Highly Sensitive Detection of miR-200c in Metastatic Lymph Nodes Using Scanning Single-Molecule Counting
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Data Source
2.2. Clinical Samples
2.3. Specimen Collection
2.4. MicroRNA Preparation and Measurement
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics
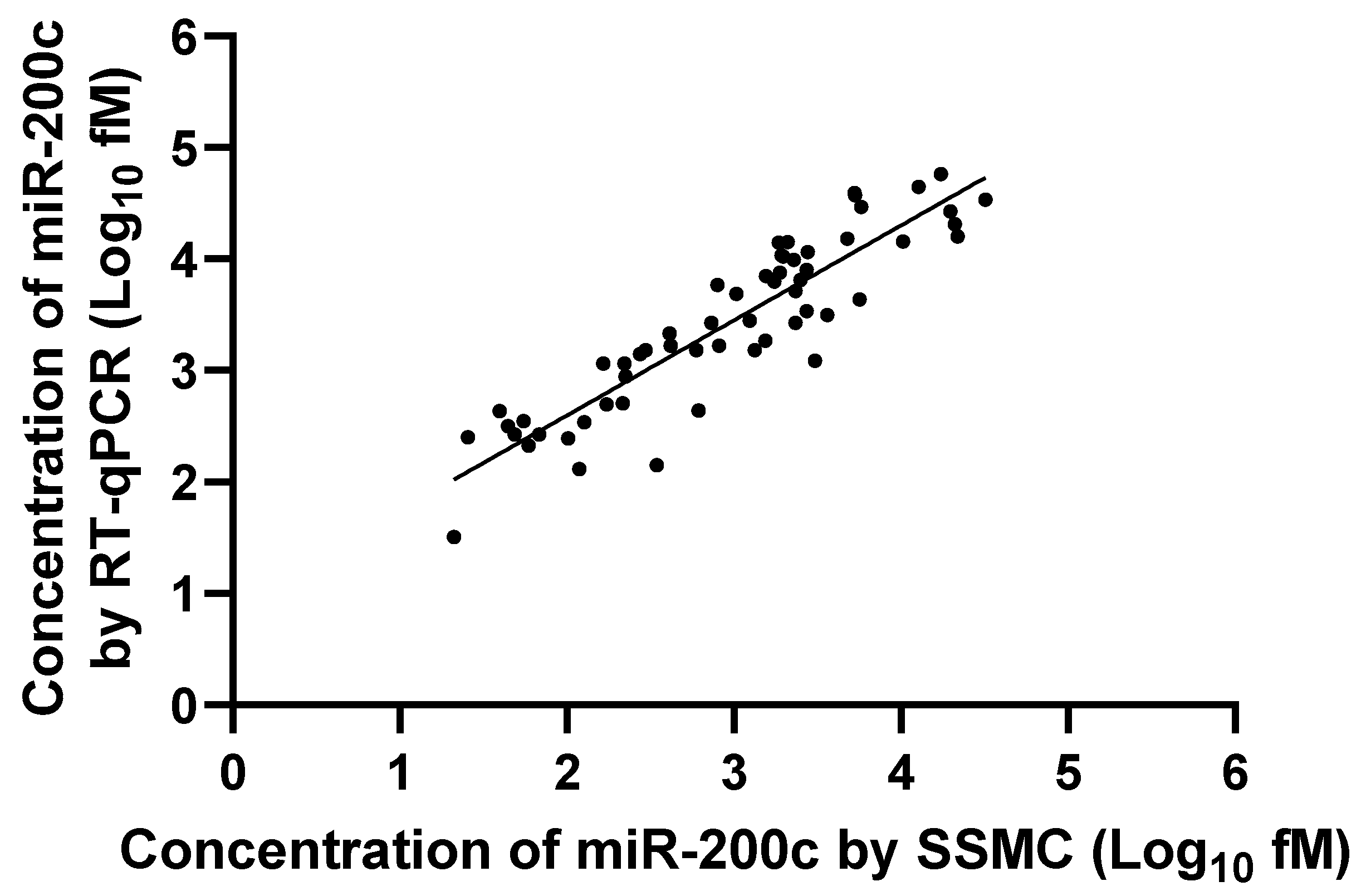
3.2. Correlation Between RT-qPCR and SSMC Results
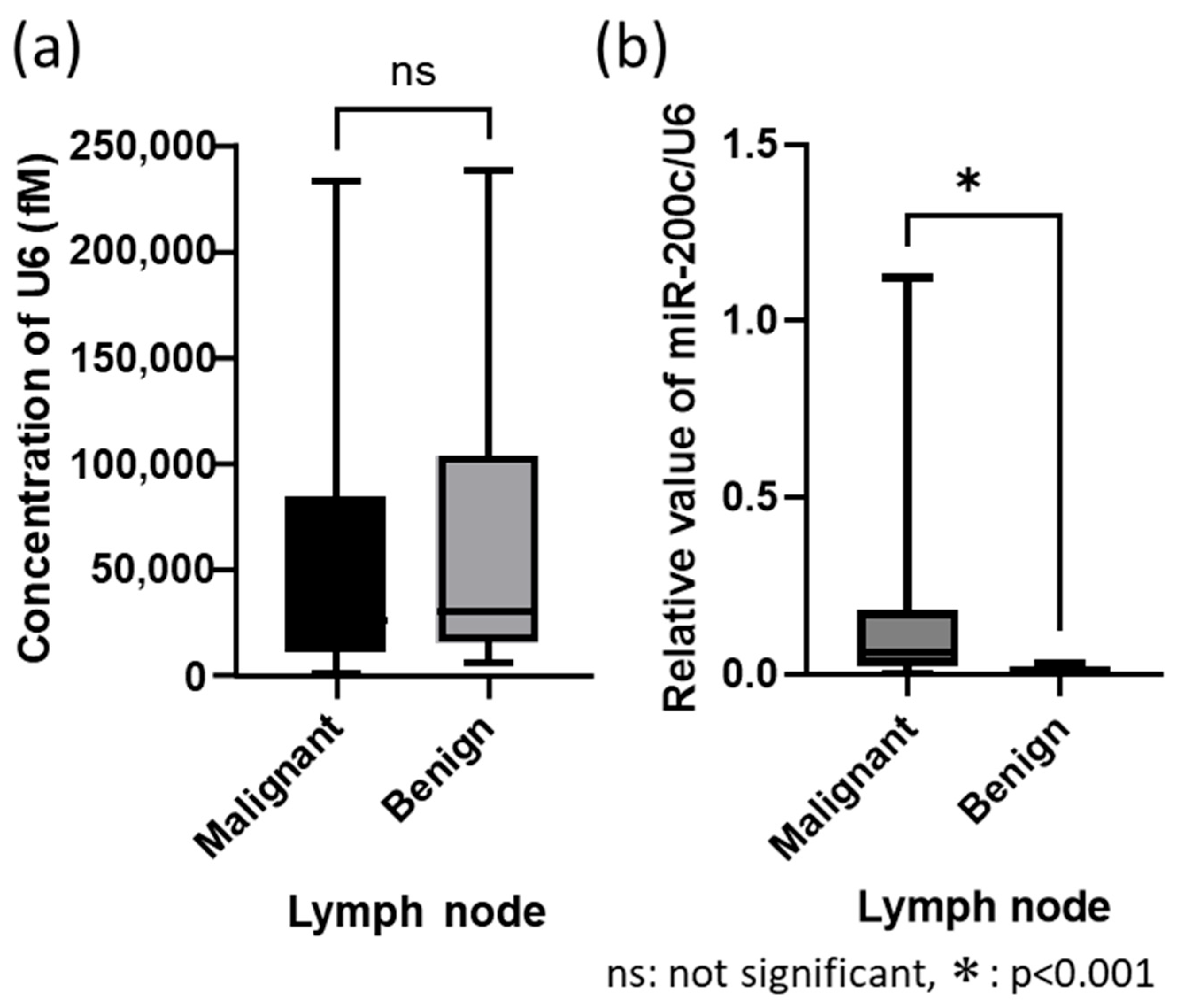
3.3. Validation Test Between with/Without Housekeeping Gene Control
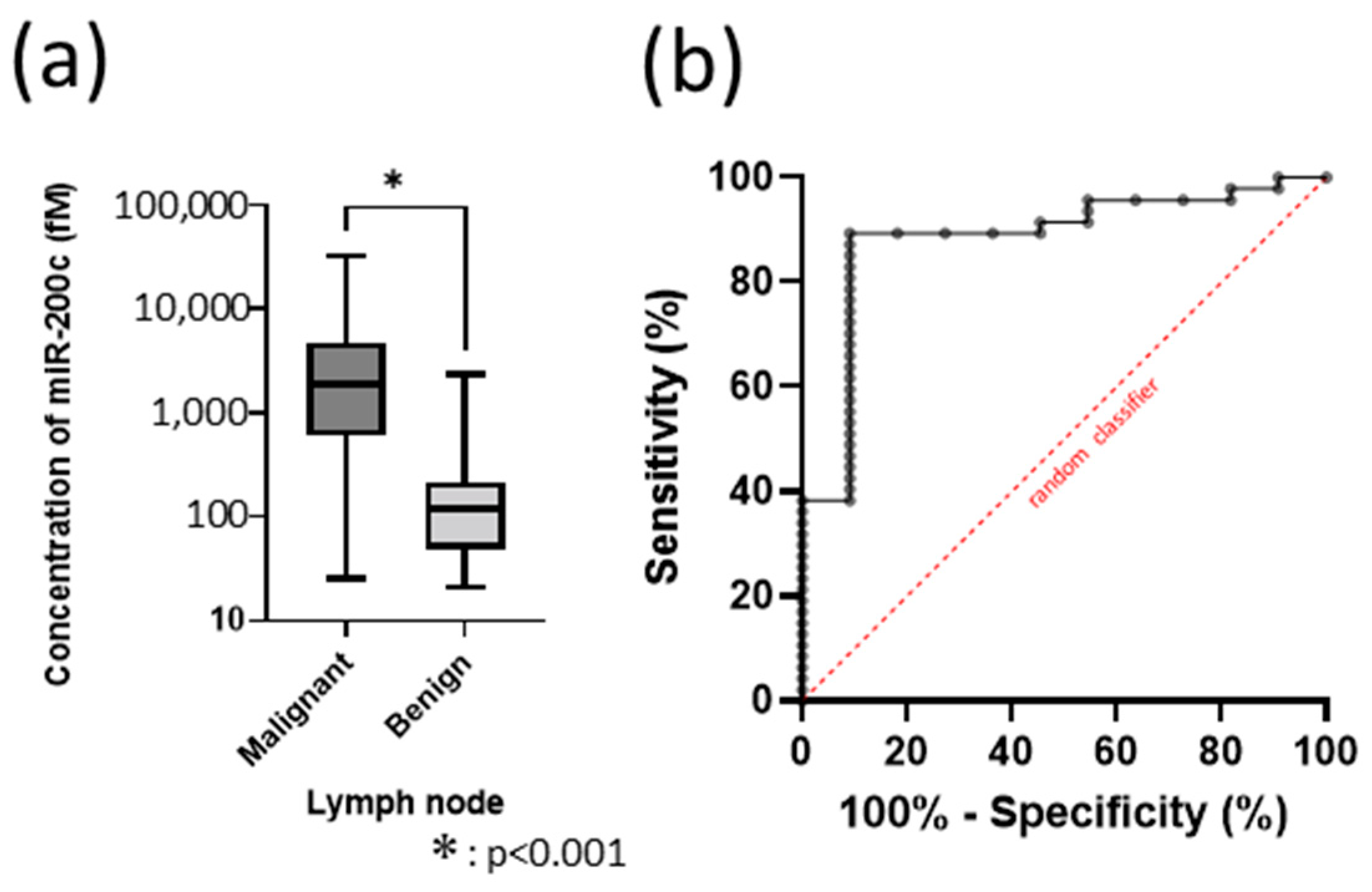
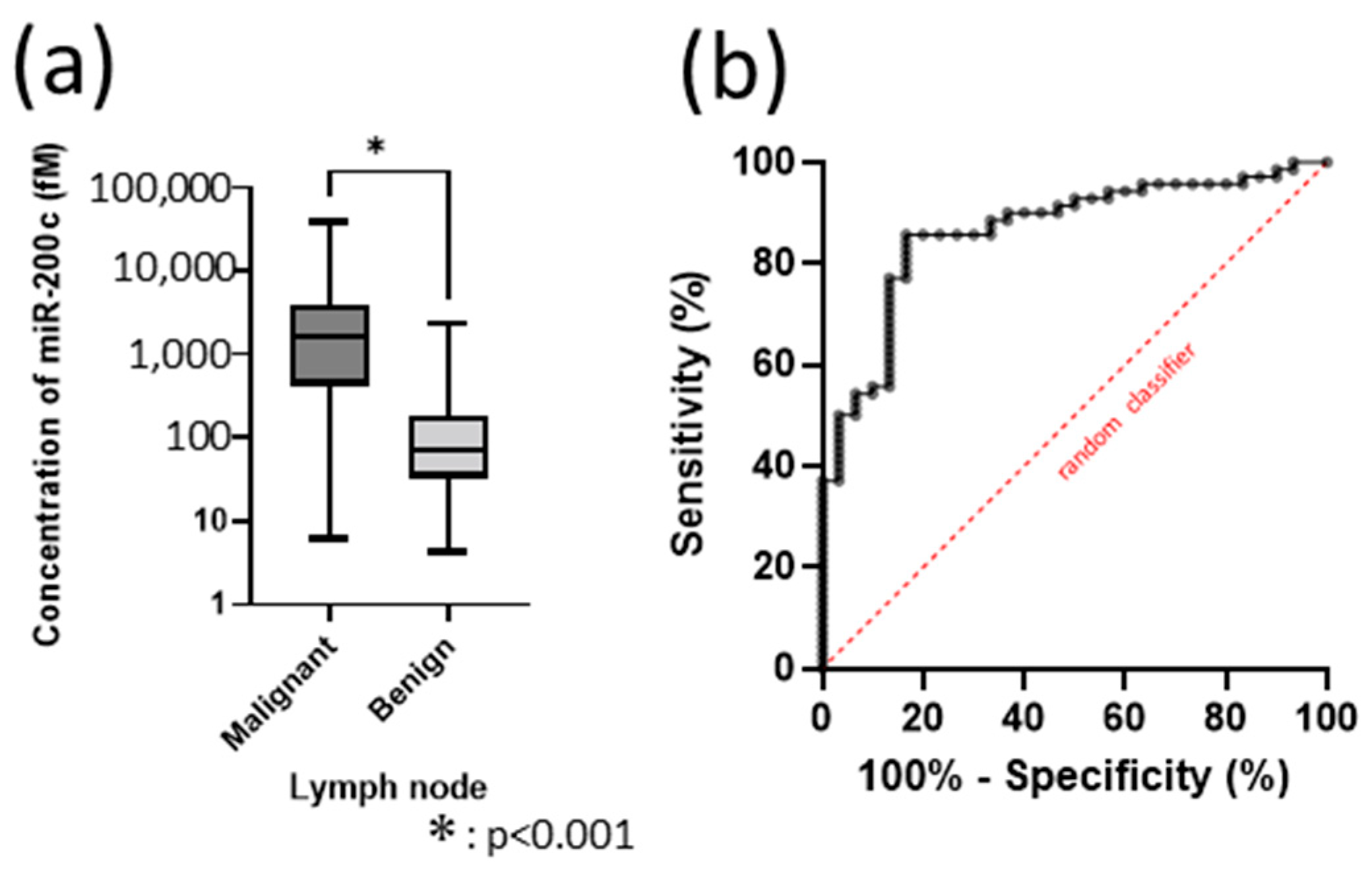
3.4. Deciding the Cut-Off Value as a Biomarker of miR-200c in 100 Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EBUS-TBNA | Endobronchial ultrasound-guided transbronchial needle aspiration |
| NPV | Negative predictive value |
| SSMC | Scanning single-molecule counting |
| RT-qPCR | quantitative real-time polymerase chain reaction |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Topalian, S.L.; Taube, J.M.; Pardoll, D.M. Neoadjuvant checkpoint blockade for cancer immunotherapy. Science 2020, 367, eaax0182. [Google Scholar] [CrossRef]
- John, A.O.; Ramnath, N. Neoadjuvant versus adjuvant systemic therapy for early-stage non-small cell lung cancer: The changing landscape due to immunotherapy. Oncologist 2023, 28, 752–764. [Google Scholar] [CrossRef]
- Rami-Porta, R.; Nishimura, K.K.; Giroux, D.J.; Detterbeck, F.; Cardillo, G.; Edwards, J.G.; Fong, K.M.; Giuliani, M.; Huang, J.; Kernstine, K.H.; et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for revision of the TNM stage groups in the forthcoming (ninth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2024, 19, 1007–1027. [Google Scholar] [CrossRef]
- Craig, C.; Johnston, J.; Goodley, P.; Bishop, P.; Al-Najjar, H.; Brown, L.; Gallagher, J.; Sundar, R.; Upperton, S.; Callister, M.; et al. What is the accuracy of clinical staging for stage III-single-station N2 NSCLC? A multi-centre UK study. JTO Clin. Res. Rep. 2024, 5, 100694. [Google Scholar] [CrossRef]
- Silvestri, G.A.; Gonzalez, A.V.; Jantz, M.A.; Margolis, M.L.; Gould, M.K.; Tanoue, L.T.; Harris, L.J.; Detterbeck, F.C. Methods for staging non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143 (Suppl. S5), e211S–e250S. [Google Scholar] [CrossRef]
- Yasufuku, K.; Chiyo, M.; Koh, E.; Moriya, Y.; Iyoda, A.; Sekine, Y.; Shibuya, K.; Iizasa, T.; Fujisawa, T. Endobronchial ultrasound guided transbronchial needle aspiration for staging of lung cancer. Lung Cancer 2005, 50, 347–354. [Google Scholar] [CrossRef]
- Yasufuku, K.; Nakajima, T.; Waddell, T.; Keshavjee, S.; Yoshino, I. Endobronchial ultrasound-guided transbronchial needle aspiration for differentiating N0 versus N1 lung cancer. Ann. Thorac. Surg. 2013, 96, 1756–1760. [Google Scholar] [CrossRef]
- Cerfolio, R.J.; Bryant, A.S.; Eloubeidi, M.A.; Frederick, P.A.; Minnich, D.J.; Harbour, K.C.; Dransfield, M.T. The true false negative rates of esophageal and endobronchial ultrasound in the staging of mediastinal lymph nodes in patients with non-small cell lung cancer. Ann. Thorac. Surg. 2010, 90, 427–434. [Google Scholar] [CrossRef]
- Sanz-Santos, J.; Serra, M.; Gallego, M.; Montón, C.; Cosio, B.; Sauleda, J.; Fernández-Villar, A.; García-Luján, R.; de Miguel, E.; Cordovilla, R.; et al. Determinants of false-negative results in non-small-cell lung cancer staging by endobronchial ultrasound-guided needle aspiration. Eur. J. Cardiothorac. Surg. 2015, 47, 642–647. [Google Scholar] [CrossRef]
- Nakajima, T.; Zamel, R.; Anayama, T.; Kimura, H.; Yoshino, I.; Keshavjee, S.; Yasufuku, K. Ribonucleic acid microarray analysis from lymph node samples obtained by endobronchial ultrasonography-guided transbronchial needle aspiration. Ann. Thorac. Surg. 2012, 94, 2097–2101. [Google Scholar] [CrossRef]
- Inage, T.; Nakajima, T.; Itoga, S.; Ishige, T.; Fujiwara, T.; Sakairi, Y.; Wada, H.; Suzuki, H.; Iwata, T.; Chiyo, M.; et al. Molecular nodal staging using miRNA expression in lung cancer patients by endobronchial ultrasound-guided transbronchial needle aspiration. Respiration 2018, 96, 267–274. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Mayer, J.; Pospisilova, S. MicroRNA isolation and stability in stored RNA samples. Biochem. Biophys. Res. Commun. 2009, 390, 1–4. [Google Scholar] [CrossRef]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef]
- Liu, A.; Xu, X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol. Biol. 2011, 724, 259–267. [Google Scholar] [CrossRef]
- Hanami, T.; Tanabe, T.; Hanashi, T.; Yamaguchi, M.; Nakata, H.; Mitani, Y.; Kimura, Y.; Soma, T.; Usui, K.; Isobe, M.; et al. Scanning single-molecule counting system for Eprobe with highly simple and effective approach. PLoS ONE 2020, 15, e0243319. [Google Scholar] [CrossRef]
- Nakata, H.; Yamaguchi, M.; Hanashi, T.; Kondo, S.; Tanabe, T. Amplification-free gene expression analysis of formalin-fixed paraffin-embedded samples using scanning single-molecule counting. Anal. Biochem. 2021, 625, 114220. [Google Scholar] [CrossRef]
- Boyd, S.D. Everything you wanted to know about small RNA but were afraid to ask. Lab. Invest. 2008, 88, 569–578. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007, 67, 7972–7976. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Carlson, J.A.; Roos, E.; Brock, G.J. Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle 2009, 8, 2064–2069. [Google Scholar] [CrossRef]
- Shen, Y.; Pan, X.; Yang, J. Gene regulation and prognostic indicators of lung squamous cell carcinoma: TCGA-derived miRNA/mRNA sequencing and DNA methylation data. J. Cell. Physiol. 2019, 234, 22896–22910. [Google Scholar] [CrossRef]
- Schliekelman, M.J.; Gibbons, D.L.; Faca, V.M.; Creighton, C.J.; Rizvi, Z.H.; Zhang, Q.; Wong, C.H.; Wang, H.; Ungewiss, C.; Ahn, Y.H.; et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011, 71, 7670–7682. [Google Scholar] [CrossRef]
- Liu, P.L.; Liu, W.L.; Chang, J.M.; Chen, Y.H.; Liu, Y.P.; Kuo, H.F.; Hsieh, C.C.; Ding, Y.S.; Chen, W.W.; Chong, I.W. MicroRNA-200c inhibits epithelial-mesenchymal transition, invasion, and migration of lung cancer by targeting HMGB1. PLoS ONE 2017, 12, e0180844. [Google Scholar] [CrossRef]
- Lei, W.; Kang, W.; Nan, Y.; Lei, Z.; Zhongdong, L.; Demin, L.; Lei, S.; Hairong, H. The downregulation of miR-200c promotes lactate dehydrogenase A expression and non-small cell lung cancer progression. Oncol. Res. 2018, 26, 1015–1022. [Google Scholar] [CrossRef]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celià-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef]
- Sulaiman, S.A.; Ab Mutalib, N.S.; Jamal, R. miR-200c regulation of metastases in ovarian cancer: Potential role in epithelial and mesenchymal transition. Front. Pharmacol. 2016, 7, 271. [Google Scholar] [CrossRef]
- Liu, X.G.; Zhu, W.Y.; Huang, Y.Y.; Ma, L.N.; Zhou, S.Q.; Wang, Y.K.; Zeng, F.; Zhou, J.H.; Zhang, Y.K. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. 2012, 29, 618–626. [Google Scholar] [CrossRef]
- Kim, M.K.; Jung, S.B.; Kim, J.S.; Roh, M.S.; Lee, J.H.; Lee, E.H.; Lee, H.W. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch. 2014, 465, 463–471. [Google Scholar] [CrossRef]
- Tejero, R.; Navarro, A.; Campayo, M.; Viñolas, N.; Marrades, R.M.; Cordeiro, A.; Ruíz-Martínez, M.; Santasusagna, S.; Molins, L.; Ramirez, J.; et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS ONE 2014, 9, e101899. [Google Scholar] [CrossRef]
- Si, L.; Tian, H.; Yue, W.; Li, L.; Li, S.; Gao, C.; Qi, L. Potential use of microRNA-200c as a prognostic marker in non-small cell lung cancer. Oncol. Lett. 2017, 14, 4325–4330. [Google Scholar] [CrossRef]
- Plönes, T.; Elze, M.; Kayser, G.; Pfeifer, D.; Burger, M.; Zissel, G. mRNA and miRNA analyses in cytologically positive endobronchial ultrasound-guided transbronchial needle aspiration: Implications for molecular staging in lung cancer patients. Cancer Cytopathol. 2014, 122, 292–298. [Google Scholar] [CrossRef]

| Patients, n | 86 |
|---|---|
| Male | 64 |
| Female | 22 |
| Median age (range), years | 70.5 (28–86) |
| Lymph nodes (total), n | 100 |
| Malignant | 70 |
| Adenocarcinoma | 45 |
| Squamous cell carcinoma | 17 |
| Non-small cell carcinoma | 1 |
| Small cell carcinoma | 7 |
| Benign | 30 |
| Malignant, n = 70 | Benign, n = 30 | |
|---|---|---|
| Purpose for EBUS-TBNA | ||
| Staging | 42 | 25 |
| Confirmation for recurrence | 22 | 5 |
| Re-biopsy for searching for gene mutation | 6 | 0 |
| Treatment | ||
| Before chemotherapy, including non-treatment | 42 | 27 |
| After chemotherapy | 28 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sata, Y.; Inage, T.; Nakajima, T.; Ito, Y.; Yoshino, I.; Suzuki, H. Highly Sensitive Detection of miR-200c in Metastatic Lymph Nodes Using Scanning Single-Molecule Counting. Cancers 2025, 17, 3133. https://doi.org/10.3390/cancers17193133
Sata Y, Inage T, Nakajima T, Ito Y, Yoshino I, Suzuki H. Highly Sensitive Detection of miR-200c in Metastatic Lymph Nodes Using Scanning Single-Molecule Counting. Cancers. 2025; 17(19):3133. https://doi.org/10.3390/cancers17193133
Chicago/Turabian StyleSata, Yuki, Terunaga Inage, Takahiro Nakajima, Yuki Ito, Ichiro Yoshino, and Hidemi Suzuki. 2025. "Highly Sensitive Detection of miR-200c in Metastatic Lymph Nodes Using Scanning Single-Molecule Counting" Cancers 17, no. 19: 3133. https://doi.org/10.3390/cancers17193133
APA StyleSata, Y., Inage, T., Nakajima, T., Ito, Y., Yoshino, I., & Suzuki, H. (2025). Highly Sensitive Detection of miR-200c in Metastatic Lymph Nodes Using Scanning Single-Molecule Counting. Cancers, 17(19), 3133. https://doi.org/10.3390/cancers17193133






