BRG1 Loss Is Frequent in Lung Cancer and Transforms Lung Epithelial Cells via Transcriptional and Epigenetic Reprograming
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. BRG1 Knockdown, Knockout, and Western Blotting
2.3. Gene Expression
2.4. DNA Methylation
2.5. In Vitro Cell Survival, Migration, and Anchorage-Independent Growth
2.6. In Vivo Tumor Growth and Drug Sensitivity
2.7. Data Analysis
3. Results
3.1. BRG1 Loss of Function Is Frequent in Lung Cancer
3.2. BRG1 Loss Transforms Immortalized Lung Epithelial Cells
3.3. BRG1-LOF Enhances CS-Induced Pre-Malignant Changes
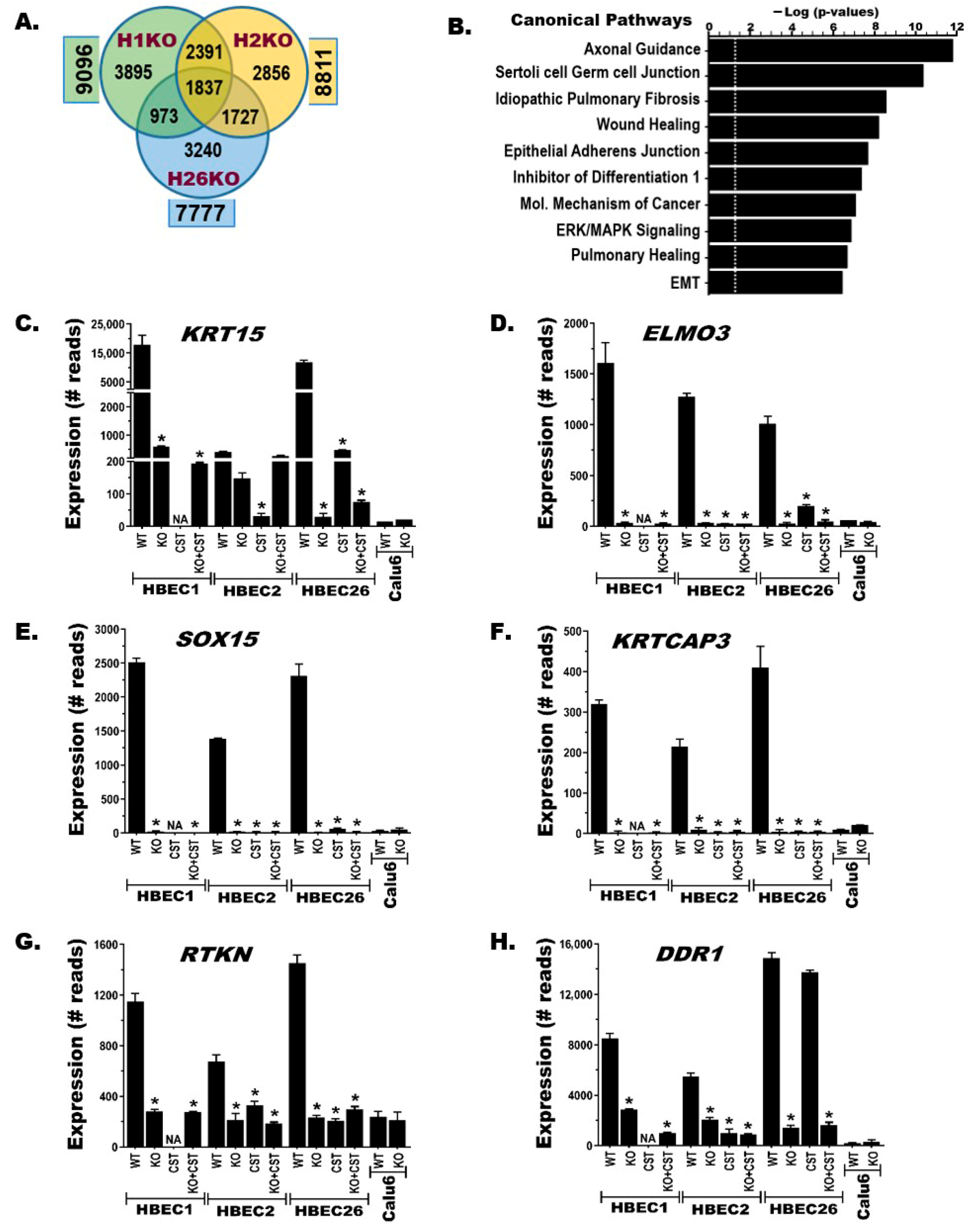
3.4. BRG1-KO Induces Transcriptome-Wide Changes
3.5. BRG1-KO Leads to Epigenome-Wide DNA Methylation Changes
3.6. BRG1-KO Promotes Epithelial-to-Mesenchymal Transition (EMT)
3.7. BRG1-LOF Creates Therapeutic Vulnerabilities to Epigenetic Drugs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BRG1 | Brahma-related gene-1, also called SMARCA4 |
| BRM | Brahma, also called SMARCA2 |
| BAF | BRG1/BRM-associated factors, also called SWI/SNF |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CS | Cigarette smoke |
| CST | Cigarette smoke-transformed |
| EMT | Epithelial-to-mesenchymal transition |
| HBECs | Human bronchial epithelial cell lines |
| LOF | Loss of function |
| NSCLC | Non-small cell lung cancer |
| SCCOHT | Small cell carcinoma of the ovary hypercalcemic type |
| SWI/SNF | Switch/Sucrose non-fermentable |
| SMARCA2 | SWI/SNF-related BAF chromatin remodeling complex subunit ATPase 2 |
| SMARCA4 | SWI/SNF-related BAF chromatin remodeling complex subunit ATPase 4 |
| SMARCA4-DTS | SMARCA4-deficient thoracic sarcoma |
References
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Helming, K.C.; Wang, X.; Roberts, C.W.M. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014, 26, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Schrock, A.B.; Kem, M.; Jessop, N.; Lee, J.; Ali, S.M.; Ross, J.S.; Lennerz, J.K.; Shaw, A.T.; Mino-Kenudson, M. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J. Thorac. Oncol. 2020, 15, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Finetti, M.A.; Grabovska, Y.; Bailey, S.; Williamson, D. Translational genomics of malignant rhabdoid tumours: Current impact and future possibilities. Semin. Cancer Biol. 2020, 61, 30–41. [Google Scholar] [CrossRef]
- Ramos, P.; Karnezis, A.N.; Craig, D.W.; Sekulic, A.; Russell, M.L.; Hendricks, W.P.; Corneveaux, J.J.; Barrett, M.T.; Shumansky, K.; Yang, Y.; et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 2014, 46, 427–429. [Google Scholar] [CrossRef]
- Rekhtman, N.; Montecalvo, J.; Chang, J.C.; Alex, D.; Ptashkin, R.N.; Ai, N.; Sauter, J.L.; Kezlarian, B.; Jungbluth, A.; Desmeules, P.; et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J. Thorac. Oncol. 2020, 15, 231–247. [Google Scholar] [CrossRef]
- Tessema, M.; Rossi, M.R.; Picchi, M.A.; Yingling, C.M.; Lin, Y.; Ramalingam, S.S.; Belinsky, S.A. Common cancer-driver mutations and their association with abnormally methylated genes in lung adenocarcinoma from never-smokers. Lung Cancer 2018, 123, 99–106. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Mittal, P.; Roberts, C.W.M. The SWI/SNF complex in cancer—Biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 2020, 17, 435–448. [Google Scholar] [CrossRef]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Tessema, M.; Weissman, B.E. The SWI/SNF Complex: A Frequently Mutated Chromatin Remodeling Complex in Cancer. Cancer Treat. Res. 2023, 190, 211–244. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Armstrong, K.; Drew, A.; Grassian, A.R.; Feldman, I.; Knutson, S.K.; Kuplast-Barr, K.; Roche, M.; Campbell, J.; Ho, P.; et al. Selective Killing of SMARCA2- and SMARCA4-deficient Small Cell Carcinoma of the Ovary, Hypercalcemic Type Cells by Inhibition of EZH2: In Vitro and In Vivo Preclinical Models. Mol. Cancer Ther. 2017, 16, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Orlando, K.A.; Douglas, A.K.; Abudu, A.; Wang, Y.; Tessier-Cloutier, B.; Su, W.; Peters, A.; Sherman, L.S.; Moore, R.; Nguyen, V.; et al. Re-expression of SMARCA4/BRG1 in small cell carcinoma of ovary, hypercalcemic type (SCCOHT) promotes an epithelial-like gene signature through an AP-1-dependent mechanism. eLife 2020, 9, e59073. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, S.Y.; Colborne, S.; Lambert, G.; Shin, C.Y.; Santos, N.D.; Orlando, K.A.; Lang, J.D.; Hendricks, W.P.D.; Bally, M.B.; et al. Histone Deacetylase Inhibitors Synergize with Catalytic Inhibitors of EZH2 to Exhibit Antitumor Activity in Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Mol. Cancer Ther. 2018, 17, 2767–2779. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Williams, R.T.; Calarco, J.P.; Miller, E.L.; Weber, C.M.; Braun, S.M.; Pulice, J.L.; Chory, E.J.; Crabtree, G.R. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017, 49, 213–222. [Google Scholar] [CrossRef]
- Stanton, B.Z.; Hodges, C.; Calarco, J.P.; Braun, S.M.; Ku, W.L.; Kadoch, C.; Zhao, K.; Crabtree, G.R. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 2017, 49, 282–288. [Google Scholar] [CrossRef]
- Fernando, T.M.; Piskol, R.; Bainer, R.; Sokol, E.S.; Trabucco, S.E.; Zhang, Q.; Trinh, H.; Maund, S.; Kschonsak, M.; Chaudhuri, S.; et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat. Commun. 2020, 11, 5551. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Bandlamudi, C.; Lavery, J.A.; Montecalvo, J.; Namakydoust, A.; Rizvi, H.; Egger, J.; Concepcion, C.P.; Paul, S.; Arcila, M.E.; et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin. Cancer Res. 2020, 26, 5701–5708. [Google Scholar] [CrossRef] [PubMed]
- Velut, Y.; Decroix, E.; Blons, H.; Alifano, M.; Leroy, K.; Petitprez, F.; Boni, A.; Garinet, S.; Biton, J.; Cremer, I.; et al. SMARCA4-deficient lung carcinoma is an aggressive tumor highly infiltrated by FOXP3+ cells and neutrophils. Lung Cancer 2022, 169, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, G.R.; Rahal, R.; Buxton, F.; Xiang, K.; McAllister, G.; Frias, E.; Bagdasarian, L.; Huber, J.; Lindeman, A.; Chen, D.; et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 3128–3133. [Google Scholar] [CrossRef] [PubMed]
- Shorstova, T.; Marques, M.; Su, J.; Johnston, J.; Kleinman, C.L.; Hamel, N.; Huang, S.; Alaoui-Jamali, M.A.; Foulkes, W.D.; Witcher, M. SWI/SNF-Compromised Cancers Are Susceptible to Bromodomain Inhibitors. Cancer Res. 2019, 79, 2761–2774. [Google Scholar] [CrossRef]
- Tagal, V.; Wei, S.; Zhang, W.; Brekken, R.A.; Posner, B.A.; Peyton, M.; Girard, L.; Hwang, T.; Wheeler, D.A.; Minna, J.D.; et al. SMARCA4-inactivating mutations increase sensitivity to Aurora kinase A inhibitor VX-680 in non-small cell lung cancers. Nat. Commun. 2017, 8, 14098. [Google Scholar] [CrossRef]
- Lissanu Deribe, Y.; Sun, Y.; Terranova, C.; Khan, F.; Martinez-Ledesma, J.; Gay, J.; Gao, G.; Mullinax, R.A.; Khor, T.; Feng, N.; et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. Nat. Med. 2018, 24, 1047–1057. [Google Scholar] [CrossRef]
- Oike, T.; Ogiwara, H.; Tominaga, Y.; Ito, K.; Ando, O.; Tsuta, K.; Mizukami, T.; Shimada, Y.; Isomura, H.; Komachi, M.; et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013, 73, 5508–5518. [Google Scholar] [CrossRef]
- Vangamudi, B.; Paul, T.A.; Shah, P.K.; Kost-Alimova, M.; Nottebaum, L.; Shi, X.; Zhan, Y.; Leo, E.; Mahadeshwar, H.S.; Protopopov, A.; et al. The SMARCA2/4 ATPase Domain Surpasses the Bromodomain as a Drug Target in SWI/SNF-Mutant Cancers: Insights from cDNA Rescue and PFI-3 Inhibitor Studies. Cancer Res. 2015, 75, 3865–3878. [Google Scholar] [CrossRef]
- Reichl, K.D.; Lee, E.C.Y.; Gopalsamy, A. Synthetic lethality: Targeting SMARCA2 ATPase in SMARCA4-deficient tumors—A review of patent literature from 2019-30 June 2023. Expert Opin. Ther. Pat. 2024, 34, 159–169. [Google Scholar] [CrossRef]
- Xue, Y.; Meehan, B.; Fu, Z.; Wang, X.Q.D.; Fiset, P.O.; Rieker, R.; Levins, C.; Kong, T.; Zhu, X.; Morin, G.; et al. SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non-small cell lung cancer. Nat. Commun. 2019, 10, 557. [Google Scholar] [CrossRef]
- Herpel, E.; Rieker, R.J.; Dienemann, H.; Muley, T.; Meister, M.; Hartmann, A.; Warth, A.; Agaimy, A. SMARCA4 and SMARCA2 deficiency in non-small cell lung cancer: Immunohistochemical survey of 316 consecutive specimens. Ann. Diagn. Pathol. 2017, 26, 47–51. [Google Scholar] [CrossRef]
- Tellez, C.S.; Grimes, M.J.; Juri, D.E.; Do, K.; Willink, R.; Dye, W.W.; Wu, G.; Picchi, M.A.; Belinsky, S.A. Flavored E-cigarette product aerosols induce transformation of human bronchial epithelial cells. Lung Cancer 2023, 179, 107180. [Google Scholar] [CrossRef]
- Gazdar, A.F.; Gao, B.; Minna, J.D. Lung cancer cell lines: Useless artifacts or invaluable tools for medical science? Lung Cancer 2010, 68, 309–318. [Google Scholar] [CrossRef]
- Ramirez, R.D.; Sheridan, S.; Girard, L.; Sato, M.; Kim, Y.; Pollack, J.; Peyton, M.; Zou, Y.; Kurie, J.M.; Dimaio, J.M.; et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004, 64, 9027–9034. [Google Scholar] [CrossRef]
- Tessema, M.; Yingling, C.M.; Picchi, M.A.; Wu, G.; Ryba, T.; Lin, Y.; Bungum, A.O.; Edell, E.S.; Spira, A.; Belinsky, S.A. ANK1 Methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer Lett. 2017, 410, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Brayer, K.J.; Frerich, C.A.; Kang, H.; Ness, S.A. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid Cystic Carcinoma. Cancer Discov. 2016, 6, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Brayer, K.J.; Mitani, Y.; Burns, E.A.; Rao, P.H.; Bell, D.; Williams, M.D.; Ferrarotto, R.; Pytynia, K.B.; El-Naggar, A.K.; et al. Oncogenic Orphan Nuclear Receptor NR4A3 Interacts and Cooperates with MYB in Acinic Cell Carcinoma. Cancers 2020, 12, 2433. [Google Scholar] [CrossRef]
- Tessema, M.; Tassew, D.D.; Yingling, C.M.; Do, K.; Picchi, M.A.; Wu, G.; Petersen, H.; Randell, S.; Lin, Y.; Belinsky, S.A.; et al. Identification of novel epigenetic abnormalities as sputum biomarkers for lung cancer risk among smokers and COPD patients. Lung Cancer 2020, 146, 189–196. [Google Scholar] [CrossRef]
- Tessema, M.; Willink, R.; Do, K.; Yu, Y.Y.; Yu, W.; Machida, E.O.; Brock, M.; Van Neste, L.; Stidley, C.A.; Baylin, S.B.; et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008, 68, 1707–1714. [Google Scholar] [CrossRef]
- Peters, T.J.; Meyer, B.; Ryan, L.; Achinger-Kawecka, J.; Song, J.; Campbell, E.M.; Qu, W.; Nair, S.; Loi-Luu, P.; Stricker, P.; et al. Characterisation and reproducibility of the HumanMethylationEPIC v2.0 BeadChip for DNA methylation profiling. BMC Genom. 2024, 25, 251. [Google Scholar] [CrossRef]
- Tessema, M.; Yingling, C.M.; Thomas, C.L.; Klinge, D.M.; Bernauer, A.M.; Liu, Y.; Dacic, S.; Siegfried, J.M.; Dahlberg, S.E.; Schiller, J.H.; et al. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene 2012, 31, 4107–4116. [Google Scholar] [CrossRef]
- Kuehl, P.J.; Yingling, C.M.; Dubose, D.; Burke, M.; Revelli, D.A.; Chen, W.; Dye, W.W.; Belinsky, S.A.; Tessema, M. Inhalation delivery dramatically improves the efficacy of topotecan for the treatment of local and distant lung cancer. Drug Deliv. 2021, 28, 767–775. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Kobayashi, E.; Kubo, T.; Kodaira, M.; Motoi, T.; Motoi, N.; Yonemori, K.; Ohe, Y.; Watanabe, S.I.; Kawai, A.; et al. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod. Pathol. 2017, 30, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Shay, J.W.; Minna, J.D. Immortalized normal human lung epithelial cell models for studying lung cancer biology. Respir. Investig. 2020, 58, 344–354. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, S.; Fleishman, J.S.; Chen, J.; Tang, H.; Chen, Z.S.; Chen, W.; Ding, M. Targeting anoikis resistance as a strategy for cancer therapy. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2024, 75, 101099. [Google Scholar] [CrossRef]
- Medina, P.P.; Romero, O.A.; Kohno, T.; Montuenga, L.M.; Pio, R.; Yokota, J.; Sanchez-Cespedes, M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum. Mutat. 2008, 29, 617–622. [Google Scholar] [CrossRef]
- Peinado, P.; Andrades, A.; Cuadros, M.; Rodriguez, M.I.; Coira, I.F.; Garcia, D.J.; Alvarez-Perez, J.C.; Balinas-Gavira, C.; Arenas, A.M.; Patino-Mercau, J.R.; et al. Comprehensive Analysis of SWI/SNF Inactivation in Lung Adenocarcinoma Cell Models. Cancers 2020, 12, 3712. [Google Scholar] [CrossRef]
- Tellez, C.S.; Juri, D.E.; Do, K.; Bernauer, A.M.; Thomas, C.L.; Damiani, L.A.; Tessema, M.; Leng, S.; Belinsky, S.A. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011, 71, 3087–3097. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B.; Durdu, S.; Burger, L.; Schubeler, D. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef]
- Belinsky, S.A.; Grimes, M.J.; Picchi, M.A.; Mitchell, H.D.; Stidley, C.A.; Tesfaigzi, Y.; Channell, M.M.; Liu, Y.; Casero, R.A., Jr.; Baylin, S.B.; et al. Combination therapy with vidaza and entinostat suppresses tumor growth and reprograms the epigenome in an orthotopic lung cancer model. Cancer Res. 2011, 71, 454–462. [Google Scholar] [CrossRef]
- Tischkowitz, M.; Huang, S.; Banerjee, S.; Hague, J.; Hendricks, W.P.D.; Huntsman, D.G.; Lang, J.D.; Orlando, K.A.; Oza, A.M.; Pautier, P.; et al. Small-Cell Carcinoma of the Ovary, Hypercalcemic Type-Genetics, New Treatment Targets, and Current Management Guidelines. Clin. Cancer Res. 2020, 26, 3908–3917. [Google Scholar] [CrossRef]
- Jones, C.A.; Tansey, W.P.; Weissmiller, A.M. Emerging Themes in Mechanisms of Tumorigenesis by SWI/SNF Subunit Mutation. Epigenet. Insights 2022, 15, 25168657221115656. [Google Scholar] [CrossRef]
- Concepcion, C.P.; Ma, S.; LaFave, L.M.; Bhutkar, A.; Liu, M.; DeAngelo, L.P.; Kim, J.Y.; Del Priore, I.; Schoenfeld, A.J.; Miller, M.; et al. Smarca4 Inactivation Promotes Lineage-Specific Transformation and Early Metastatic Features in the Lung. Cancer Discov. 2022, 12, 562–585. [Google Scholar] [CrossRef]
- Orvis, T.; Hepperla, A.; Walter, V.; Song, S.; Simon, J.; Parker, J.; Wilkerson, M.D.; Desai, N.; Major, M.B.; Hayes, D.N.; et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 2014, 74, 6486–6498. [Google Scholar] [CrossRef]
- Berlin, M.; Cantley, J.; Bookbinder, M.; Bortolon, E.; Broccatelli, F.; Cadelina, G.; Chan, E.W.; Chen, H.; Chen, X.; Cheng, Y.; et al. PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models. J. Med. Chem. 2024, 67, 1262–1313. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tessema, M.; Yingling, C.M.; Phillips, L.M.; Do, K.; Picchi, M.A.; Willink, R.; Belinsky, S.A. BRG1 Loss Is Frequent in Lung Cancer and Transforms Lung Epithelial Cells via Transcriptional and Epigenetic Reprograming. Cancers 2025, 17, 3092. https://doi.org/10.3390/cancers17183092
Tessema M, Yingling CM, Phillips LM, Do K, Picchi MA, Willink R, Belinsky SA. BRG1 Loss Is Frequent in Lung Cancer and Transforms Lung Epithelial Cells via Transcriptional and Epigenetic Reprograming. Cancers. 2025; 17(18):3092. https://doi.org/10.3390/cancers17183092
Chicago/Turabian StyleTessema, Mathewos, Christin M. Yingling, Loryn M. Phillips, Kieu Do, Maria A. Picchi, Randy Willink, and Steven A. Belinsky. 2025. "BRG1 Loss Is Frequent in Lung Cancer and Transforms Lung Epithelial Cells via Transcriptional and Epigenetic Reprograming" Cancers 17, no. 18: 3092. https://doi.org/10.3390/cancers17183092
APA StyleTessema, M., Yingling, C. M., Phillips, L. M., Do, K., Picchi, M. A., Willink, R., & Belinsky, S. A. (2025). BRG1 Loss Is Frequent in Lung Cancer and Transforms Lung Epithelial Cells via Transcriptional and Epigenetic Reprograming. Cancers, 17(18), 3092. https://doi.org/10.3390/cancers17183092




