Methylation Analyses in Liquid Biopsy of Lung Cancer Patients: A Novel and Intriguing Approach Against Resistance to Target Therapies and Immunotherapies
Simple Summary
Abstract
1. Introduction
2. Aberrant DNA Methylation in Cancer
3. cfDNA Methylation Analysis Methods
3.1. Bisulfite Conversion-Cased Methods
3.2. Bisulfite Conversion-Free Methods
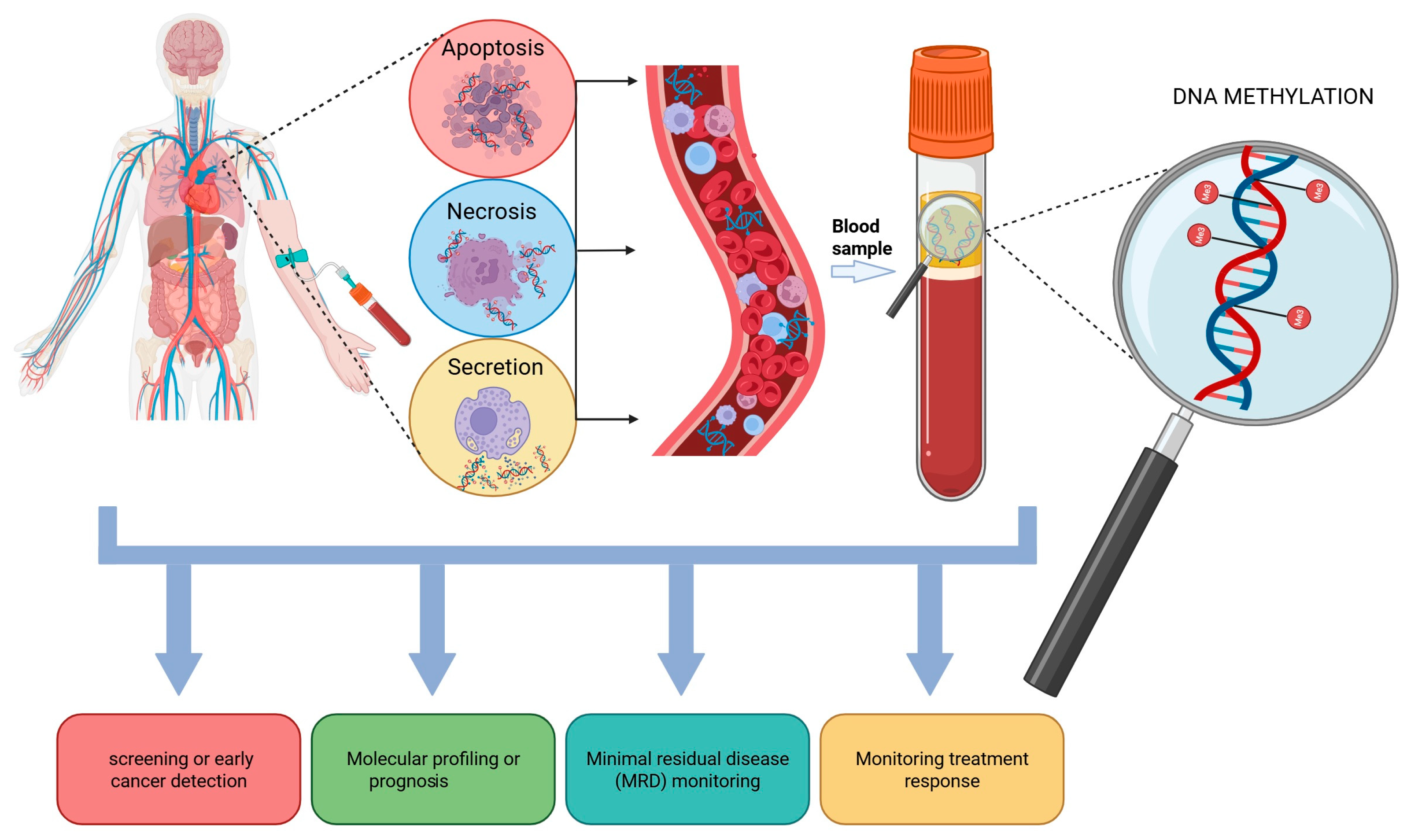
4. cf/ctDNA Origin and Clinical Applications in Lung Cancer
ctDNA Methylation as Biomarkers in Lung Cancer
5. Aberrant DNA Methylation in NSCLC Resistance
5.1. Aberrant Methylation and ctDNA Methylation Linked to KRAS G12Ci Resistance
5.2. ct(f)DNA Methylation Linked to TKI Resistance
5.3. Aberrant Methylation and ctDNA Methylation Linked to ICI-Resistance
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Vrba, L.; Oshiro, M.M.; Kim, S.S.; Garland, L.L.; Placencia, C.; Mahadevan, D.; Nelson, M.A.; Futscher, B.W. DNA methylation biomarkers discovered in silico detect cancer in liquid biopsies from non-small cell lung cancer patients. Epigenetics 2019, 15, 419–430. [Google Scholar] [CrossRef]
- Esteller, M.; Dawson, M.A.; Kadoch, C.; Rassool, F.V.; Jones, P.A.; Baylin, S.B. The Epigenetic Hallmarks of Cancer. Cancer Discov. 2024, 14, 1783–1809. [Google Scholar] [CrossRef] [PubMed]
- Duruisseaux, M.; Esteller, M. Lung cancer epigenetics: From knowledge to applications. Semin. Cancer Biol. 2018, 51, 116–128. [Google Scholar] [CrossRef]
- Bjaanæs, M.M.; Fleischer, T.; Halvorsen, A.R.; Daunay, A.; Busato, F.; Solberg, S.; Jørgensen, L.; Kure, E.; Edvardsen, H.; Børresen-Dale, A.L.; et al. Genome-wide DNA methylation analyses in lung adenocarcinomas: Association with EGFR, KRAS and TP53 mutation status, gene expression and prognosis. Mol. Oncol. 2016, 10, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Davalos, V.; Esteller, M. Cancer epigenetics in clinical practice. CA Cancer J. Clin. 2023, 73, 376–424. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, F.P.; Sparaneo, A.; Muscarella, L.A. Monitoring EGFR-lung cancer evolution: A possible beginning of a “methylation era” in TKI resistance prediction. Front. Oncol. 2023, 13, 1137384. [Google Scholar] [CrossRef]
- Santarpia, M.; Liguori, A.; Karachaliou, N.; Gonzalez-Cao, M.; Daffinà, M.G.; D’AVeni, A.; Marabello, G.; Altavilla, G.; Rosell, R. Osimertinib in the treatment of non-small-cell lung cancer: Design, development and place in therapy. Lung Cancer Targets Ther. 2017, 8, 109–125. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Role of Mammalian DNA Methyltransferases in Development. Annu. Rev. Biochem. 2020, 89, 135–158. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. In Epigenetics: Development and Disease; Kundu, T.K., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 289–317. [Google Scholar] [CrossRef]
- Yang, X.; Han, H.; De Carvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef]
- Straussman, R.; Nejman, D.; Roberts, D.; Steinfeld, I.; Blum, B.; Benvenisty, N.; Simon, I.; Yakhini, Z.; Cedar, H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009, 16, 564–571. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Sung, K.W.K.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Almouzni, G.; Cedar, H. Maintenance of Epigenetic Information. Cold Spring Harb. Perspect. Biol. 2016, 8, a019372. [Google Scholar] [CrossRef]
- Baxter, E.; Windloch, K.; Gannon, F.; Lee, J.S. Epigenetic regulation in cancer progression. Cell Biosci. 2014, 4, 45. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.; Gehrke, C.; Kuo, K.; Ehrlich, M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988, 48, 1159–1161. [Google Scholar] [PubMed]
- Gama-Sosa, M.A.; Slagel, V.A.; Trewyn, R.W.; Oxenhandler, R.; Kuo, K.C.; Gehrke, C.W.; Ehrlich, M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983, 11, 6883–6894. [Google Scholar] [CrossRef] [PubMed]
- Alagia, A.; Gullerova, M. The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine. Front. Cell Dev. Biol. 2022, 10, 915685. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene Silencing in Cancer in Association with Promoter Hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef]
- Greger, V.; Passarge, E.; Höpping, W.; Messmer, E.; Horsthemke, B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum. Genet. 1989, 83, 155–158. [Google Scholar] [CrossRef]
- Sproul, D.; Kitchen, R.R.; Nestor, C.E.; Dixon, J.M.; Sims, A.H.; Harrison, D.J.; Ramsahoye, B.H.; Meehan, R.R. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol. 2012, 13, R84. [Google Scholar] [CrossRef]
- Li, E.; Beard, C.; Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 1993, 366, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. Aberrant Epigenetic Landscape in Cancer: How Cellular Identity Goes Awry. Dev. Cell. 2010, 19, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Gaudet, F.; Waghmare, A.; Jaenisch, R. Chromosomal Instability and Tumors Promoted by DNA Hypomethylation. Science 2003, 300, 455. [Google Scholar] [CrossRef] [PubMed]
- Karpf, A.R.; Matsui, S.-I. Genetic Disruption of Cytosine DNA Methyltransferase Enzymes Induces Chromosomal Instability in Human Cancer Cells. Cancer Res. 2005, 65, 8635–8639. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D.; et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011, 43, 768–775. [Google Scholar] [CrossRef]
- Hon, G.C.; Hawkins, R.D.; Caballero, O.L.; Lo, C.; Lister, R.; Pelizzola, M.; Valsesia, A.; Ye, Z.; Kuan, S.; Edsall, L.E.; et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2011, 22, 246–258. [Google Scholar] [CrossRef]
- Li, J.; Xu, C.; Lee, H.J.; Ren, S.; Zi, X.; Zhang, Z.; Wang, H.; Yu, Y.; Yang, C.; Gao, X.; et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020, 580, 93–99. [Google Scholar] [CrossRef]
- Brinkman, A.B.; Nik-Zainal, S.; Simmer, F.; Rodríguez-González, F.G.; Smid, M.; Alexandrov, L.B.; Butler, A.; Martin, S.; Davies, H.; Glodzik, D.; et al. Partially methylated domains are hypervariable in breast cancer and fuel widespread CpG island hypermethylation. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer: Current status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 1–36. [Google Scholar] [CrossRef]
- Luo, H.; Wei, W.; Ye, Z.; Zheng, J.; Xu, R.-H. Liquid Biopsy of Methylation Biomarkers in Cell-Free DNA. Trends Mol. Med. 2021, 27, 482–500. [Google Scholar] [CrossRef] [PubMed]
- Ezegbogu, M.; Wilkinson, E.; Reid, G.; Rodger, E.J.; Brockway, B.; Russell-Camp, T.; Kumar, R.; Chatterjee, A. Cell-free DNA methylation in the clinical management of lung cancer. Trends Mol. Med. 2024, 30, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Zhang, Z.; Zheng, J.; Zhang, W.; Cai, J. 5-Hydroxymethylcytosine modifications in circulating cell-free DNA: Frontiers of cancer detection, monitoring, and prognostic evaluation. Biomark. Res. 2025, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; He, B.; Yi, C.; Peng, J. Liquid biopsies: DNA methylation analyses in circulating cell-free DNA. J. Genet. Genom. 2018, 45, 185–192. [Google Scholar] [CrossRef]
- Soto, J.; Rodriguez-Antolin, C.; Vallespín, E.; Carpeño, J.d.C.; de Caceres, I.I. The impact of next-generation sequencing on the DNA methylation–based translational cancer research. Transl. Res. 2016, 169, 1–18.e1. [Google Scholar] [CrossRef]
- Patil, V.; Ward, R.L.; Hesson, L.B. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 2014, 9, 823–828. [Google Scholar] [CrossRef]
- Nair, S.S.; Luu, P.-L.; Qu, W.; Maddugoda, M.; Huschtscha, L.; Reddel, R.; Chenevix-Trench, G.; Toso, M.; Kench, J.G.; Horvath, L.G.; et al. Guidelines for whole genome bisulphite sequencing of intact and FFPET DNA on the Illumina HiSeq X Ten. Epigenet. Chromatin 2018, 11, 1–20. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, H.; An, K.; Liu, Z.; Hai, L.; Li, R.; Zhou, Y.; Zhao, W.; Jia, Y.; Wu, N.; et al. Whole-genome bisulfite sequencing analysis of circulating tumour DNA for the detection and molecular classification of cancer. Clin. Transl. Med. 2022, 12, e1014. [Google Scholar] [CrossRef]
- Huang, J.; Wang, L. Cell-Free DNA Methylation Profiling Analysis—Technologies and Bioinformatics. Cancers 2019, 11, 1741. [Google Scholar] [CrossRef]
- Yong, W.-S.; Hsu, F.-M.; Chen, P.-Y. Profiling genome-wide DNA methylation. Epigenet. Chromatin. 2016, 9, 26. [Google Scholar] [CrossRef]
- Widschwendter, M.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Ryan, A.; Gentry-Maharaj, A.; Zikan, M.; Cibula, D.; Eichner, J.; et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Zikan, M.; Wahl, B.; Lempiäinen, H.; Paprotka, T.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Eichner, J.; et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Lu, P.; Zhou, X.; Liao, Y.; Liu, X.; Li, J.; Wang, W.; Wang, J.; Wen, L.; Fu, W.; et al. Genome-Scale Methylation Analysis of Circulating Cell-Free DNA in Gastric Cancer Patients. Clin. Chem. 2021, 68, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Li, J.; Guo, H.; Liu, X.; Zheng, S.; Zhang, D.; Zhu, W.; Qu, J.; Guo, L.; Du, D.; et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res. 2015, 25, 1250–1264. [Google Scholar] [CrossRef]
- Sigalotti, L.; Covre, A.; Colizzi, F.; Fratta, E. Quantitative Methylation-Specific PCR: A Simple Method for Studying Epigenetic Modifications of Cell-Free DNA. Methods Mol. Biol. 2019, 1909, 137–162. [Google Scholar] [CrossRef]
- Estival, A.; Sanz, C.; Ramirez, J.-L.; Velarde, J.M.; Domenech, M.; Carrato, C.; De Las Peñas, R.; Gil-Gil, M.; Sepúlveda, J.; Armengol, R.; et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 2019, 9, 11125. [Google Scholar] [CrossRef]
- Huang, G.; Krocker, J.D.; Kirk, J.L.; Merwat, S.N.; Ju, H.; Soloway, R.D.; Wieck, L.R.; Li, A.; Okorodudu, A.O.; Petersen, J.R.; et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin. Chem. Lab. Med. CCLM 2014, 52, 899–909. [Google Scholar] [CrossRef]
- van Ginkel, J.H.; Huibers, M.M.H.; van Es, R.J.J.; de Bree, R.; Willems, S.M. Droplet digital PCR for detection and quantification of circulating tumor DNA in plasma of head and neck cancer patients. BMC Cancer 2017, 17, 428. [Google Scholar] [CrossRef]
- Liu, L.; Toung, J.; Jassowicz, A.; Vijayaraghavan, R.; Kang, H.; Zhang, R.; Kruglyak, K.; Huang, H.; Hinoue, T.; Shen, H.; et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018, 29, 1445–1453. [Google Scholar] [CrossRef]
- Stackpole, M.L.; Zeng, W.; Li, S.; Liu, C.-C.; Zhou, Y.; He, S.; Yeh, A.; Wang, Z.; Sun, F.; Li, Q.; et al. Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat. Commun. 2022, 13, 5566. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, W.; Liu, B.; Krysan, K.; Dubinett, S.M. Noninvasive Lung Cancer Subtype Classification Using Tumor-Derived Signatures and cfDNA Methylome. Cancer Res. Commun. 2024, 4, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Kresse, S.H.; Brandt-Winge, S.; Pharo, H.; Flatin, B.T.B.; Jeanmougin, M.; Vedeld, H.M.; Lind, G.E. Evaluation of commercial kits for isolation and bisulfite conversion of circulating cell-free tumor DNA from blood. Clin. Epigenet.. 2023, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.; Arribas, C.; Esteller, M. Validation of a DNA Methylation Microarray for 850,000 CpG Sites of the Human Genome Enriched in Enhancer Sequences. Epigenomics 2015, 8, 389–399. [Google Scholar] [CrossRef]
- Gallardo-Gómez, M.; Moran, S.; de la Cadena, M.P.; Martínez-Zorzano, V.S.; Rodríguez-Berrocal, F.J.; Rodríguez-Girondo, M.; Esteller, M.; Cubiella, J.; Bujanda, L.; Castells, A.; et al. A new approach to epigenome-wide discovery of non-invasive methylation biomarkers for colorectal cancer screening in circulating cell-free DNA using pooled samples. Clin. Epigenet. 2018, 10, 53. [Google Scholar] [CrossRef]
- Cheruba, E.; Viswanathan, R.; Wong, P.-M.; Womersley, H.J.; Han, S.; Tay, B.; Lau, Y.; Gan, A.; Poon, P.S.Y.; Skanderup, A.; et al. Heat selection enables highly scalable methylome profiling in cell-free DNA for noninvasive monitoring of cancer patients. Sci. Adv. 2022, 8, eabn4030. [Google Scholar] [CrossRef]
- Diep, D.; Plongthongkum, N.; Gore, A.; Fung, H.-L.; Shoemaker, R.; Zhang, K. Library-free methylation sequencing with bisulfite padlock probes. Nat. Methods 2012, 9, 270–272. [Google Scholar] [CrossRef]
- Xu, R.-H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef]
- Xu, W.; Lu, J.; Zhao, Q.; Wu, J.; Sun, J.; Han, B.; Zhao, X.; Kang, Y. Genome-Wide Plasma Cell-Free DNA Methylation Profiling Identifies Potential Biomarkers for Lung Cancer. Dis. Markers 2019, 2019, 4108474. [Google Scholar] [CrossRef]
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Alaiwi, S.A.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 2020, 26, 1041–1043. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Zhao, Q.; Wang, Z.; Lu, S.; Kang, Y.; Jin, G.; Tian, J. Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer. Front. Genet. 2020, 11, 596078. [Google Scholar] [CrossRef]
- Huang, J.; Soupir, A.C.; Wang, L. Cell-free DNA methylome profiling by MBD-seq with ultra-low input. Epigenetics 2021, 17, 239–252. [Google Scholar] [CrossRef]
- Lleshi, E.; Milne-Clark, T.; Yu, H.L.; Martin, H.W.; Hanson, R.; Lach, R.; Rossi, S.H.; Riediger, A.L.; Görtz, M.; Sültmann, H.; et al. Prostate cancer detection through unbiased capture of methylated cell-free DNA. iScience 2024, 27, 110330. [Google Scholar] [CrossRef]
- Behrouzi, R.; Clipson, A.; Simpson, K.L.; Blackhall, F.; Rothwell, D.G.; Dive, C.; Mouliere, F. Cell-free and extrachromosomal DNA profiling of small cell lung cancer. Trends Mol. Med. 2024, 31, 64–78. [Google Scholar] [CrossRef]
- Shao, J.; Wang, S.; West-Szymanski, D.; Karpus, J.; Shah, S.; Ganguly, S.; Smith, J.; Zu, Y.; He, C.; Li, Z. Cell-free DNA 5-hydroxymethylcytosine is an emerging marker of acute myeloid leukemia. Sci. Rep. 2022, 12, 12410. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-X.; Szulwach, K.E.; Fu, Y.; Dai, Q.; Yi, C.; Li, X.; Li, Y.; Chen, C.-H.; Zhang, W.; Jian, X.; et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011, 29, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Lu, X.; Shih, A.H.; Nie, J.; You, Q.; Xu, M.M.; Melnick, A.M.; Levine, R.L.; He, C. A Highly Sensitive and Robust Method for Genome-wide 5hmC Profiling of Rare Cell Populations. Mol. Cell. 2016, 63, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017, 27, 1243–1257. [Google Scholar] [CrossRef]
- Kwon, H.-J.; Shin, S.H.; Kim, H.H.; Min, N.Y.; Lim, Y.; Joo, T.-W.; Lee, K.J.; Jeong, M.-S.; Kim, H.; Yun, S.-Y.; et al. Advances in methylation analysis of liquid biopsy in early cancer detection of colorectal and lung cancer. Sci. Rep. 2023, 13, 13502. [Google Scholar] [CrossRef]
- Ko, K.; Kananazawa, Y.; Yamada, T.; Kakinuma, D.; Matsuno, K.; Ando, F.; Kuriyama, S.; Matsuda, A.; Yoshida, H. Methylation status and long-fragment cell-free DNA are prognostic biomarkers for gastric cancer. Cancer Med. 2021, 10, 2003–2012. [Google Scholar] [CrossRef]
- Jensen, S.Ø.; Øgaard, N.; Ørntoft, M.-B.W.; Rasmussen, M.H.; Bramsen, J.B.; Kristensen, H.; Mouritzen, P.; Madsen, M.R.; Madsen, A.H.; Sunesen, K.G.; et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer—A clinical biomarker discovery and validation study. Clin. Epigenet. 2019, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, X.; Zhang, H.; Wang, K.; He, J. Cell-free circulating tumor DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget 2017, 8, 75742–75755. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.; Zahalka, T.; Fechner, G.; Müller, S.C.; Ellinger, J. Serum DNA hypermethylation in patients with kidney cancer: Results of a prospective study. Anticancer Res. 2013, 33, 4651–4656. [Google Scholar]
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation of the SHOX2 / PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 2017, 12, 77–84. [Google Scholar] [CrossRef]
- Skrypkina, I.; Tsyba, L.; Onyshchenko, K.; Morderer, D.; Kashparova, O.; Nikolaienko, O.; Panasenko, G.; Vozianov, S.; Romanenko, A.; Rynditch, A. Concentration and Methylation of Cell-Free DNA from Blood Plasma as Diagnostic Markers of Renal Cancer. Dis. Markers 2016, 2016, 3693096. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, J.W.; Kristensen, S.S.; Tholstrup, D.; Harsløf, L.B.S.; Pedersen, O.B.; Brown, P.D.N.; Grønbæk, K. Aberrant methylation of cell-free circulating DNA in plasma predicts poor outcome in diffuse large B cell lymphoma. Clin. Epigenet. 2016, 8, 95. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C R Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2020, 124, 345–358. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Angeles, A.K.; Janke, F.; Bauer, S.; Christopoulos, P.; Riediger, A.L.; Sültmann, H. Liquid Biopsies beyond Mutation Calling: Genomic and Epigenomic Features of Cell-Free DNA in Cancer. Cancers 2021, 13, 5615. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rodriguez, I.R.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Papadopoulos, N. Pathophysiology of ctDNA Release into the Circulation and Its Characteristics: What Is Important for Clinical Applications. Recent Results Cancer Res. 2020, 215, 163–180. [Google Scholar] [CrossRef]
- Warton, K.; Lin, V.; Navin, T.; Armstrong, N.J.; Kaplan, W.; Ying, K.; Gloss, B.; Mangs, H.; Nair, S.S.; Hacker, N.F.; et al. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genom. 2014, 15, 476. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.M.; Spellman, P.T.; Gray, J. Circulating-tumor DNA as an early detection and diagnostic tool. Curr. Opin. Genet. Dev. 2017, 42, 14–21. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta BBA Rev. Cancer. 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Khier, S.; Lohan, L. Kinetics of Circulating Cell-Free DNA for Biomedical Applications: Critical Appraisal of the Literature. Futur. Sci. OA 2018, 4, FSO295. [Google Scholar] [CrossRef]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.; Kinzler, K.; Pantel, K.; Alix-Panabières, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid bi-opsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Van Seventer, E.E.; Siravegna, G.; Hartwig, A.V.; Jaimovich, A.; He, Y.; Kanter, K.; Fish, M.G.; Fosbenner, K.D.; Miao, B.; et al. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin. Cancer Res. 2021, 27, 5586–5594. [Google Scholar] [CrossRef]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Mason, P.; Carbone, D.; Cushman, R.; Waggoner, A. The importance of inorganic phosphate in regulation of energy me-tabolism of Streptococcus lactis. J. Biol. Chem. 1981, 256, 1861–1866. [Google Scholar] [CrossRef]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef]
- Moran, S.; Martínez-Cardús, A.; Sayols, S.; Musulén, E.; Balañá, C.; Estival-Gonzalez, A.; Moutinho, C.; Heyn, H.; Diaz-Lagares, A.; de Moura, M.C.; et al. Epigenetic profiling to classify cancer of unknown primary: A multicentre, retrospective analysis. Lancet Oncol. 2016, 17, 1386–1395. [Google Scholar] [CrossRef]
- Feng, H.; Jin, P.; Wu, H. Disease prediction by cell-free DNA methylation. Brief. Bioinform. 2018, 20, 585–597. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e296. [Google Scholar] [CrossRef]
- Borg, M.; Wen, S.W.C.; Andersen, R.F.; Timm, S.; Hansen, T.F.; Hilberg, O. Methylated Circulating Tumor DNA in Blood as a Tool for Diagnosing Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 3959. [Google Scholar] [CrossRef] [PubMed]
- Maffeo, D.; Rina, A.; Serio, V.B.; Markou, A.; Powrózek, T.; Constâncio, V.; Nunes, S.P.; Jerónimo, C.; Calvo, A.; Mari, F.; et al. The Evidence Base for Circulating Tumor DNA-Methylation in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2024, 16, 3641. [Google Scholar] [CrossRef] [PubMed]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.W.Y.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA Methylation as a Noninvasive Epigenetic Biomarker for the Detection of Cancer. Dis. Markers 2017, 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Jones, A.; Evans, I.; Reisel, D.; Dillner, J.; Sundström, K.; Steyerberg, E.W.; Vergouwe, Y.; Wegwarth, O.; Rebitschek, F.G.; et al. Epigenome-based cancer risk prediction: Rationale, opportunities and challenges. Nat. Rev. Clin. Oncol. 2018, 15, 292–309. [Google Scholar] [CrossRef]
- Kontic, M.; Markovic, F. Use of DNA methylation patterns for early detection and management of lung cancer: Are we there yet? Oncol. Res. 2025, 33, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Mastoraki, S.; Balgkouranidou, I.; Tsaroucha, E.; Klinakis, A.; Georgoulias, V.; Lianidou, E. KMT2C promoter methylation in plasma-circulating tumor DNA is a prognostic biomarker in non-small cell lung cancer. Mol. Oncol. 2020, 15, 2412–2422. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology. npj Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef]
- Tian, F.-M.; Zhong, C.-Y.; Wang, X.-N.; Meng, Y. PDE3A is hypermethylated in cisplatin resistant non-small cell lung cancer cells and is a modulator of chemotherapy response. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2635–2641. [Google Scholar] [PubMed]
- Zhao, J.; Xue, X.; Fu, W.; Dai, L.; Jiang, Z.; Zhong, S.; Deng, B.; Yin, J. Epigenetic activation of FOXF1 confers cancer stem cell properties to cisplatin resistant non small cell lung cancer. Int. J. Oncol. 2020, 56, 1083–1092. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Yang, J.; Li, B.; Wang, J. CDH13 promoter methylation regulates cisplatin resistance of non-small cell lung cancer cells. Oncol. Lett. 2018, 16, 5715–5722. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, H.; Gan, Y.; Cui, C.; Zhang, B.; Gu, L.; Zhou, J.; Zhu, G.; Deng, D. P16 Methylation Leads to Paclitaxel Re-sistance of Advanced Non-Small Cell Lung Cancer. J. Cancer 2019, 10, 1726–1733. [Google Scholar] [CrossRef]
- Li, X.-Y.; Wu, J.-Z.; Cao, H.-X.; Ma, R.; Zhong, Y.-J.; Feng, J.-F. Blockade of DNA methylation enhances the therapeutic effect of gefitinib in non-small cell lung cancer cells. Oncol. Rep. 2013, 29, 1975–1982. [Google Scholar] [CrossRef]
- Noro, R.; Gemma, A.; Miyanaga, A.; Kosaihira, S.; Minegishi, Y.; Nara, M.; Kokubo, Y.; Seike, M.; Kataoka, K.; Matsuda, K.; et al. PTEN inactivation in lung cancer cells and the effect of its recovery on treatment with epidermal growth factor receptor tyrosine kinase inhibitors. Int. J. Oncol. 2007, 31, 1157–1163. [Google Scholar]
- Su, S.-F.; Liu, C.-H.; Cheng, C.-L.; Ho, C.-C.; Yang, T.-Y.; Chen, K.-C.; Hsu, K.-H.; Tseng, J.-S.; Chen, H.-W.; Chang, G.-C.; et al. Genome-Wide Epigenetic Landscape of Lung Adenocarcinoma Links HOXB9 DNA Methylation to Intrinsic EGFR-TKI Resistance and Heterogeneous Responses. JCO Precis. Oncol. 2021, 5, 418–431. [Google Scholar] [CrossRef]
- Niu, X.; Liu, F.; Zhou, Y.; Zhou, Z.; Zhou, D.; Wang, T.; Li, Z.; Ye, X.; Yu, Y.; Weng, X.; et al. Genome-wide DNA Methyla-tion Analysis Reveals GABBR2 as a Novel Epigenetic Target for EGFR 19 Deletion Lung Adenocarcinoma with Induction Erlotinib Treatment. Clin. Cancer Res. 2017, 23, 5003–5014. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, Y.; Duan, J.; Bai, H.; Wang, Z.; Wei, L.; Zhao, J.; Zhuo, M.; Wang, S.; Yang, L.; et al. DNA Methylation status of Wnt antagonist SFRP5 can predict the response to the EGFR-tyrosine kinase inhibitor therapy in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2012, 31, 80. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Liggett, T.E.; Melnikov, A.A.; Monitto, C.L.; Kusuke, D.; Shiga, K.; Kobayashi, T.; Horii, A.; Chatterjee, A.; Levenson, V.V.; et al. Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle 2012, 11, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, Z.-G.; Gao, B.; Shao, G.-G.; Li, G.-H. 5-Aza-CdR can reverse gefitinib resistance caused by DAPK gene promoter methylation in lung adenocarcinoma cells. Int. J. Clin. Exp. Pathol. 2015, 8, 12961. [Google Scholar] [PubMed]
- Terai, H.; Soejima, K.; Yasuda, H.; Sato, T.; Naoki, K.; Ikemura, S.; Arai, D.; Ohgino, K.; Ishioka, K.; Hamamoto, J.; et al. Long-term exposure to gefitinib induces acquired resistance through DNA methylation changes in the EGFR-mutant PC9 lung cancer cell line. Int. J. Oncol. 2014, 46, 430–436. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Zhang, L.; Xu, W.; Li, J.; Liu, Y.; Zeng, X.; Zhong, M.; Zhu, Y. The Multi-Omics Analysis of Key Genes Regulating EGFR-TKI Resistance, Immune Infiltration, SCLC Transformation in EGFR-Mutant NSCLC. J. Inflamm. Res. 2022, 15, 649–667. [Google Scholar] [CrossRef]
- Hou, T.; Ma, J.; Hu, C.; Zou, F.; Jiang, S.; Wang, Y.; Han, C.; Zhang, Y. Decitabine reverses gefitinib resistance in PC9 lung adenocarcinoma cells by demethylation of RASSF1A and GADD45β promoter. Int. J. Clin. Exp. Pathol. 2019, 12, 4002–4010. [Google Scholar]
- Zhao, M.; Zhang, Y.; Li, J.; Li, X.; Cheng, N.; Wang, Q.; Cai, W.; Zhao, C.; He, Y.; Chang, J.; et al. Histone deacetylation, as opposed to promoter methylation, results in epigenetic BIM silencing and resistance to EGFR TKI in NSCLC. Oncol. Lett. 2017, 15, 1089–1096. [Google Scholar] [CrossRef]
- Jia, X.; Tian, J.; Chen, P.; Dong, J.; Li, L.; Chen, D.; Zhang, J.; Liao, D.; He, Z.; Luo, K. Methylation-modulated PFTK1 reg-ulates gefitinib resistance via Wnt/β-catenin signaling in EGFR mutant non-small-cell lung cancer cells. Commun. Biol. 2024, 7, 1649. [Google Scholar] [CrossRef]
- Kim, J.Y.; Choi, J.K.; Jung, H. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin. Epigenet. 2020, 12, 119. [Google Scholar] [CrossRef]
- Duruisseaux, M.; Martínez-Cardús, A.; Calleja-Cervantes, M.E.; Moran, S.; de Moura, M.C.; Davalos, V.; Piñeyro, D.; Sanchez-Cespedes, M.; Girard, N.; Brevet, M.; et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: A multicentre, retrospective analysis. Lancet Respir. Med. 2018, 6, 771–781. [Google Scholar] [CrossRef]
- Tew, B.Y.; Durand, J.K.; Bryant, K.L.; Hayes, T.K.; Peng, S.; Tran, N.L.; Gooden, G.C.; Buckley, D.N.; Der, C.J.; Baldwin, A.S.; et al. Genome-wide DNA methylation analysis of KRAS mutant cell lines. Sci. Rep. 2020, 10, 10149. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.-W.-C.; Andersen, R.-F.; Petersen, L.-M.; Hager, H.; Hilberg, O.; Jakobsen, A.; Hansen, T.-F. Comparison of Mutated KRAS and Methylated HOXA9 Tumour-Specific DNA in Advanced Lung Adenocarcinoma. Cancers 2020, 12, 3728. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Ye, J.; Chen, Y.; Lizaso, A.; Huang, L.; Shi, L.; Su, J.; Han-Zhang, H.; Chuai, S.; Li, L.; et al. Parallel serial assessment of somatic mutation and methylation profile from circulating tumor DNA predicts treatment response and impending disease progression in osimertinib-treated lung adenocarcinoma patients. Transl. Lung Cancer Res. 2019, 8, 1016–1028. [Google Scholar] [CrossRef]
- Ntzifa, A.; Londra, D.; Rampias, T.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. DNA Methylation Analysis in Plasma Cell-Free DNA and Paired CTCs of NSCLC Patients before and after Osimertinib Treatment. Cancers 2021, 13, 5974. [Google Scholar] [CrossRef]
- Ntzifa, A.; Marras, T.; Kallergi, G.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Comprehensive liquid biopsy analysis for monitoring NSCLC patients under second-line osimertinib treatment. Front. Oncol. 2024, 14, 1435537. [Google Scholar] [CrossRef]
- Shen, Z.; Chen, C.; Sun, J.; Huang, J.; Liu, S. The status of WIF1 methylation in cell-free DNA is associated with the in-susceptibility for gefitinib in the treatment of lung cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 2239–2248. [Google Scholar] [CrossRef]
- Nguyen, H.-N.; Cao, N.-P.T.; Van Nguyen, T.-C.; Le, K.N.D.; Nguyen, D.T.; Nguyen, Q.-T.T.; Nguyen, T.-H.T.; Van Ngu-yen, C.; Le, H.T.; Nguyen, M.-L.T.; et al. Liquid biopsy uncovers distinct patterns of DNA methylation and copy number changes in NSCLC patients with different EGFR-TKI resistant mutations. Sci. Rep. 2021, 11, 16436. [Google Scholar] [CrossRef]
- Fujimoto, M.; Yasuda, H.; Arai, E.; Nakajima, M.; Takata, S.; Morikawa, K.; Tanaka, H.; Itani, H.; Honda, T.; Horiuchi, K.; et al. Plasma cell-free DNA methylation profile before afatinib treatment is associated with progression-free and overall survival of patients with epidermal growth factor receptor gene mutation-positive non-small cell lung cancer. Clin. Epigenet. 2025, 17, 63. [Google Scholar] [CrossRef]
- El Zarif, T.; Meador, C.B.; Qiu, X.; Seo, J.-H.; Davidsohn, M.P.; Savignano, H.; Lakshminarayanan, G.; McClure, H.M.; Canniff, J.; Fortunato, B.; et al. Detecting Small Cell Transformation in Patients with Advanced EGFR Mutant Lung Ade-nocarcinoma through Epigenomic cfDNA Profiling. Clin. Cancer Res. 2024, 30, 3798–3811. [Google Scholar] [CrossRef]
- Janke, F.; Angeles, A.K.; Riediger, A.L.; Bauer, S.; Reck, M.; Stenzinger, A.; Schneider, M.A.; Muley, T.; Thomas, M.; Christopoulos, P.; et al. Longitudinal monitoring of cell-free DNA methylation in ALK-positive non-small cell lung cancer patients. Clin. Epigenet. 2022, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Li, X.; Gao, Y.; Guo, S.; Sun, D.; Zhou, H.; Sun, Y.; Wang, P.; Zhi, H.; Bai, J.; et al. MeImmS: Predict Clinical Benefit of Anti-PD-1/PD-L1 Treatments Based on DNA Methylation in Non-small Cell Lung Cancer. Front. Genet. 2021, 12, 676449. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Kim, H.S.; Kim, J.Y.; Sun, J.-M.; Ahn, J.S.; Ahn, M.-J.; Park, K.; Esteller, M.; Lee, S.-H.; Choi, J.K. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun. 2019, 10, 4278. [Google Scholar] [CrossRef] [PubMed]
- Guler, G.D.; Ning, Y.; Coruh, C.; Mognol, G.P.; Phillips, T.; Nabiyouni, M.; Hazen, K.; Scott, A.; Volkmuth, W.; Levy, S. Plasma cell-free DNA hydroxymethylation profiling reveals anti-PD-1 treatment response and resistance biology in non-small cell lung cancer. J. Immunother. Cancer 2024, 12, e008028. [Google Scholar] [CrossRef]
- Shao, J.; Xu, Y.; Olsen, R.J.; Kasparian, S.; Sun, K.; Mathur, S.; Zhang, J.; He, C.; Chen, S.-H.; Bernicker, E.H.; et al. 5-Hydroxymethylcytosine in Cell-Free DNA Predicts Immunotherapy Response in Lung Cancer. Cells 2024, 13, 715. [Google Scholar] [CrossRef]
- Hsiao, A.; Woodward, B.; Ye, P.; Varga, M.G.; Altaie, G.; Lu, K.; Searle, N.; Viens, R.; Langpap, S.; Li, Z.; et al. Brief Report: Methylation-Based ctDNA Serial Monitoring Correlates With Immunotherapy Response in NSCLC. Clin. Lung Cancer 2024, 26, 72–77. [Google Scholar] [CrossRef]
- Prelaj, A.; Miskovic, V.; Zanitti, M.; Trovo, F.; Genova, C.; Viscardi, G.; Rebuzzi, S.; Mazzeo, L.; Provenzano, L.; Kosta, S.; et al. Artificial intelligence for predictive biomarker discovery in immuno-oncology: A systematic review. Ann. Oncol. 2023, 35, 29–65. [Google Scholar] [CrossRef]
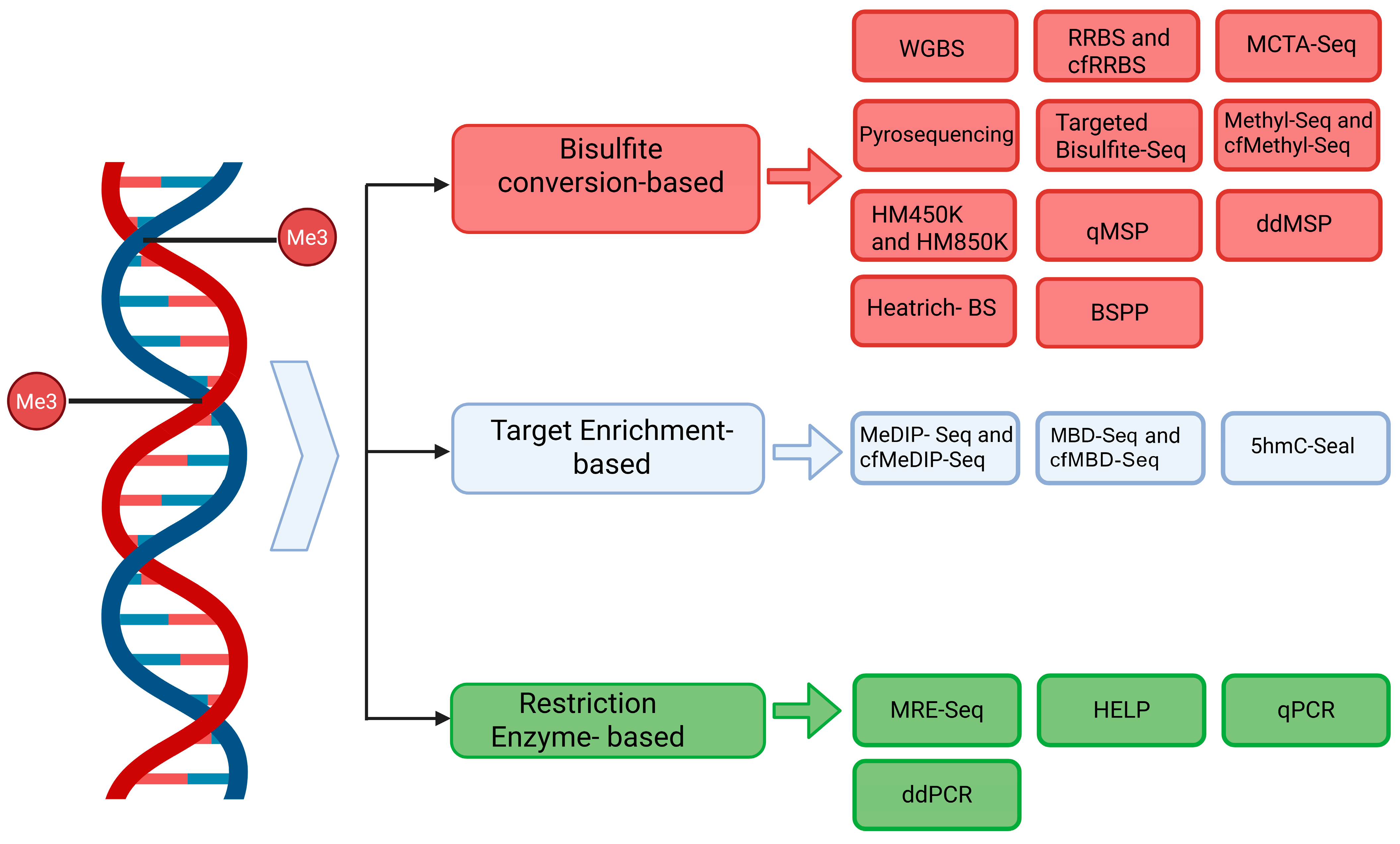
| Method | Technology Category | Target | Advantages | Disadvantages | Input | Refs |
|---|---|---|---|---|---|---|
| WGBS | Bisulfite-based | Genome-wide | Base-resolution, unbiased coverage | DNA degradation, high cost | ~125–250 pg | [39,40] |
| RRBS and cfRRBS | Bisulfite-based | CpG-rich regions | Cost-effective, single-base resolution | Requires high-quality DNA, limited coverage | ≥10 ng | [43,44] |
| MCTA-Seq | Bisulfite-based | CGGCGG-rich CpGs | High sensitivity, very low input | Sequence bias, limited regions | ~7.5 pg | [45,46] |
| Pyrosequencing | Bisulfite-based | Targeted CpGs | Quantitative, real-time analysis | Low sensitivity (<5%), not suitable for rare cfDNA | ~10–50 ng | [48,77] |
| Targeted Bisulfite-Seq | Bisulfite-based | Selected regions | High resolution, scalable | Complex primer/probe design | 50 ng | [51] |
| Methyl-Seq and cfMethyl-Seq | Bisulfite-based | CpG-rich cfDNA | Preserves cfDNA, UMIs for accuracy | Still requires bisulfite, potential loss | ~5–10 ng | [52,53,54] |
| HM450K and HM850K arrays | Bisulfite-based | Predesigned CpG panel | Hotspot methylation with high accuracy | Low genome-wide coverage | ~10 ng | [55,56] |
| qMSP | Bisulfite-based | Specific DMRs | Low input, high sensitivity | Locus-specific, limited multiplexing | ~20–100 ng | [33,47] |
| ddMSP | Bisulfite-based | Specific DMRs | Ultra-sensitive, quantifies rare methylation events, suitable for cfDNA | Locus-specific, limited to known biomarkers, complex setup | ~10–50 ng | [50] |
| Heatrich-BS | Bisulfite-based | CpG-dense cfDNA | Enrichment for CpG regions, efficient workflow | Not truly genome-wide | ~5–10 ng | [57] |
| BSPP | Bisulfite-based | Specific loci (targeted CpGs) | High specificity via padlock probes, applicable | Complex design, requires optimization for each locus | ~10–50 ng | [58,59] |
| MeDIP-Seq and cfMeDIP-Seq | Target enrichment-based | Methylated DNA | Preserves DNA, low input | Background noise, lower resolution | 1–10 ng | [60,61,62] |
| 5hmC-Seal | Target enrichment-based | 5hmC | Detects epigenetic variants (5hmC), high sensitivity | Complex protocol, enrichment adds cost | ~1–5 ng | [67,68,69] |
| MRE-Seq | Restriction enzyme-based | Unmethylated CpG sites | No bisulfite, methylation-sensitive digestion | Not ideal for cfDNA, low resolution | ~10–50 ng | [70] |
| HELP | Restriction enzyme-based | Specific CpG sites | No bisulfite, relatively simple | Low genome coverage, not ideal for cfDNA | ~10–50 ng | [71] |
| qPCR | Restriction enzyme-based | Probe-based PCR | Ultra-low input, fast turnaround | Very limited coverage, high false-positive/negative risk | ~10–50 ng | [64,75,76] |
| ddPCR | Restriction enzyme-based | Probe-based PCR | High sensitivity and precision, ideal for rare allele detection in cfDNA | Limited to known loci, low multiplexing capacity | ~10–50 ng | [72,73] |
| References | Target | Population and Therapy | Biological Effects |
|---|---|---|---|
| [135] | EGFR T790M mutation | 8 pts with IV stage LUAD osimertinib (post-1L) | Methylation levels are higher in ctDNA of pts with detectable somatic mutations than in pts without somatic mutations. The decrease in methylation levels and maxAF reflects treatment efficacy and the increase reflects PD. |
| [136] | EGFR mutation | 42 pts with IV stage LUAD osimertinib (post-1L) | A significant increase in methylation is found for at least one of the 9 tested genes at PD compared to baseline. Difference trend in PFS is shown between pts who are positive for DNA methylation of at least one gene at PD and those who are negative. |
| [137,138] | EGFR mutation | 27 pts with IV stage LUAD osimertinib (post-1L) | The increase in methylation is found for at least one of the nine tested genes at PD compared with baseline in both plasma cfDNA and paired CTC analysis. |
| [138] | EGFR mutation | Pts with IV stage LUAD gefitinib (1L) | Methylation level of WIF1 promoter is lower in the cfDNA of pts with a complete or partial response to gefitinib. Pts with hypomethylated WIF1 have better PFS and OS. |
| [139] | EGFR mutation | 122 pts with III-IV stage LUAD gefitinib, erlotinib, afatinib | Higher hypomethylation is found in cases with on-target resistances compared with those with off-target mutations. Hipo-methylation and CNA correlate with the duration of response only in EGFR amplification cases. |
| [140] | EGFR mutation | 103 pts with III-IV stage LUAD afatinib (1L) | cfDNA methylation levels are correlated with PFS are clustered in the cadherin, Wnt and EGFR signalling pathways. Pre-afatinib levels of CEP170 and CHCHD6 cfDNA methylation are associated with both PFS and OS. Pre-afatinib and post-afatinib levels of SLC9A3R2 and INTS1 cfDNA methylation correlate with bone metastasis. |
| [141] | EGFR mutation | 32 pts with IV stage LUAD EGFR-TKI | Histone modifications, DNA methylation, and chromatin accessibility allow discrimination between cfDNA samples from pts with tSCLC and EGFR-mutated LAUD. |
| [142] | ALK-rearranged | 21 pts with IV stage LUAD crizotinib, ceritinib, alectinib, brigatinib, lorlatinib | Higher 5-mC scores is associated with shorter OS. 5-mC scores can predict treatment response and PD. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trombetta, D.; Delcuratolo, M.D.; Fabrizio, F.P.; Delli Muti, F.; Rossi, A.; Centonza, A.; Guerra, F.P.; Sparaneo, A.; Piazzolla, M.; Parente, P.; et al. Methylation Analyses in Liquid Biopsy of Lung Cancer Patients: A Novel and Intriguing Approach Against Resistance to Target Therapies and Immunotherapies. Cancers 2025, 17, 3021. https://doi.org/10.3390/cancers17183021
Trombetta D, Delcuratolo MD, Fabrizio FP, Delli Muti F, Rossi A, Centonza A, Guerra FP, Sparaneo A, Piazzolla M, Parente P, et al. Methylation Analyses in Liquid Biopsy of Lung Cancer Patients: A Novel and Intriguing Approach Against Resistance to Target Therapies and Immunotherapies. Cancers. 2025; 17(18):3021. https://doi.org/10.3390/cancers17183021
Chicago/Turabian StyleTrombetta, Domenico, Marco Donatello Delcuratolo, Federico Pio Fabrizio, Francesco Delli Muti, Antonio Rossi, Antonella Centonza, Francesco Pio Guerra, Angelo Sparaneo, Michele Piazzolla, Paola Parente, and et al. 2025. "Methylation Analyses in Liquid Biopsy of Lung Cancer Patients: A Novel and Intriguing Approach Against Resistance to Target Therapies and Immunotherapies" Cancers 17, no. 18: 3021. https://doi.org/10.3390/cancers17183021
APA StyleTrombetta, D., Delcuratolo, M. D., Fabrizio, F. P., Delli Muti, F., Rossi, A., Centonza, A., Guerra, F. P., Sparaneo, A., Piazzolla, M., Parente, P., & Muscarella, L. A. (2025). Methylation Analyses in Liquid Biopsy of Lung Cancer Patients: A Novel and Intriguing Approach Against Resistance to Target Therapies and Immunotherapies. Cancers, 17(18), 3021. https://doi.org/10.3390/cancers17183021







