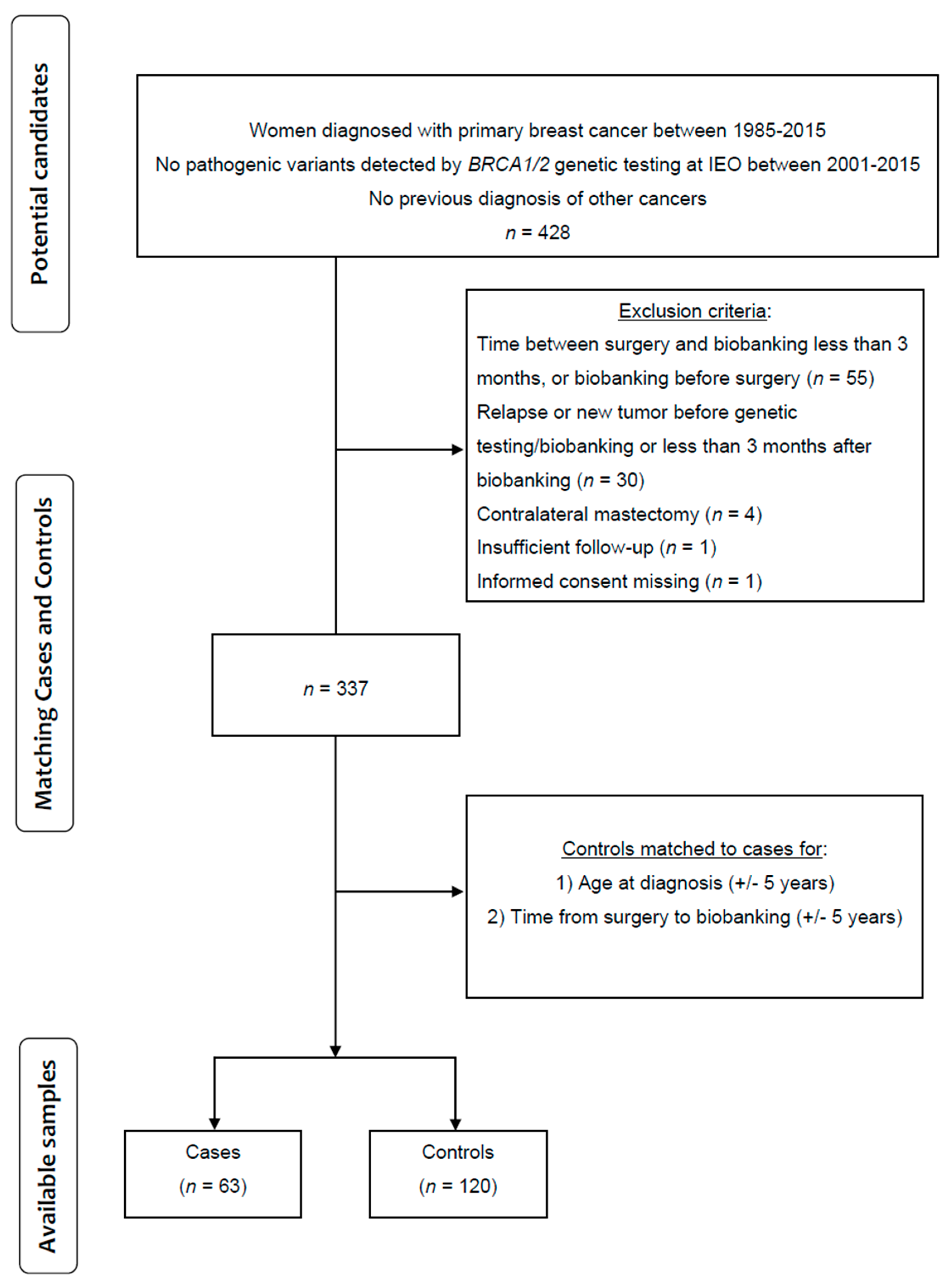
2.1. Study Design and Patient Characteristics
2.2. DNA Methylation Assessment
2.2.1. DNA Methylation Data Pre-Processing
2.2.2. Differentially Methylated Analysis
2.2.3. Deriving Methylation-Based Surrogates
3. Results
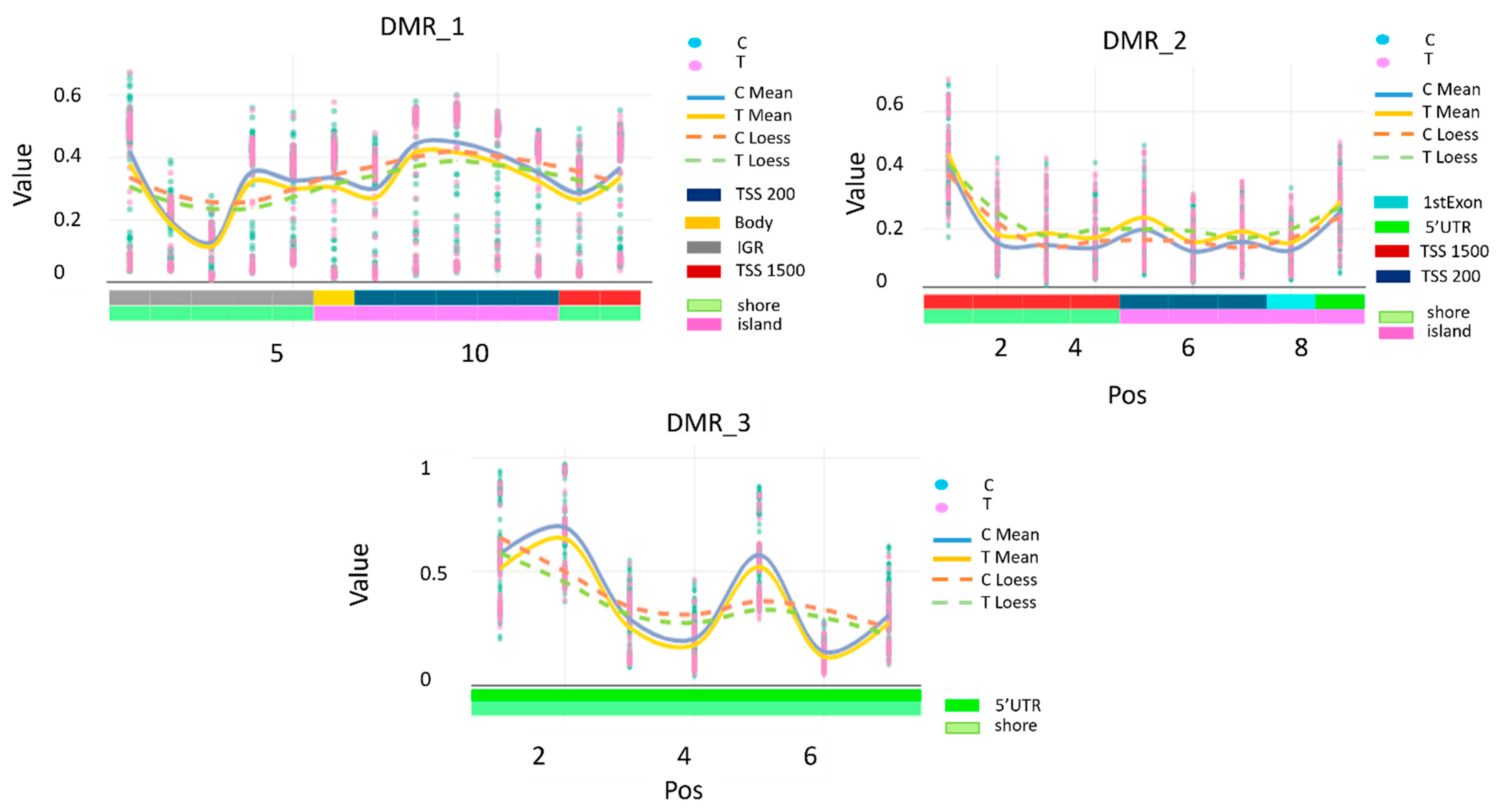
Epigenome-Wide Differentially Methylated Probes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lüönd, F.; Tiede, S.; Christofori, G. Breast Cancer as an Example of Tumour Heterogeneity and Tumour Cell Plasticity during Malignant Progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Yoshida, R. Hereditary Breast and Ovarian Cancer (HBOC): Review of Its Molecular Characteristics, Screening, Treatment, and Prognosis. Breast Cancer 2021, 28, 1167–1180. [Google Scholar] [CrossRef]
- Joo, J.E.; Dowty, J.G.; Milne, R.L.; Wong, E.M.; Dugué, P.-A.; English, D.; Hopper, J.L.; Goldgar, D.E.; Giles, G.G.; Southey, M.C.; et al. Heritable DNA Methylation Marks Associated with Susceptibility to Breast Cancer. Nat. Commun. 2018, 9, 867. [Google Scholar] [CrossRef]
- Xu, Z.; Sandler, D.P.; Taylor, J.A. Blood DNA Methylation and Breast Cancer: A Prospective Case-Cohort Analysis in the Sister Study. J. Natl. Cancer Inst. 2020, 112, 87–94. [Google Scholar] [CrossRef]
- Severi, G.; Southey, M.C.; English, D.R.; Jung, C.; Lonie, A.; McLean, C.; Tsimiklis, H.; Hopper, J.L.; Giles, G.G.; Baglietto, L. Epigenome-Wide Methylation in DNA from Peripheral Blood as a Marker of Risk for Breast Cancer. Breast Cancer Res. Treat. 2014, 148, 665–673. [Google Scholar] [CrossRef]
- van Veldhoven, K.; Polidoro, S.; Baglietto, L.; Severi, G.; Sacerdote, C.; Panico, S.; Mattiello, A.; Palli, D.; Masala, G.; Krogh, V.; et al. Epigenome-Wide Association Study Reveals Decreased Average Methylation Levels Years before Breast Cancer Diagnosis. Clin. Epigenetics 2015, 7, 67. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Eccles, M.R. Epigenetic Drivers of Tumourigenesis and Cancer Metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef]
- Fackler, M.J.; Cho, S.; Cope, L.; Gabrielson, E.; Visvanathan, K.; Wilsbach, K.; Meir-Levi, D.; Lynch, C.F.; Marks, J.; Geradts, J.; et al. DNA Methylation Markers Predict Recurrence-Free Interval in Triple-Negative Breast Cancer. NPJ Breast Cancer 2020, 6, 3. [Google Scholar] [CrossRef]
- Garg, P.; Singhal, G.; Horne, D.; Salgia, R.; Singhal, S.S. Metabolic Reprogramming in Breast Cancer: Pathways Driving Progression, Drug Resistance, and Emerging Therapeutics. Biochim. Biophys. Acta Rev. Cancer. 2025, 1880, 189396. [Google Scholar] [CrossRef]
- Wishart, D.S. Is Cancer a Genetic Disease or a Metabolic Disease? eBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef]
- Bui, V.N.V.; Daugaard, T.F.; Sorensen, B.S.; Nielsen, A.L. Expression of the Non-Coding RNA Nc886 Facilitates the Development of Tyrosine Kinase Inhibitor Resistance in EGFR-Mutated Non-Small-Cell Lung Cancer Cells. Biochem. Biophys. Res. Commun. 2024, 731, 150395. [Google Scholar] [CrossRef]
- Vietri, M.; D’elia, G.; Benincasa, G.; Ferraro, G.; Caliendo, G.; Nicoletti, G.; Napoli, C. DNA Methylation and Breast Cancer: A Way Forward (Review). Int. J. Oncol. 2021, 59, 98. [Google Scholar] [CrossRef]
- Aleyasin, S.A.; Moradi, A.; Abolhasani, N.; Abdollahi, M. Investigating FGFR2 Gene as a Blood-Based Epigenetic Biomarker in Gastric Cancer. Mol. Biol. Rep. 2024, 51, 253. [Google Scholar] [CrossRef]
- Wang, T.; Li, P.; Qi, Q.; Zhang, S.; Xie, Y.; Wang, J.; Liu, S.; Ma, S.; Li, S.; Gong, T.; et al. A Multiplex Blood-Based Assay Targeting DNA Methylation in PBMCs Enables Early Detection of Breast Cancer. Nat. Commun. 2023, 14, 4724. [Google Scholar] [CrossRef]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical Evaluation of the Illumina MethylationEPIC BeadChip Microarray for Whole-Genome DNA Methylation Profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.J.; Butcher, L.M.; Feber, A.; Teschendorff, A.E.; Chakravarthy, A.R.; Wojdacz, T.K.; Beck, S. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics 2014, 30, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA Methylation Arrays as Surrogate Measures of Cell Mixture Distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef]
- Hillary, R.F.; Marioni, R.E. MethylDetectR: A Software for Methylation-Based Health Profiling. Wellcome Open Res. 2021, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Raitoharju, E.; Rajić, S.; Marttila, S. Non-Coding 886 (Nc886/VtRNA2-1), the Epigenetic Odd Duck—Implications for Future Studies. Epigenetics 2024, 19, 2332819. [Google Scholar] [CrossRef]
- Carpenter, B.L.; Zhou, W.; Madaj, Z.; DeWitt, A.K.; Ross, J.P.; Grønbæk, K.; Liang, G.; Clark, S.J.; Molloy, P.L.; Jones, P.A. Mother–Child Transmission of Epigenetic Information by Tunable Polymorphic Imprinting. Proc. Natl. Acad. Sci. USA 2018, 115, E11970–E11977. [Google Scholar] [CrossRef]
- Marttila, S.; Tamminen, H.; Rajić, S.; Mishra, P.P.; Lehtimäki, T.; Raitakari, O.; Kähönen, M.; Kananen, L.; Jylhävä, J.; Hägg, S.; et al. Methylation Status of VTRNA2-1/Nc886 Is Stable across Populations, Monozygotic Twin Pairs and in Majority of Tissues. Epigenomics 2022, 14, 1105–1124. [Google Scholar] [CrossRef]
- Marttila, S.; Viiri, L.E.; Mishra, P.P.; Kühnel, B.; Matias-Garcia, P.R.; Lyytikäinen, L.-P.; Ceder, T.; Mononen, N.; Rathmann, W.; Winkelmann, J.; et al. Methylation Status of Nc886 Epiallele Reflects Periconceptional Conditions and Is Associated with Glucose Metabolism through Nc886 RNAs. Clin. Epigenetics 2021, 13, 143. [Google Scholar] [CrossRef]
- Raitoharju, E.; Marttila, S. Commentary on “Epigenome-Wide Analysis across the Development Span of Pediatric Acute Lymphoblastic Leukemia: Backtracking to Birth”. Mol. Cancer 2025, 24, 8. [Google Scholar] [CrossRef]
- Pan, B.; Yu, J.; Liu, X. Upregulation of MiR-886 Indicates Poor Prognosis and Promotes Tumour Progression of Prostate Cancer. Andrologia 2022, 54, e14296. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Lee, Y.S. Nc886, an RNA Polymerase III-Transcribed Noncoding RNA Whose Expression Is Dynamic and Regulated by Intriguing Mechanisms. Int. J. Mol. Sci. 2023, 24, 8533. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xiao, L.; Wen, S.J.; Yu, T.; Sharma, S.; Chung, H.K.; Warner, B.; Mallard, C.G.; Rao, J.N.; Gorospe, M.; et al. Small Noncoding Vault RNA2 -1 Disrupts Gut Epithelial Barrier Function via Interaction with HuR. EMBO Rep. 2023, 24, e54925. [Google Scholar] [CrossRef] [PubMed]
- Shrout, M.R.; Madison, A.A.; Renna, M.E.; Alfano, C.M.; Povoski, S.P.; Lipari, A.M.; Agnese, D.M.; Carson, W.E.; Malarkey, W.B.; Bailey, M.T.; et al. The Gut Connection: Intestinal Permeability as a Pathway from Breast Cancer Survivors’ Relationship Satisfaction to Inflammation across Treatment. Brain Behav. Immun. 2022, 100, 145–154. [Google Scholar] [CrossRef]
- Jang, J.J.; Kang, D.; Lee, Y.-S.; Lee, Y.S. The Versatile Roles of Nc886, a Fascinating and Peculiar Regulatory Non-Coding RNA, in Cancer. Int. J. Mol. Sci. 2024, 25, 10825. [Google Scholar] [CrossRef]
- Gallo, S.; Kong, E.; Ferro, I.; Polacek, N. Small but Powerful: The Human Vault RNAs as Multifaceted Modulators of Pro-Survival Characteristics and Tumorigenesis. Cancers 2022, 14, 2787. [Google Scholar] [CrossRef]
- Aghajani Mir, M. Vault RNAs (VtRNAs): Rediscovered Non-Coding RNAs with Diverse Physiological and Pathological Activities. Genes. Dis. 2024, 11, 772–787. [Google Scholar] [CrossRef]
- Acevedo, N.; Scala, G.; Merid, S.K.; Frumento, P.; Bruhn, S.; Andersson, A.; Ogris, C.; Bottai, M.; Pershagen, G.; Koppelman, G.H.; et al. DNA Methylation Levels in Mononuclear Leukocytes from the Mother and Her Child Are Associated with IgE Sensitization to Allergens in Early Life. Int. J. Mol. Sci. 2021, 22, 801. [Google Scholar] [CrossRef]
- Char, R.; Pierre, P. The RUFYs, a Family of Effector Proteins Involved in Intracellular Trafficking and Cytoskeleton Dynamics. Front. Cell Dev. Biol. 2020, 8, 779. [Google Scholar] [CrossRef]
- Kitagishi, Y.; Matsuda, S. RUFY, Rab and Rap Family Proteins Involved in a Regulation of Cell Polarity and Membrane Trafficking. Int. J. Mol. Sci. 2013, 14, 6487–6498. [Google Scholar] [CrossRef]
- Li, J.; Hu, K.; Huang, J.; Zhou, L.; Yan, Y.; Xu, Z. A Pancancer Analysis of the Expression Landscape and Clinical Relevance of Fibroblast Growth Factor Receptor 2 in Human Cancers. Front. Oncol. 2021, 11, 644854. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-W.; Lu, R.J.-H.; Jayasinghe, R.G.; Foltz, S.M.; Porta-Pardo, E.; Geffen, Y.; Wendl, M.C.; Lazcano, R.; Kolodziejczak, I.; Song, Y.; et al. Integrative Multi-Omic Cancer Profiling Reveals DNA Methylation Patterns Associated with Therapeutic Vulnerability and Cell-of-Origin. Cancer Cell 2023, 41, 1567–1585.e7. [Google Scholar] [CrossRef] [PubMed]
- Kaszak, I.; Witkowska-Piłaszewicz, O.; Niewiadomska, Z.; Dworecka-Kaszak, B.; Ngosa Toka, F.; Jurka, P. Role of Cadherins in Cancer—A Review. Int. J. Mol. Sci. 2020, 21, 7624. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.L.; Kim, A.C.; Hens, J.R. The Role and Function of Cadherins in the Mammary Gland. Breast Cancer Res. 2012, 14, 203. [Google Scholar] [CrossRef]
- Ku, S.-C.; Liu, H.-L.; Su, C.-Y.; Yeh, I.-J.; Yen, M.-C.; Anuraga, G.; Ta, H.D.K.; Chiao, C.-C.; Xuan, D.T.M.; Prayugo, F.B.; et al. Comprehensive Analysis of Prognostic Significance of Cadherin (CDH) Gene Family in Breast Cancer. Aging 2022, 14, 8498–8567. [Google Scholar] [CrossRef]
- Xu, M.; Liu, C.; Pu, L.; Lai, J.; Li, J.; Ning, Q.; Liu, X.; Deng, S. Systemic Analysis of the Expression Levels and Prognosis of Breast Cancer-Related Cadherins. Exp. Biol. Med. 2021, 246, 1706–1720. [Google Scholar] [CrossRef]
- Berndt, B.; Haverkampf, S.; Reith, G.; Keil, S.; Niggemann, B.; Zänker, K.S.; Dittmar, T. Fusion of CCL21 Non-Migratory Active Breast Epithelial and Breast Cancer Cells Give Rise to CCL21 Migratory Active Tumor Hybrid Cell Lines. PLoS ONE 2013, 8, e63711. [Google Scholar] [CrossRef]
- Rizeq, B.; Malki, M.I. The Role of CCL21/CCR7 Chemokine Axis in Breast Cancer Progression. Cancers 2020, 12, 1036. [Google Scholar] [CrossRef] [PubMed]
- Papa, V.; Pezzino, V.; Costantino, A.; Belfiore, A.; Giuffrida, D.; Frittitta, L.; Vannelli, G.B.; Brand, R.; Goldfine, I.D.; Vigneri, R. Elevated Insulin Receptor Content in Human Breast Cancer. J. Clin. Investig. 1990, 86, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.Q.; Jung, S.; Nguyen, H.A.; Lee, B.; Vu, S.H.; Myagmarjav, D.; Eum, H.H.; Lee, H.-O.; Jo, T.; Choi, Y.; et al. The Regulation of Insulin Receptor/Insulin-like Growth Factor 1 Receptor Ratio, an Important Factor for Breast Cancer Prognosis, by TRIP-Br1. J. Hematol. Oncol. 2022, 15, 82. [Google Scholar] [CrossRef] [PubMed]
| (A) | |||||
| Cases | Controls | Total | p-Values | ||
| Total | 63 | 120 | 183 | ||
| Age at the test | Median (Q1, Q3) | 42 (37, 49) | 45 (38, 50) | 183 | 0.43 |
| BMI | Median (Q1, Q3) | 22.1 (20.2, 24) | 22.1 (20.2, 24) | 182 | 0.69 |
| BMI at 18 years old | Median (Q1, Q3) | 20 (18, 21) | 19 (18, 20) | 177 | 0.04 |
| Time from surgery to blood draw (months) | Median (Q1, Q3) | 12.5 (8.2, 29.2) | 21.5 (9.4, 52.7) | 183 | 0.085 |
| Oral contraceptive | No | 24 (39.34) | 37 (60.66) | 61 | 0.32 |
| Yes | 36 (31.3) | 79 (68.7) | 115 | ||
| Missing | 7 | ||||
| Pregnancies | No | 15 (29.41) | 36 (70.59) | 51 | 0.39 |
| Yes | 48 (36.36) | 84 (63.64) | 132 | ||
| n. of pregnancies | No | 15 (29.41) | 36 (70.59) | 51 | 0.64 |
| One | 17 (34.69) | 32 (65.31) | 49 | ||
| More than one | 31 (37.35) | 52 (62.65) | 83 | ||
| menopausal status | Pre | 48 (33.8) | 94 (66.2) | 142 | 0.65 |
| Peri | 0 (0) | 1 (100) | 1 | ||
| Post | 14 (38.89) | 22 (61.11) | 36 | ||
| Missing | 40 | ||||
| Smoking | Former | 15 (31.91) | 32 (68.09) | 47 | 0.68 |
| No | 43 (36.75) | 74 (63.25) | 117 | ||
| Yes | 5 (27.78) | 13 (72.22) | 18 | ||
| Missing | 1 | ||||
| Family history | No | 0 (0) | 2 (100) | 2 | 0.26 |
| Breast only | 59 (33.91) | 115 (66.09) | 174 | ||
| Breast and ovary | 4 (57.14) | 3 (42.86) | 7 | ||
| Family history—First degree breast | 0 | 21 (43.75) | 27 (56.25) | 48 | 0.19 |
| 1 | 34 (31.19) | 75 (68.81) | 109 | ||
| 2 | 7 (28) | 18 (72) | 25 | ||
| 3 | 1 (100) | 0 (0) | 1 | ||
| Family history—First degree ovary | 0 | 61 (34.08) | 118 (65.92) | 179 | 0.61 |
| 1 | 2 (50) | 2 (50) | 4 | ||
| Family history—Second degree breast | 0 | 20 (28.99) | 49 (71.01) | 69 | 0.39 |
| 1 | 32 (35.56) | 58 (64.44) | 90 | ||
| 2 | 9 (42.86) | 12 (57.14) | 21 | ||
| 3 | 2 (66.67) | 1 (33.33) | 3 | ||
| Family history—Second degree ovary | 0 | 61 (34.08) | 118 (65.92) | 179 | 0.61 |
| 1 | 2 (50) | 2 (50) | 4 | ||
| (B) | |||||
| Cases | Controls | Total | p-Values | ||
| Total | 63 | 120 | 183 | ||
| Histotype | In situ | 9 (40.91) | 13 (59.09) | 22 | 0.85 |
| Ductal | 48 (34.29) | 92 (65.71) | 140 | ||
| Lobular | 4 (30.77) | 9 (69.23) | 13 | ||
| Other | 2 (25.00) | 6 (75.00) | 8 | ||
| Molecular subtype | Luminal A/B | 40 (35.71) | 72 (64.29) | 112 | 0.69 |
| Luminal B Her2+ | 7 (31.82) | 15 (68.18) | 22 | ||
| Her2+ | 5 (31.25) | 11 (68.75) | 16 | ||
| TN | 2 (18.18) | 9 (81.82) | 11 | ||
| Missing | 9 | 13 | 22 | ||
| pN | Negative | 25 (25.00) | 75 (75.00) | 100 | 0.004 * |
| Positive | 32 (46.38) | 37 (53.62) | 69 | ||
| Px | 6 (42.86) | 8 (57.14) | 14 | ||
| pT | Is | 8 (38.10) | 13 (61.90) | 21 | 0.045 |
| 1 | 32 (27.59) | 84 (72.41) | 116 | ||
| 2 | 14 (43.75) | 18 (56.25) | 32 | ||
| 3 | 8 (61.54) | 5 (38.46) | 13 | ||
| Missing | 1 | 1 | |||
| DMRindex | Chromosome | Cpg Probe ID | Position | Strand | Type | Gene | Feature | Cgi |
|---|---|---|---|---|---|---|---|---|
| DMR_1 | 5 | cg07158503 | 135415693 | R | II | IGR | shore | |
| cg04515200 | 135415762 | F | II | IGR | shore | |||
| cg13581155 | 135415781 | F | II | IGR | shore | |||
| cg11608150 | 135415948 | R | I | IGR | shore | |||
| cg06478886 | 135416029 | R | II | IGR | shore | |||
| cg04481923 | 135416205 | R | II | VTRNA2–1 | Body | island | ||
| cg18678645 | 135416331 | R | II | VTRNA2–1 | TSS200 | island | ||
| cg06536614 | 135416381 | F | I | VTRNA2–1 | TSS200 | island | ||
| cg25340688 | 135416398 | F | I | VTRNA2–1 | TSS200 | island | ||
| cg26896946 | 135416405 | F | I | VTRNA2–1 | TSS200 | island | ||
| cg00124993 | 135416412 | F | I | VTRNA2–1 | TSS200 | island | ||
| cg08745965 | 135416529 | F | II | VTRNA2–1 | TSS1500 | shore | ||
| DMR_2 | 5 | cg19626725 | 178986131 | F | II | RUFY1 | TSS1500 | shore |
| cg00080972 | 178986291 | F | II | RUFY1 | TSS1500 | shore | ||
| cg21226059 | 178986404 | F | II | RUFY1 | TSS1500 | shore | ||
| cg14820908 | 178986412 | F | II | RUFY1 | TSS1500 | shore | ||
| cg02136620 | 178986620 | F | II | RUFY1 | TSS200 | island | ||
| cg09060608 | 178986726 | R | II | RUFY1 | TSS200 | island | ||
| cg05457628 | 178986728 | R | I | RUFY1 | TSS200 | island | ||
| cg22764044 | 178986830 | F | II | RUFY1 | 1stExon | island | ||
| cg26516362 | 178986906 | F | I | RUFY1 | 5′UTR | island | ||
| DMR_3 | 10 | cg06791446 | 123355268 | F | II | FGFR2 | 5′UTR | shore |
| cg25052156 | 123355454 | F | II | FGFR2 | 5′UTR | shore | ||
| cg22633036 | 123355576 | R | II | FGFR2 | 5′UTR | shore | ||
| cg11430259 | 123355748 | R | I | FGFR2 | 5′UTR | shore | ||
| cg02210151 | 123356041 | R | II | FGFR2 | 5′UTR | shore | ||
| cg17681491 | 123356205 | R | II | FGFR2 | 5′UTR | shore | ||
| cg18566515 | 123356236 | R | I | FGFR2 | 5′UTR | shore |
| Mechanism | Genes Involved | Biological Impact |
|---|---|---|
| Cell Migration | RUFY1, (CCL21), (CDH4) | Immune cell recruitment, cancer invasion |
| Growth Signalling | FGFR2, (IR) | Proliferation, metabolic dysregulation |
| Adhesion/Junctions | vtRNA2–1, (CDH4) | EMT, barrier integrity |
| Trafficking | RUFY1 | Receptor recycling, signalling modulation |
| Glucose regulation | RUFY1, vtRNA2–1, (IR) | Glucose regulation—Warburg effect |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polidoro, S.; Johansson, H.; Cugliari, G.; Guerrieri-Gonzaga, A.; Aristarco, V.; Macis, D.; Calvello, M.; Marabelli, M.; Feroce, I.; Serrano, D.; et al. Impact of Epigenome-Wide Methylation and Breast Cancer Recurrence in Women Tested Negative for BRCA Genes: The Breast Methylation Risk (BREMERI) Study. Cancers 2025, 17, 3132. https://doi.org/10.3390/cancers17193132
Polidoro S, Johansson H, Cugliari G, Guerrieri-Gonzaga A, Aristarco V, Macis D, Calvello M, Marabelli M, Feroce I, Serrano D, et al. Impact of Epigenome-Wide Methylation and Breast Cancer Recurrence in Women Tested Negative for BRCA Genes: The Breast Methylation Risk (BREMERI) Study. Cancers. 2025; 17(19):3132. https://doi.org/10.3390/cancers17193132
Chicago/Turabian StylePolidoro, Silvia, Harriet Johansson, Giovanni Cugliari, Aliana Guerrieri-Gonzaga, Valentina Aristarco, Debora Macis, Mariarosaria Calvello, Monica Marabelli, Irene Feroce, Davide Serrano, and et al. 2025. "Impact of Epigenome-Wide Methylation and Breast Cancer Recurrence in Women Tested Negative for BRCA Genes: The Breast Methylation Risk (BREMERI) Study" Cancers 17, no. 19: 3132. https://doi.org/10.3390/cancers17193132
APA StylePolidoro, S., Johansson, H., Cugliari, G., Guerrieri-Gonzaga, A., Aristarco, V., Macis, D., Calvello, M., Marabelli, M., Feroce, I., Serrano, D., Cagnacci, S., Zanzottera, C., Fava, F., Bellerba, F., Bonanni, B., & Gandini, S. (2025). Impact of Epigenome-Wide Methylation and Breast Cancer Recurrence in Women Tested Negative for BRCA Genes: The Breast Methylation Risk (BREMERI) Study. Cancers, 17(19), 3132. https://doi.org/10.3390/cancers17193132







