The Prognostic Value of a Nomogram Model Based on Tumor Immune Markers and Clinical Factors for Adult Primary Glioma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
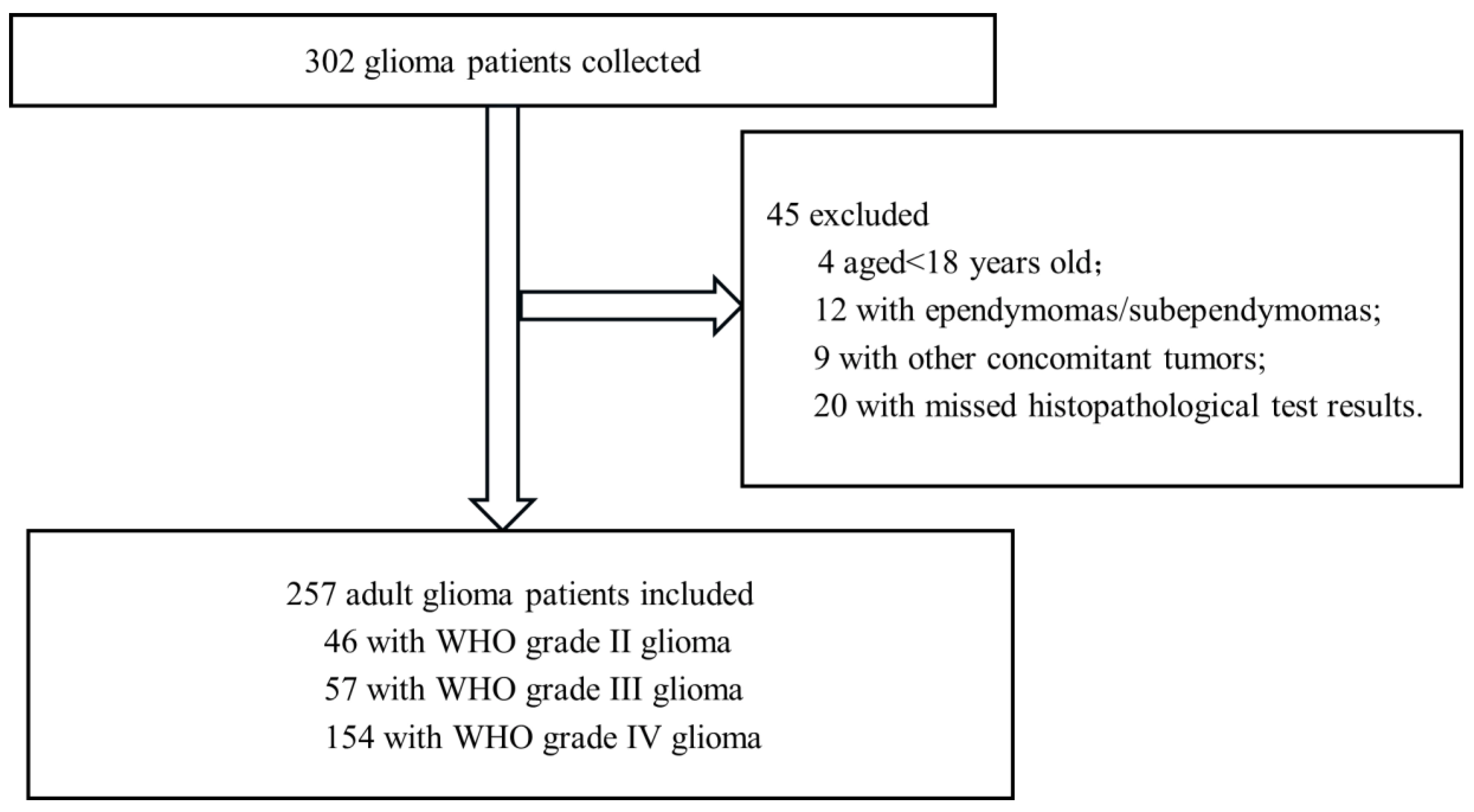
2.1. Study Design and Participants
2.2. Histopathological Detection Methods
2.3. WHO Grading Criteria for Glioma
2.4. Treatment of Glioma
2.5. Follow-Up
2.6. Statistical Analysis
3. Results
3.1. Comparison of Characteristics Among Adult Glioma Patients with Different WHO Grades
3.2. Treatment Plan
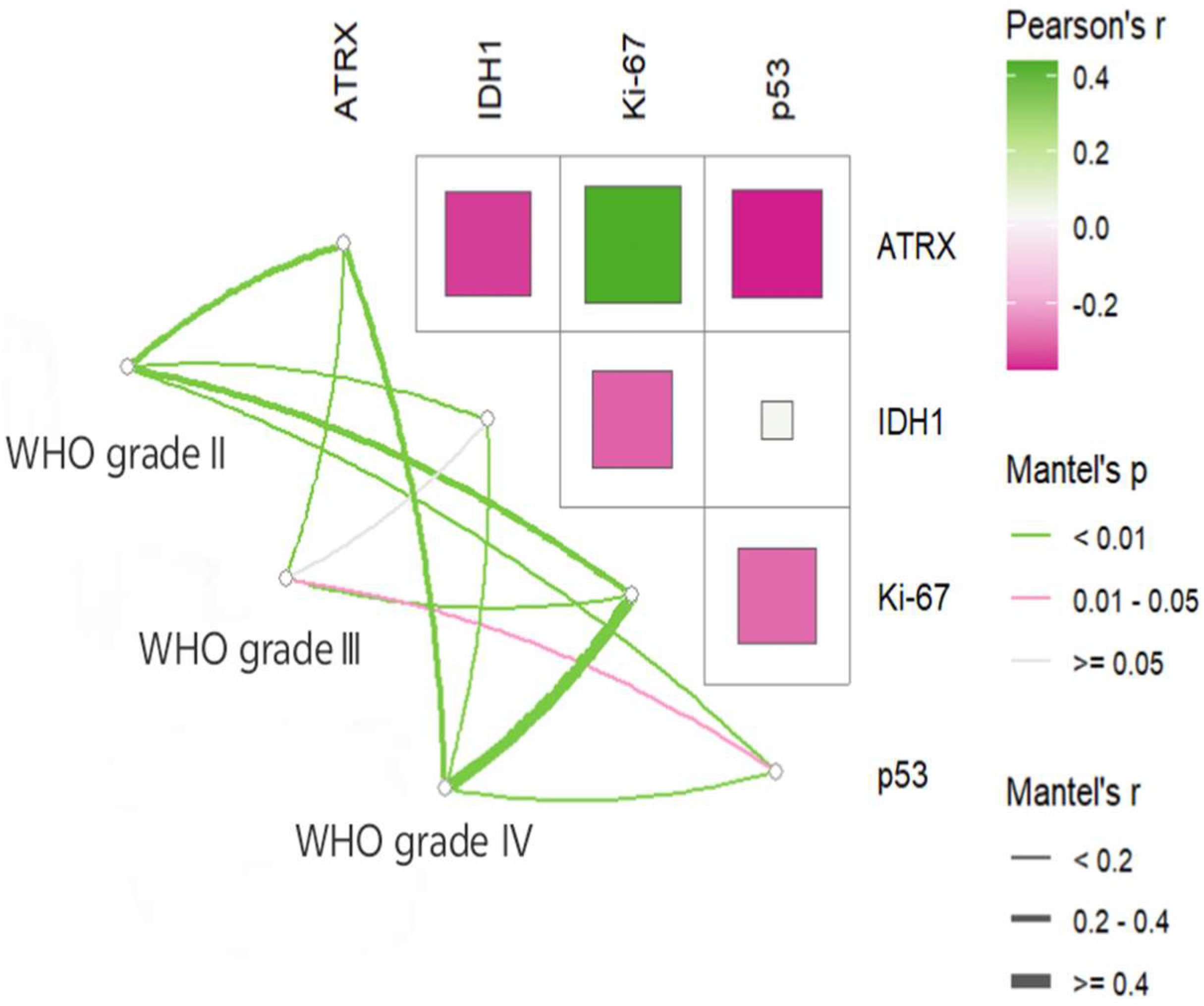
3.3. Mutation and Correlation Analysis of Common Immune Markers Across Glioma Grades
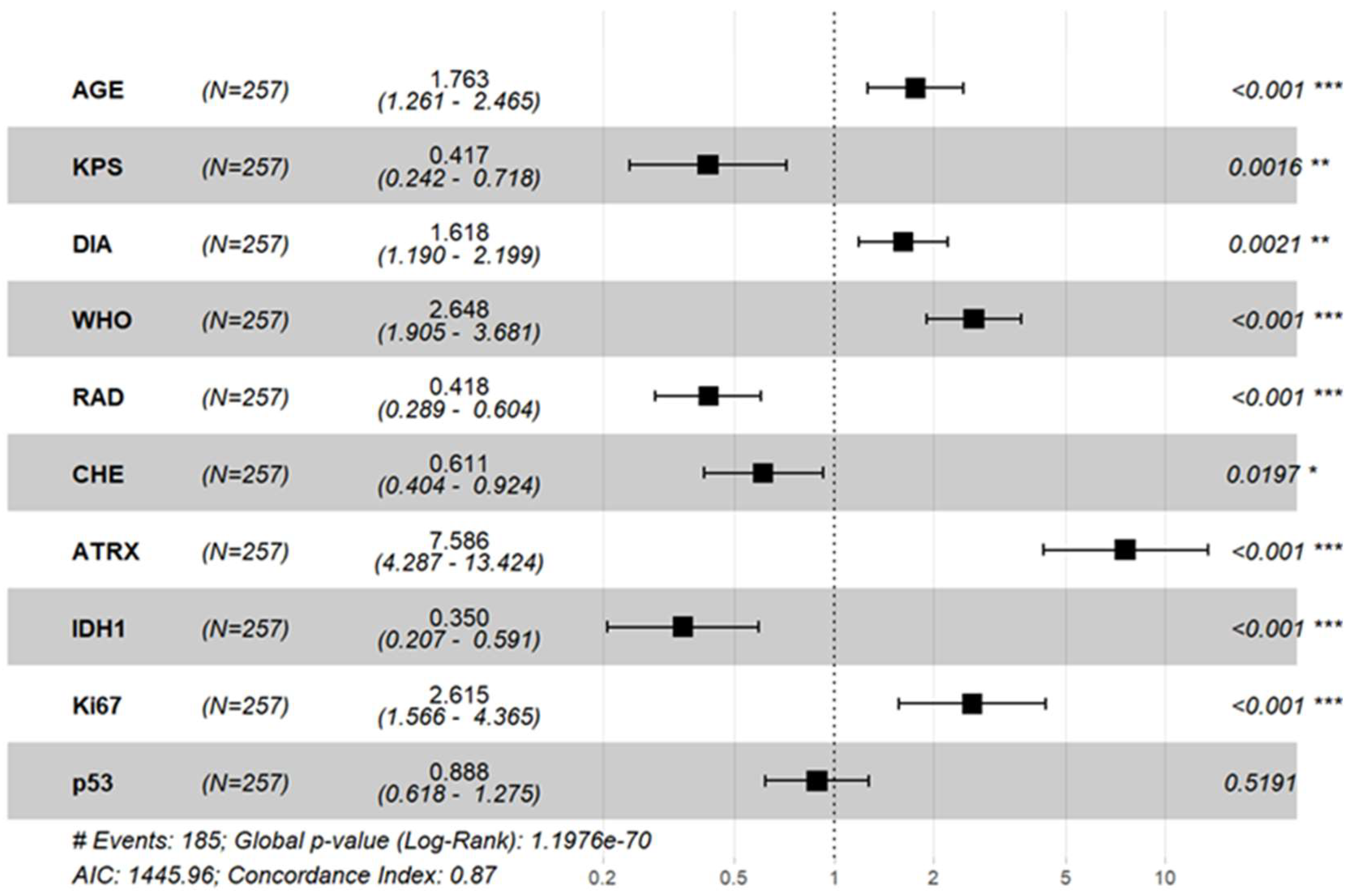
3.4. Cox Regression Analysis of Prognostic Factors
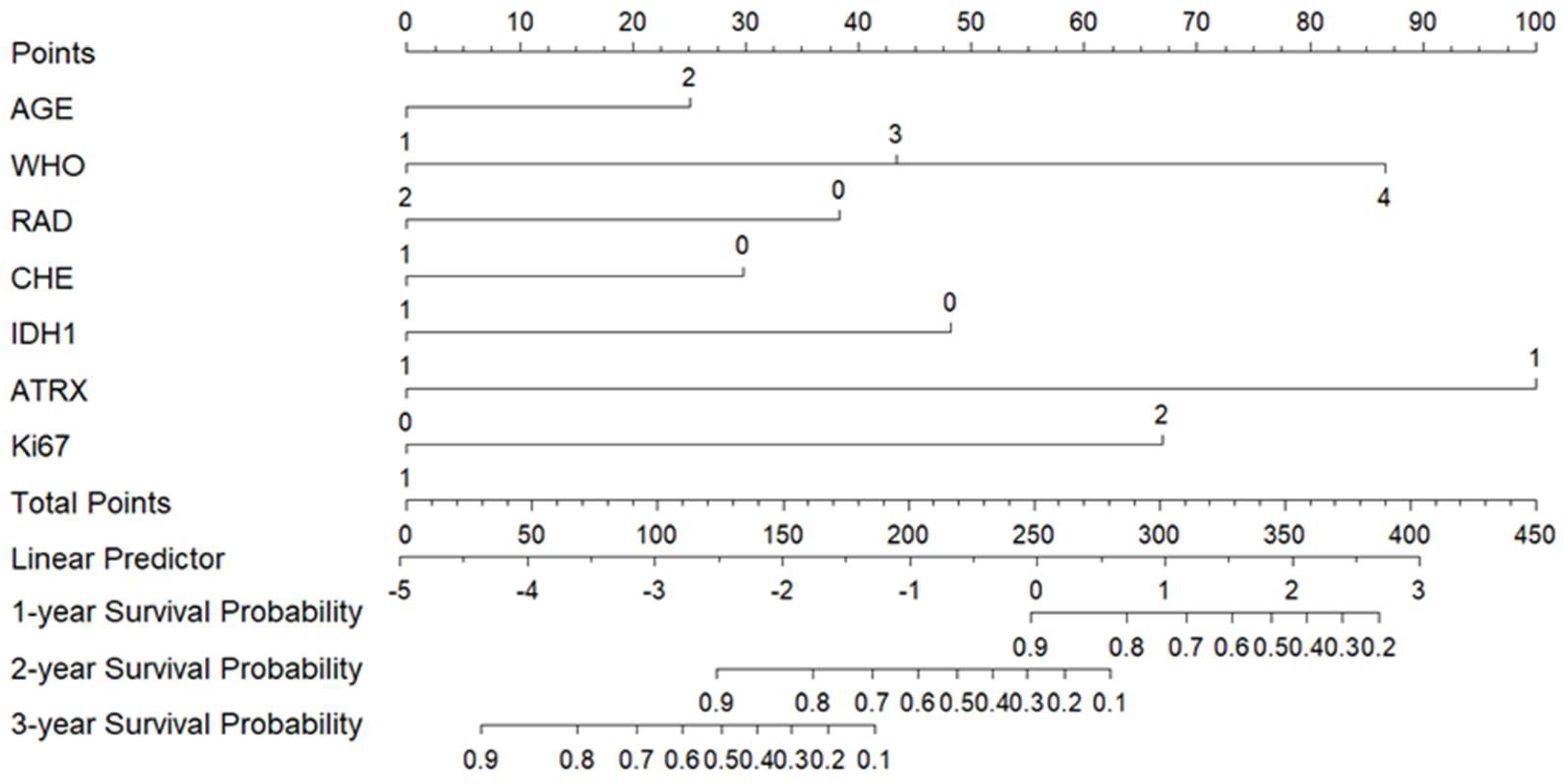
3.5. Construction and Validation of a Nomogram Prognostic Model
3.5.1. Construction of the Nomogram Prognostic Model
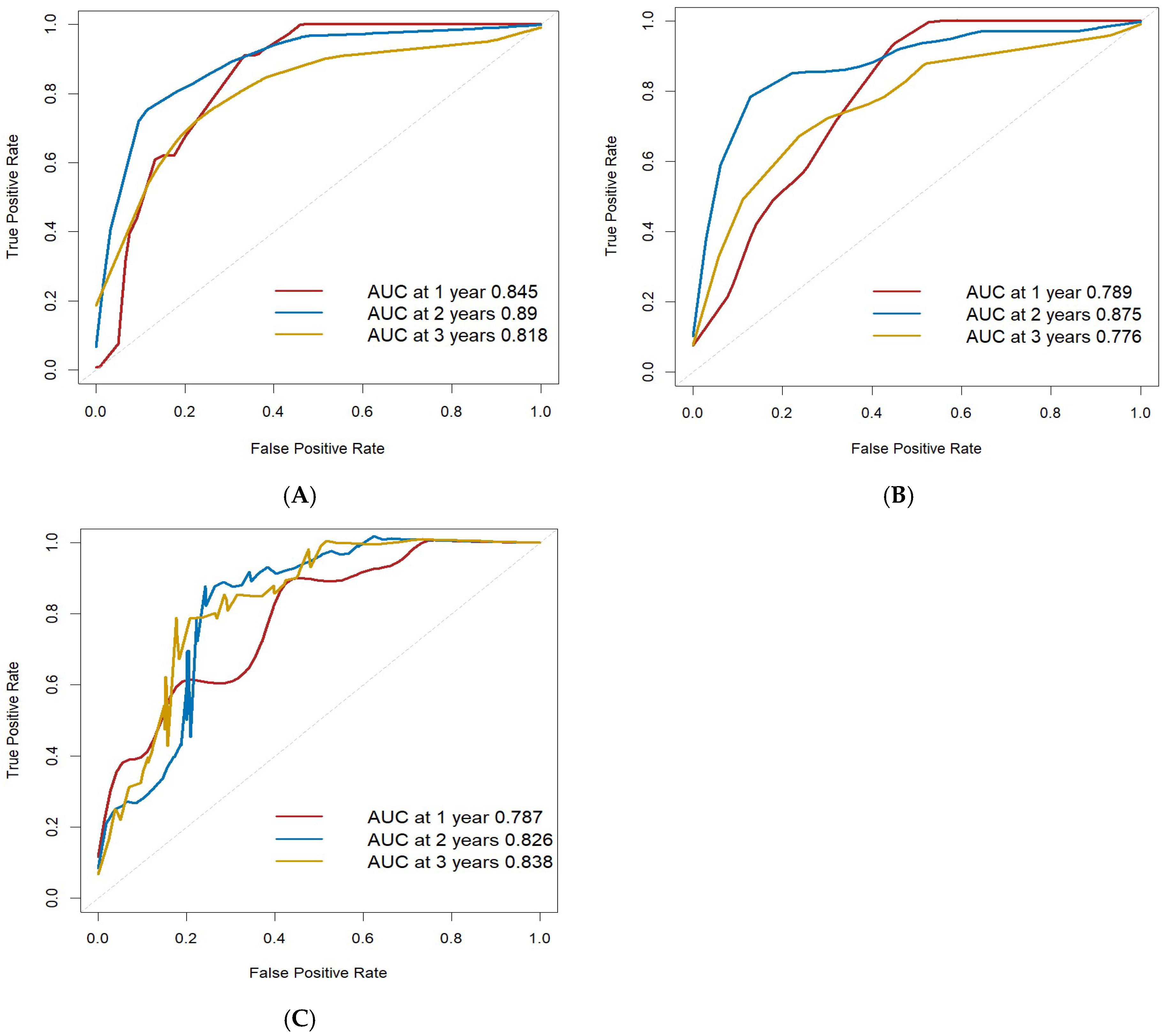
3.5.2. Validation and Evaluation of the Nomogram Prognostic Model
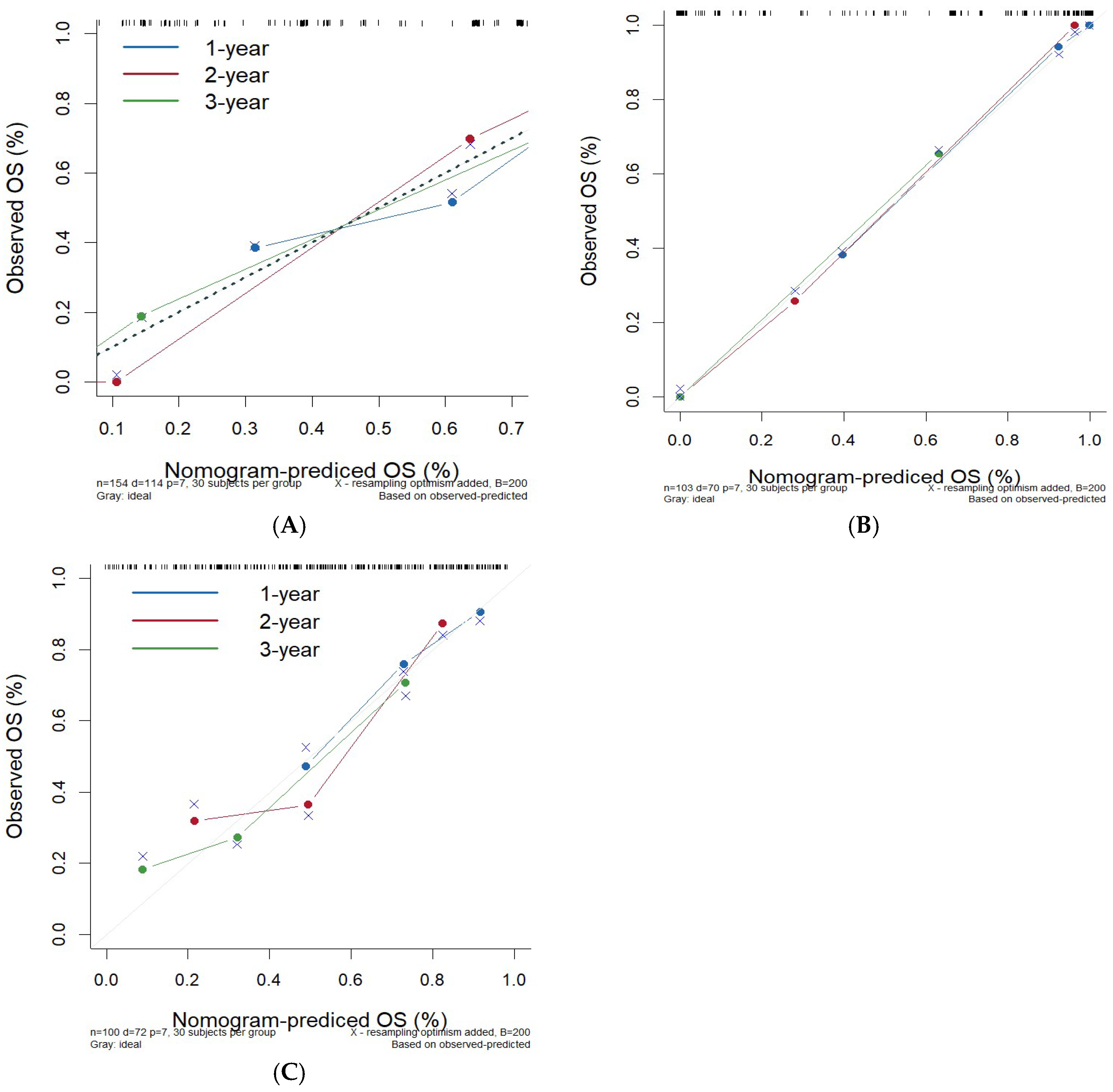
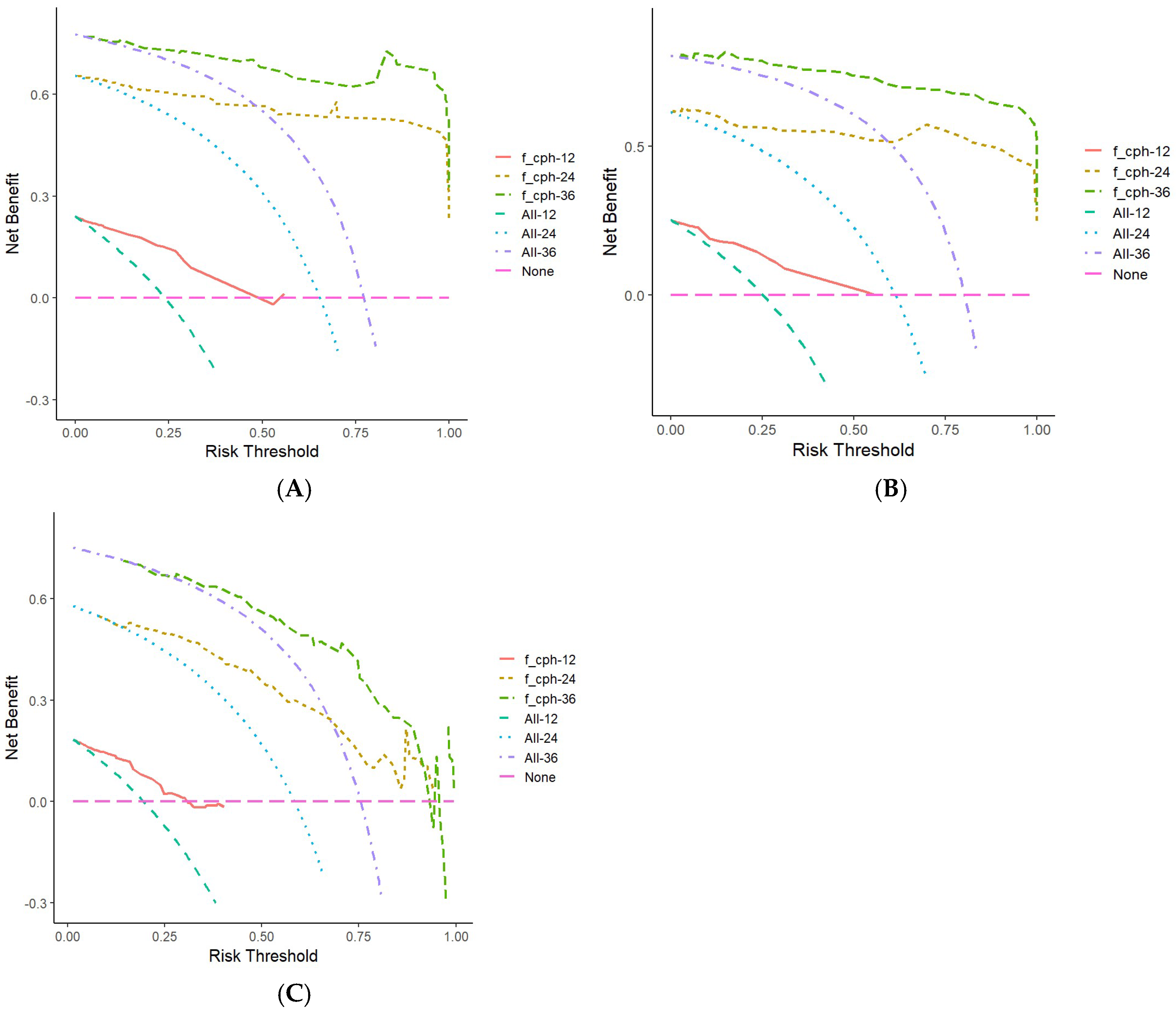
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weller, M.; Wen, P.Y.; Chang, S.M.; Dirven, L.; Lim, M.; Monje, M.; Reifenberger, G. Glioma. Nat. Rev. Dis. Primers 2024, 10, 33. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016–2020. Neuro-Oncol. 2023, 25 (Suppl. S2), iv1–iv99. [Google Scholar] [CrossRef]
- Wang, L.; Piao, Y. Application and challenges of molecular testing in pathological diagnosis of central nervous system tumors. J. Clin. Exp. Pathol. 2024, 40, 455–457. [Google Scholar]
- Tang, F.; Wang, D.W.; Xi, C.; Yang, J.Z.; Liu, Z.Y.; Yu, D.H.; Wang, Z.F.; Li, Z.Q. Local and Systemic Effects of IDH Mutations on Primary Glioma Patients. Immunology 2023, 169, 503–514. [Google Scholar] [CrossRef]
- Li, B.; Li, H.; Zhang, L.; Ren, T.; Meng, J. Expression Analysis of Human Glioma Susceptibility Gene and P53 in Human Glioma and Its Clinical Significance Based on Bioinformatics. Ann. Transl. Med. 2023, 11, 53. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, Y.; Yang, C.; Zhang, X.; Xu, C.; Qiao, X.; Xu, J.; Tian, S.; Fang, C.; Kang, C. Omics-Based Integrated Analysis Identified ATRX as a Biomarker Associated with Glioma Diagnosis and Prognosis. Cancer Biol. Med. 2019, 16, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Dahlrot, R.H.; Bangsø, J.A.; Petersen, J.K.; Rosager, A.M.; Sørensen, M.D.; Reifenberger, G.; Hansen, S.; Kristensen, B.W. Prognostic Role of Ki-67 in Glioblastomas Excluding Contribution from Non-Neoplastic Cells. Sci. Rep. 2021, 11, 17918. [Google Scholar] [CrossRef]
- Li, G.; Li, L.; Li, Y.; Qian, Z.; Wu, F.; He, Y.; Jiang, H.; Li, R.; Wang, D.; Zhai, Y.; et al. An MRI Radiomics Approach to Predict Survival and Tumour-Infiltrating Macrophages in Gliomas. Brain 2022, 145, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Nakase, T.; Guerra, G.A.; Ostrom, Q.T.; Ge, T.; Melin, B.S.; Wrensch, M.; Wiencke, J.K.; Jenkins, R.B.; Eckel-Passow, J.E.; Glioma International Case-Control Study (GICC); et al. Genome-Wide Polygenic Risk Scores Predict Risk of Glioma and Molecular Subtypes. Neuro-Oncol. 2024, 26, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, J.; Lou, J. Expression levels and clinical significance of Ki-67 and IDH1-R132H in gliomas. Guangdong Med. J. 2024, 45, 1039–1042. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Mijderwijk, H.J.; Nieboer, D.; Incekara, F.; Berger, K.; Steyerberg, E.W.; van den Bent, M.J.; Reifenberger, G.; Hänggi, D.; Smits, M.; Senft, C.; et al. Development and External Validation of a Clinical Prediction Model for Survival in Patients with IDH Wild-Type Glioblastoma. J. Neurosurg. 2022, 137, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Nam, D.H.; Ram, Z.; Poon, W.S.; Wang, J.; Boldbaatar, D.; Mao, Y.; Ma, W.; Mao, Q.; You, Y.; et al. Clinical Practice Guidelines for the Management of Adult Diffuse Gliomas. Cancer Lett. 2021, 499, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 Classification of Central Nervous System Tumors and Implications for Therapy for Adult-Type Gliomas: A Review. JAMA Oncol. 2022, 8, 1493–1501. [Google Scholar] [CrossRef]
- Synhaeve, N.E.; Van Den Bent, M.J.; French, P.J.; Dinjens, W.N.M.; Atmodimedjo, P.N.; Kros, J.M.; Verdijk, R.; Dirven, C.M.F.; Dubbink, H.J. Clinical Evaluation of a Dedicated next Generation Sequencing Panel for Routine Glioma Diagnostics. Acta Neuropathol. Commun. 2018, 6, 126. [Google Scholar] [CrossRef]
- Kan, L.K.; Drummond, K.; Hunn, M.; Williams, D.; O’Brien, T.J.; Monif, M. Potential Biomarkers and Challenges in Glioma Diagnosis, Therapy and Prognosis. BMJ Neurol. Open 2020, 2, e000069. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Lu, M.; Wen, P.Y.; Taylor, J.W.; Maher, E.A.; Arrillaga-Romany, I.; Peters, K.B.; Ellingson, B.M.; Rosenblum, M.K.; Chun, S.; et al. Vorasidenib and Ivosidenib in IDH1-Mutant Low-Grade Glioma: A Randomized, Perioperative Phase 1 Trial. Nat. Med. 2023, 29, 615–622. [Google Scholar] [CrossRef]
- Mellinghoff, I.K.; Van den Bent, M.J.; Blumenthal, D.T.; Touat, M.; Peters, K.B.; Clarke, J.; Mendez, J.; Yust-Katz, S.; Welsh, L.; Mason, W.P.; et al. Vorasidenib in IDH1- or IDH2-Mutant Low-Grade Glioma. N. Engl. J. Med. 2023, 389, 589–601. [Google Scholar] [CrossRef]
- Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; Morozova, O.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar]
- Olar, A.; Sulman, E.P. Molecular Markers in Low-Grade Glioma-Toward Tumor Reclassification. Semin. Radiat. Oncol. 2015, 25, 155–163. [Google Scholar] [CrossRef]
- Hu, C.; Wang, K.; Damon, C.; Fu, Y.; Ma, T.; Kratz, L.; Lal, B.; Ying, M.; Xia, S.; Cahill, D.P.; et al. ATRX Loss Promotes Immunosuppressive Mechanisms in IDH1 Mutant Glioma. Neuro-Oncol. 2022, 24, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Murnyak, B.; Huang, L.E. Association of TP53 Alteration with Tissue Specificity and Patient Outcome of IDH1-Mutant Glioma. Cells 2021, 10, 2116. [Google Scholar] [CrossRef] [PubMed]
- Hoyos, D.; Zappasodi, R.; Schulze, I.; Sethna, Z.; de Andrade, K.C.; Bajorin, D.F.; Bandlamudi, C.; Callahan, M.K.; Funt, S.A.; Hadrup, S.R.; et al. Fundamental immune-oncogenicity trade-offs define driver mutation fitness. Nature 2022, 606, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, A.; Park, S.H.; Seo, J.W.; Park, C.K. Immunohistochemical Analysis of ATRX, IDH1 and P53 in Glioblastoma and Their Correlations with Patient Survival. J. Korean Med. Sci. 2016, 31, 1208–1214. [Google Scholar] [CrossRef]
- Nguyen, D.N.; Heaphy, C.M.; De Wilde, R.F.; Orr, B.A.; Odia, Y.; Eberhart, C.G.; Meeker, A.K.; Rodriguez, F.J. Molecular and Morphologic Correlates of the Alternative Lengthening of Telomeres Phenotype in High-Grade Astrocytomas. Brain Pathol. 2013, 23, 237–243. [Google Scholar] [CrossRef]
- Newcomb, E.W.; Cohen, H.; Lee, S.R.; Bhalla, S.K.; Bloom, J.; Hayes, R.L.; Miller, D.C. Survival of Patients with Glioblastoma Multiforme Is Not Influenced by Altered Expression of P16, P53, EGFR, MDM2 or Bcl-2 Genes. Brain Pathol. 1998, 8, 655–667. [Google Scholar] [CrossRef]
- Priambada, D.; Thohar Arifin, M.; Saputro, A.; Muzakka, A.; Karlowee, V.; Sadhana, U.; Bakhtiar, Y.; Prihastomo, K.T.; Risdianto, A.; Brotoarianto, H.K.; et al. Immunohistochemical Expression of IDH1, ATRX, Ki67, GFAP, and Prognosis in Indonesian Glioma Patients. Int. J. Gen. Med. 2023, 16, 393–403. [Google Scholar] [CrossRef]
- Yang, Z.; Ling, F.; Ruan, S.; Hu, J.; Tang, M.; Sun, X.; Long, W. Clinical and Prognostic Implications of 1p/19q, IDH, BRAF, MGMT Promoter, and TERT Promoter Alterations, and Expression of Ki-67 and P53 in Human Gliomas. Cancer Manag. Res. 2021, 13, 8755–8765. [Google Scholar] [CrossRef]
- Malueka, R.G.; Dwianingsih, E.K.; Bayuangga, H.F.; Panggabean, A.S.; Argo, I.W.; Donurizki, A.D.; Shaleh, S.; Wicaksono, A.S.; Dananjoyo, K.; Asmedi, A.; et al. Clinicopathological Features and Prognosis of Indonesian Patients with Gliomas with IDH Mutation: Insights into Its Significance in a Southeast Asian Population. Asian Pac. J. Cancer Prev. APJCP 2020, 21, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Hervey-Jumper, S.L.; Zhang, Y.; Phillips, J.J.; Morshed, R.A.; Young, J.S.; McCoy, L.; Lafontaine, M.; Luks, T.; Ammanuel, S.; Kakaizada, S.; et al. Interactive Effects of Molecular, Therapeutic, and Patient Factors on Outcome of Diffuse Low-Grade Glioma. J. Clin. Oncol. 2023, 41, 2029–2042. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Ding, W.; Wang, J.; Zhou, B.; Li, Y.; Jiang, W.; Pan, H.; Gu, J.; Sun, X. Application of Intraoperative Radiotherapy for Malignant Glioma. Cancer Radiother. 2023, 27, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Zhou, D.; Shen, H.; Ma, C.; Wu, D.; Hou, L.; Wang, H.; Xu, T. Development of a prognostic model related to homologous recombination deficiency in glioma based on multiple machine learning. Front. Immunol. 2024, 15, 1452097. [Google Scholar] [CrossRef]
- Huang, D.; Gao, T.; Zhang, Y.; Lyu, X.; Liu, S.; Chen, Y.; Su, C.; Hu, W.; Lv, Y. A Study on Prognosis of Diffuse Glioma Based on Clinical Factors and Magnetic Resonance Imaging Radiomics. World Neurosurg. 2024, 186, e514–e530. [Google Scholar] [CrossRef]
| n | WHO II | WHO III | WHO IV | χ2 | p Value | |
|---|---|---|---|---|---|---|
| Gender | ||||||
| male | 143 | 23 | 35 | 85 | 1.372 | 0.503 |
| female | 114 | 23 | 22 | 69 | ||
| Age (y) | ||||||
| <60 | 137 | 39 | 38 | 60 | 35.130 | <0.001 |
| ≥60 | 120 | 7 | 19 | 94 | ||
| Smoking history | ||||||
| yes | 86 | 16 | 22 | 48 | 1.075 | 0.584 |
| no | 171 | 30 | 35 | 106 | ||
| History of head trauma | ||||||
| yes | 20 | 4 | 3 | 13 | 0.651 | 0.722 |
| no | 237 | 42 | 54 | 141 | ||
| KPS score | ||||||
| ≥80 | 237 | 46 | 56 | 135 | 11.221 | 0.004 |
| <80 | 20 | 0 | 1 | 19 | ||
| Diameter (cm) | ||||||
| <5 | 157 | 32 | 34 | 91 | 1.699 | 0.428 |
| ≥5 | 100 | 14 | 23 | 63 | ||
| Symptom | ||||||
| intracranial hypertension | 100 | 15 | 22 | 63 | 29.090 | <0.001 |
| neurological dysfunction | 100 | 10 | 22 | 68 | ||
| epilepsy | 39 | 16 | 10 | 13 | ||
| mental disorders | 8 | 1 | 0 | 7 | ||
| no obvious symptoms | 10 | 4 | 3 | 3 | ||
| Tumor distribution | ||||||
| left side | 123 | 24 | 27 | 72 | 3.695 | 0.718 |
| right side | 97 | 15 | 25 | 57 | ||
| bilateral | 21 | 4 | 4 | 13 | ||
| middle | 16 | 3 | 1 | 12 | ||
| Tumor location | ||||||
| frontal lobe | 101 | 28 | 24 | 49 | 25.461 | 0.001 |
| temporal lobe | 63 | 4 | 11 | 48 | ||
| parietooccipital lobe | 53 | 8 | 8 | 37 | ||
| insular lobe | 14 | 0 | 6 | 8 | ||
| others | 26 | 6 | 8 | 12 | ||
| Surgical type | ||||||
| subtotal resection | 154 | 33 | 30 | 91 | 5.371 | 0.251 |
| partial resection | 80 | 10 | 23 | 47 | ||
| stereotactic biopsy | 23 | 3 | 4 | 16 | ||
| Postoperative chemotherapy | ||||||
| yes | 204 | 32 | 43 | 129 | 5.058 | 0.080 |
| no | 53 | 14 | 14 | 25 | ||
| Postoperative radiotherapy | ||||||
| yes | 185 | 3 | 12 | 57 | 18.090 | <0.001 |
| no | 72 | 43 | 45 | 97 | ||
| ATRX | ||||||
| positive | 171 | 10 | 26 | 135 | 83.536 | <0.001 |
| negative | 86 | 36 | 31 | 19 | ||
| IDH1 | ||||||
| positive | 62 | 24 | 17 | 21 | 30.038 | <0.001 |
| negative | 195 | 22 | 40 | 133 | ||
| p53 | ||||||
| positive | 106 | 29 | 31 | 46 | 21.304 | <0.001 |
| negative | 151 | 17 | 26 | 108 | ||
| Ki67 | ||||||
| weakly positive | 84 | 42 | 32 | 10 | 134.111 | <0.001 |
| Strong positive | 173 | 4 | 25 | 144 | ||
| OS | |||
|---|---|---|---|
| HR | 95%CI | p Value | |
| Gender (male/female) | 0.913 | 0.683–1.220 | 0.540 |
| Age (<60/≥60) | 3.502 | 2.555–4.800 | <0.001 |
| Smoking history (no/yes) | 0.799 | 0.585–1.092 | 0.159 |
| KPS score(<80/≥80) | 0.114 | 0.069–0.188 | <0.001 |
| Diameter (<5 cm/≥5 cm) | 1.514 | 1.129–2.030 | 0.006 |
| History of head trauma (no/yes) | 1.145 | 0.674–1.944 | 0.616 |
| WHO grade | |||
| WHO II | 1 * | 0 * | |
| WHO III | 3.477 | 1.784–6.775 | <0.001 |
| WHO IV | 14.739 | 7.995–27.170 | <0.001 |
| Postoperative radiotherapy (no/yes) | 0.281 | 0.204–0.388 | <0.001 |
| Postoperative chemotherapy (no/yes) | 0.739 | 0.512–1.068 | 0.107 |
| ATRX (−/+) | 11.042 | 7.083–17.213 | <0.001 |
| IDH1 (−/+) | 0.221 | 0.140–0.350 | <0.001 |
| p53 (−/+) | 0.475 | 0.349–0.646 | <0.001 |
| Ki67 (weak+/strong+) | 6.611 | 4.402–9.929 | <0.001 |
| Training Set (n = 154) | Validation Set (n = 103) | CGGA (n = 100) | χ2 | p | |
|---|---|---|---|---|---|
| Gender | |||||
| male | 91 (59.1%) | 50 (48.5%) | 61 (61.0%) | 3.898 | 0.142 |
| female | 63 (40.9%) | 53 (51.1%) | 39 (39.0%) | ||
| Age (year) | |||||
| <60 | 78 (50.6%) | 56 (54.4%) | 52 (52.0%) | 0.343 | 0.843 |
| ≥60 | 76 (49.4%) | 47 (45.6%) | 48 (48.0%) | ||
| WHO grade | |||||
| WHO II | 28 (18.2%) | 19 (18.4%) | 18 (18.0%) | 1.038 | 0.904 |
| WHO III | 31 (20.1%) | 26 (25.2%) | 22 (22.0%) | ||
| WHO IV | 95 (61.7%) | 58 (56.3%) | 60 (60.0%) | ||
| Postoperative radiotherapy | |||||
| yes | 112 (72.7%) | 75 (72.8%) | 89 (89.0%) | 10.821 | 0.004 |
| no | 42 (27.3%) | 28 (27.2%) | 11 (11.0%) | ||
| Postoperative chemotherapy | |||||
| yes | 121 (78.6%) | 77 (74.8%) | 69 (69.0%) | 2.946 | 0.229 |
| no | 33 (21.4%) | 26 (25.2%) | 31 (31.0%) | ||
| ATRX | |||||
| negative | 45 (29.2%) | 38 (36.9%) | 37 (37.0%) | 2.342 | 0.310 |
| positive | 109 (70.8%) | 65 (63.1%) | 63 (63.0%) | ||
| IDH1 | |||||
| negative | 113 (73.4%) | 86 (71.8%) | 70 (70.0%) | 0.344 | 0.842 |
| positive | 41 (26.6%) | 171 (28.2%) | 30 (30.0%) | ||
| Ki-67 | |||||
| weak+ | 50 (32.5%) | 35 (34.0%) | 30 (30.0%) | 0.376 | 0.829 |
| strong+ | 104 (67.5%) | 68 (66.0%) | 70 (70.0%) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Zhang, Z.; Zhao, Y.; Liu, Y.; Yuan, J.; Wang, Y.; Li, J. The Prognostic Value of a Nomogram Model Based on Tumor Immune Markers and Clinical Factors for Adult Primary Glioma. Cancers 2025, 17, 3043. https://doi.org/10.3390/cancers17183043
Wen J, Zhang Z, Zhao Y, Liu Y, Yuan J, Wang Y, Li J. The Prognostic Value of a Nomogram Model Based on Tumor Immune Markers and Clinical Factors for Adult Primary Glioma. Cancers. 2025; 17(18):3043. https://doi.org/10.3390/cancers17183043
Chicago/Turabian StyleWen, Junpeng, Ziling Zhang, Yan Zhao, Yingzi Liu, Jiangwei Yuan, Yuxiang Wang, and Juan Li. 2025. "The Prognostic Value of a Nomogram Model Based on Tumor Immune Markers and Clinical Factors for Adult Primary Glioma" Cancers 17, no. 18: 3043. https://doi.org/10.3390/cancers17183043
APA StyleWen, J., Zhang, Z., Zhao, Y., Liu, Y., Yuan, J., Wang, Y., & Li, J. (2025). The Prognostic Value of a Nomogram Model Based on Tumor Immune Markers and Clinical Factors for Adult Primary Glioma. Cancers, 17(18), 3043. https://doi.org/10.3390/cancers17183043







