Hemopexin Suppresses Hepatocellular Carcinoma via TNF-α-Mediated Mitochondrial Apoptosis
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data Acquisition and Processing
2.2. Gene Set Function Enrichment Analysis and Tumor Microenvironment Analysis
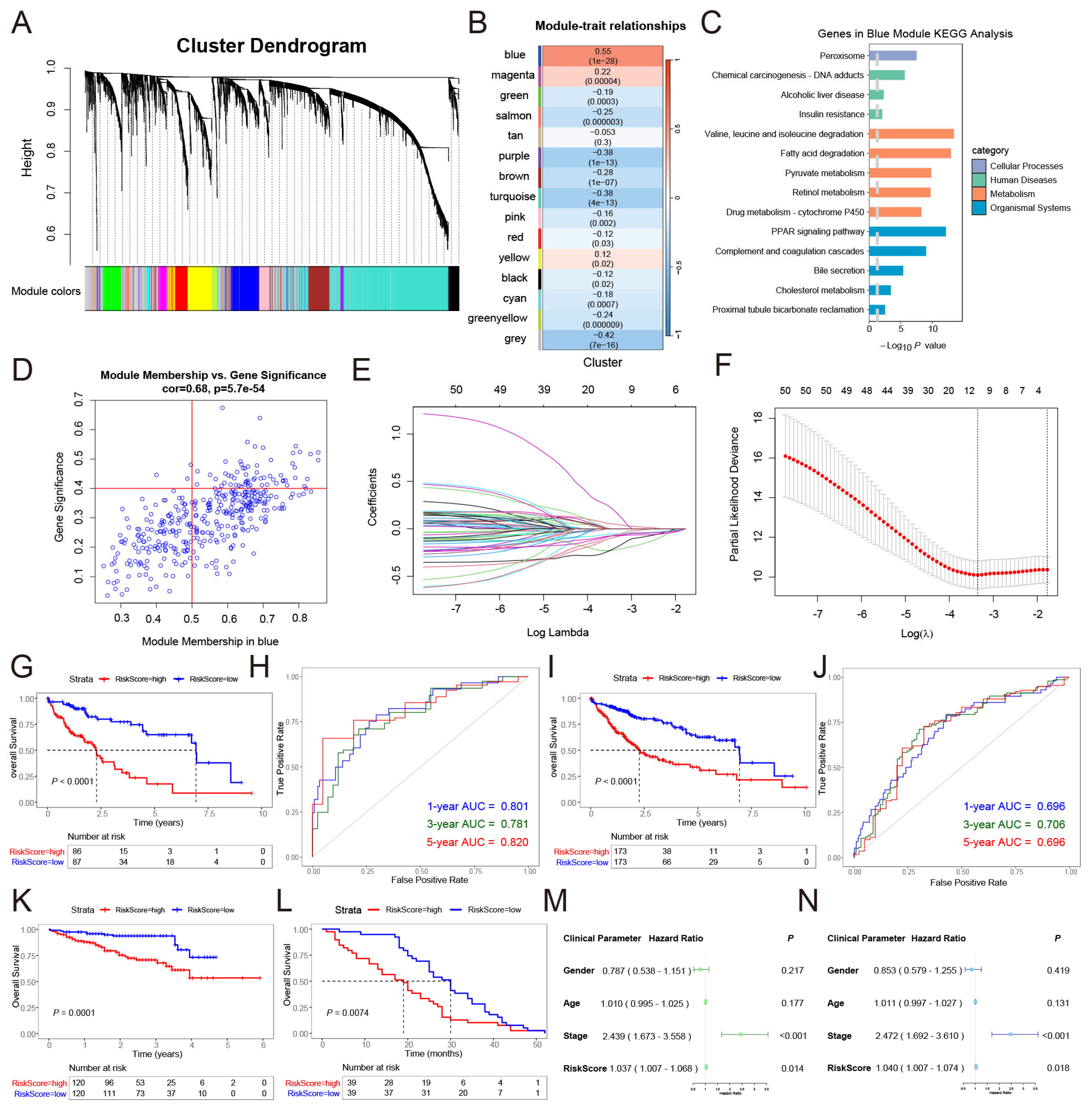
2.3. Molecular Subtyping of Fibrinolysis in HCC and Weighted Gene Co-Expression Network Analysis
2.4. Construction of the Risk Signature
2.5. Animals and Xenograft Tumor Model
2.6. Cell Lines and Cell Culture
2.7. Lentivirus Packaging Procedure and Infection
2.8. RNA Extraction and qRT-PCR
2.9. Western Blotting
2.10. CCK-8 Assay
2.11. Clone Formation Assay
2.12. EdU Cell Proliferation Assay
2.13. Wound Healing Assay
2.14. Flow Cytometry Apoptosis Assay
2.15. Mitochondrial Membrane Potential Assay Kit with JC-1 Analysis
2.16. TUNEL Apoptosis Assay
2.17. H&E Staining and Immunohistochemistry Analysis
2.18. RNA-seq Analysis
2.19. Statistical Analysis
3. Results
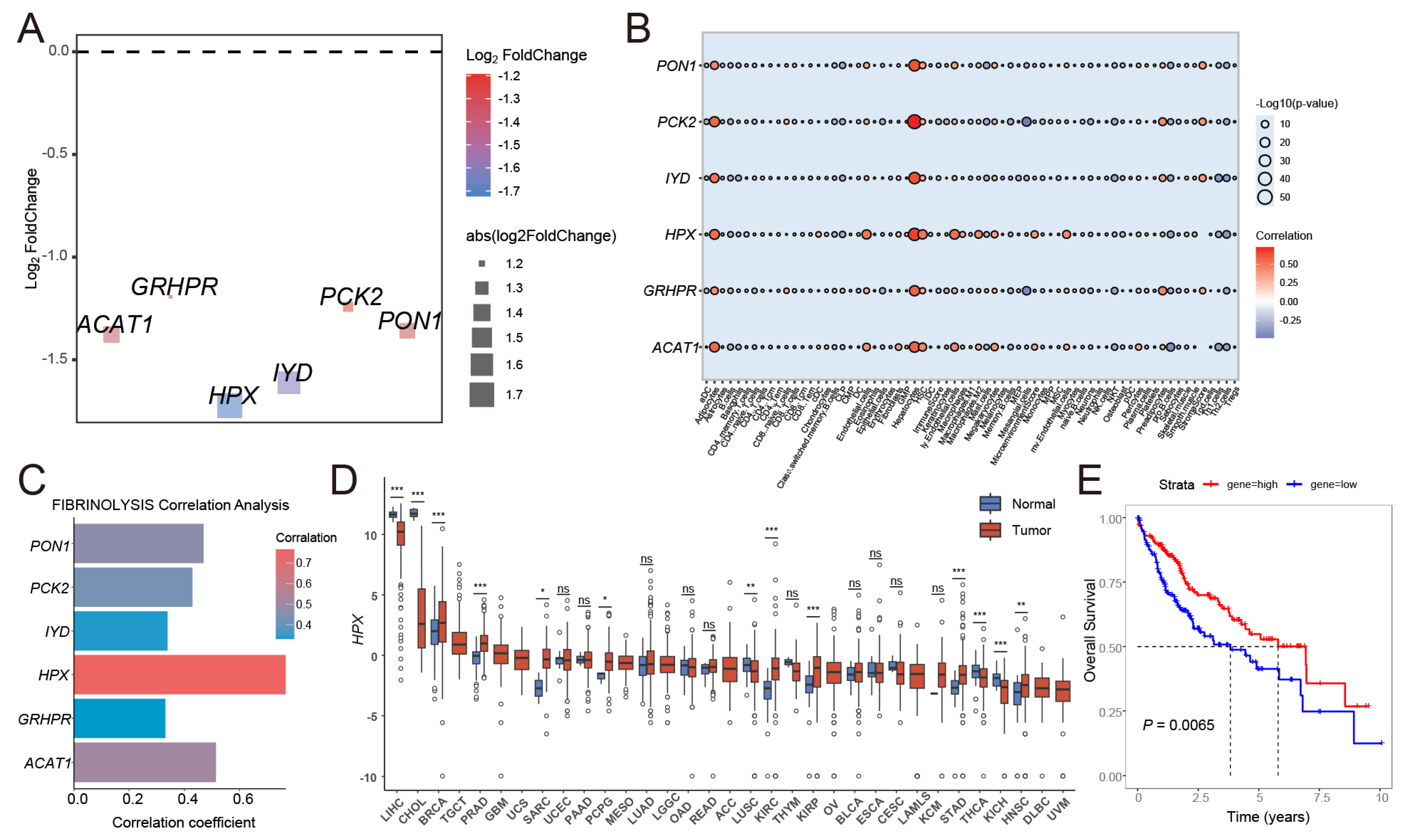
3.1. Analysis of the Biological Role of Fibrinolysis in Pan-Cancer
3.2. Construction and Clinical Analysis of Fibrinolysis Molecular Subtypes in HCC
3.3. A Risk Signature Was Constructed Based on the Fibrinolysis Molecular Subtype of HCC
3.4. The Key Gene HPX in HCC Was Identified Through the Risk Signature
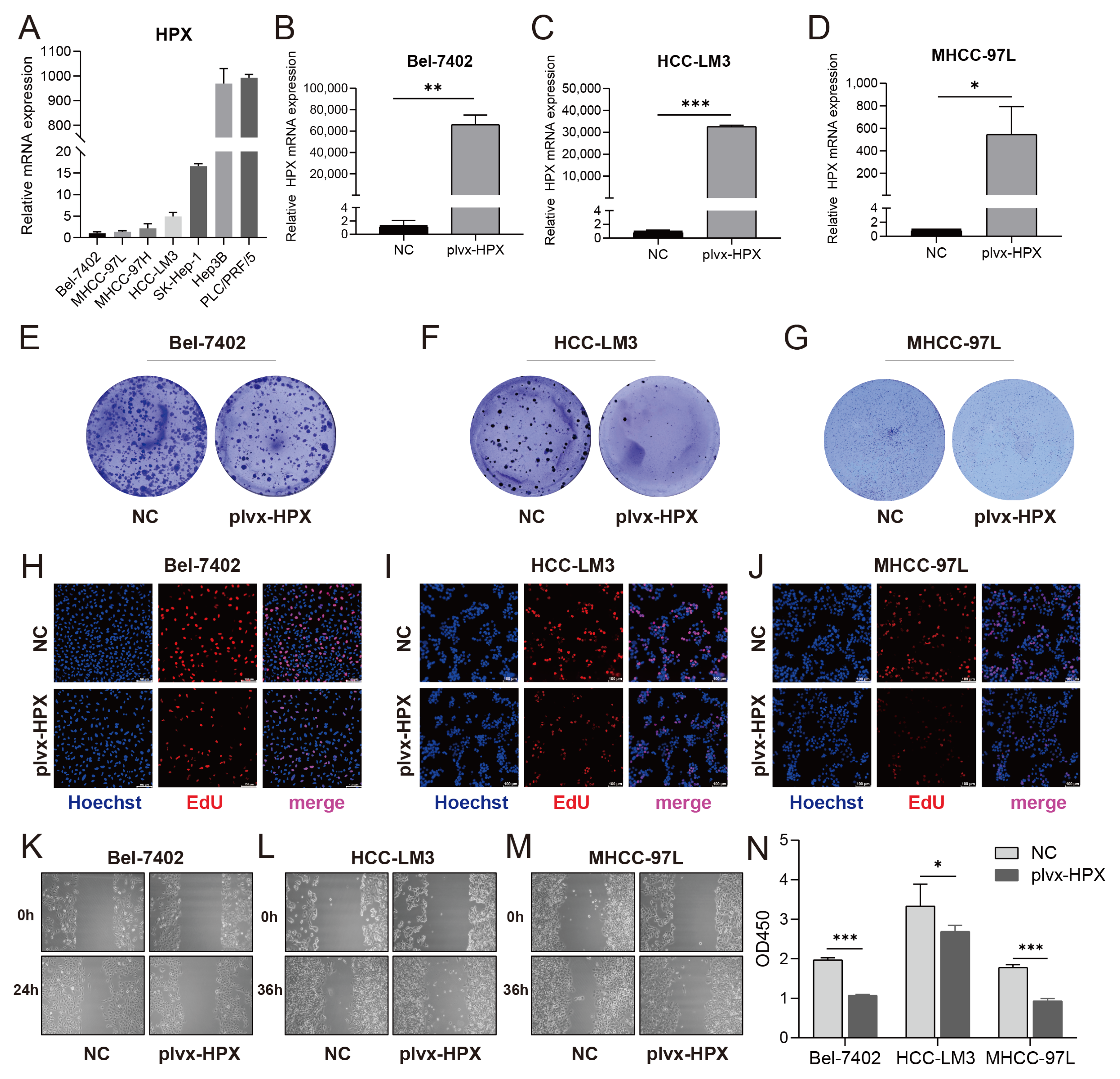
3.5. HPX Overexpression Suppressed Proliferation and Migration in HCC Cells
3.6. HPX Overexpression Promoted Apoptosis Through Mitochondrial Pathway Activation
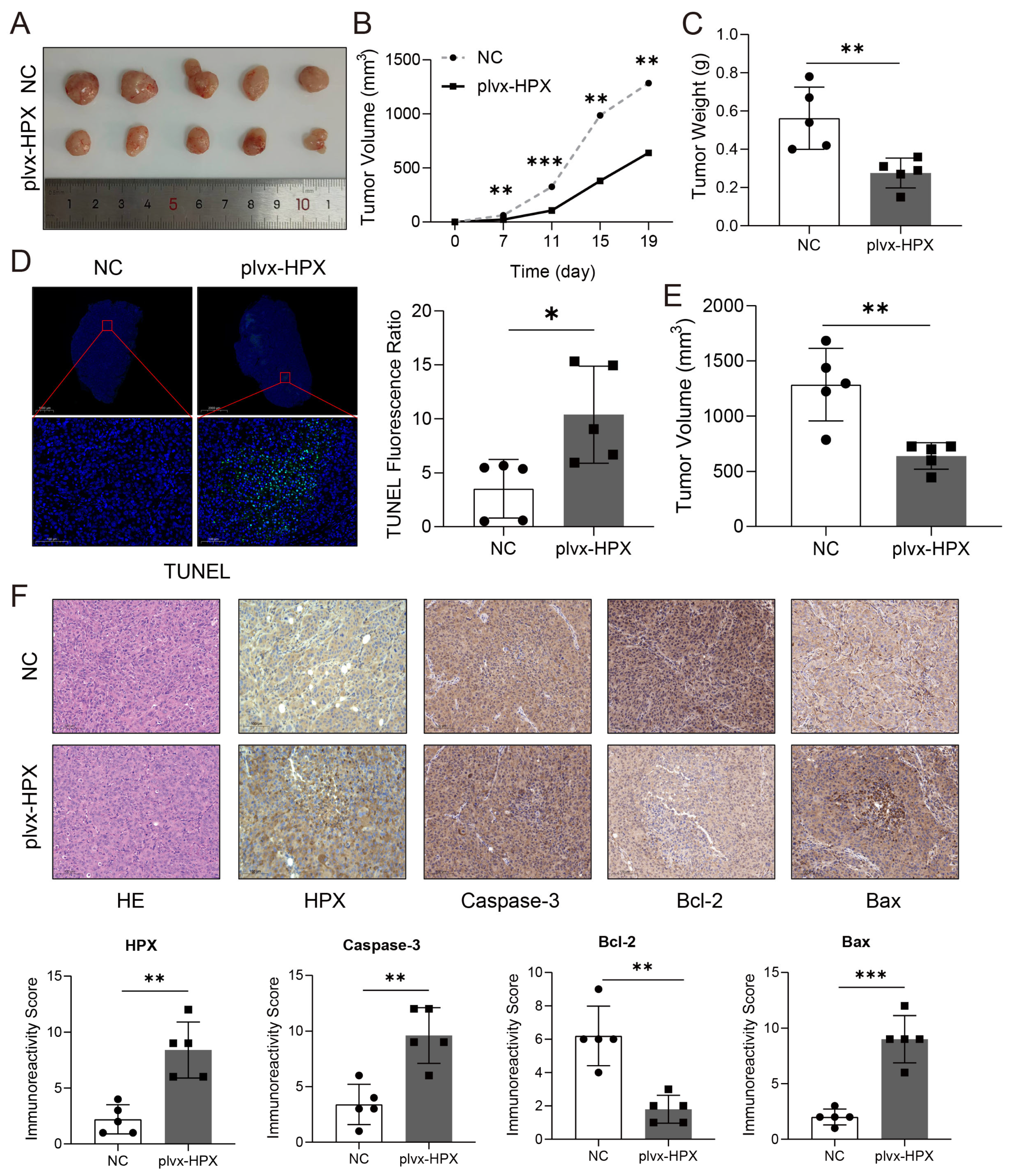
3.7. HPX Overexpression Inhibited Tumor Growth and Promoted Apoptosis In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACAT1 | Acetyl-CoA Acetyltransferase 1 |
| AUC | Area Under the Curve |
| ATCC | American Type Culture Collection |
| Bax | BCL2-Associated X Protein |
| Bcl-2 | B-Cell Lymphoma 2 |
| CCK-8 | Cell Counting Kit-8 |
| CNV | Copy Number Variation |
| COX | Cyclooxygenase |
| DAB | 3,3’-Diaminobenzidine |
| DAPI | 4′,6-Diamidino-2-Phenylindole |
| DD | D-Dimer |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DEGs | Differentially Expressed Genes |
| ECM | Extracellular Matrix |
| EdU | 5-Ethynyl-2’-Deoxyuridine |
| EGFR | Epidermal Growth Factor Receptor |
| ERK | Extracellular Signal-Regulated Kinase |
| ESTIMATE | Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data |
| FBG | Fibrinogen |
| FBS | Fetal Bovine Serum |
| FpA | Fibrinopeptide A |
| FpB | Fibrinopeptide B |
| FPKM | Fragments Per Kilobase of transcript per Million mapped reads |
| GC | Gastric Cancer |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| GSVA | Gene Set Variation Analysis |
| H&E | Hematoxylin and Eosin |
| HCC | Hepatocellular Carcinoma |
| HPX | Hemopexin |
| IHC | Immunohistochemistry |
| ICGC | International Cancer Genome Consortium |
| IYD | Iodotyrosine Deiodinase |
| JC-1 | 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine Iodide |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| MEK | Mitogen-Activated Protein Kinase Kinase |
| MEM | Minimum Essential Medium |
| NMF | Non-Negative Matrix Factorization |
| PAI-1 | Plasminogen Activator Inhibitor-1 |
| PBS | Phosphate-Buffered Saline |
| PCK2 | Phosphoenolpyruvate Carboxykinase 2 |
| PEI | Polyethylenimine |
| PON1 | Paraoxonase 1 |
| PVDF | Polyvinylidene Fluoride |
| RIPA | Radioimmunoprecipitation Assay |
| ROC | Receiver Operating Characteristic |
| SDS-PAGE | Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis |
| TCGA | The Cancer Genome Atlas |
| TF | Tissue Factor |
| TNF-α | Tumor Necrosis Factor-Alpha |
| TUNEL | Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling |
| uPA | Urokinase-Type Plasminogen Activator |
| uPAR | Urokinase-Type Plasminogen Activator Receptor |
| VEGF | Vascular Endothelial Growth Factor |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| xCell | A Gene Signature-Based Method for Immune Cell Type Identification |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Huang, X.; Lin, D.; Huang, X.; Lin, S.; Luo, S.; Xu, X.; Weng, X. Long-term trend of future Cancer onset: A model-based prediction of Cancer incidence and onset age by region and gender. Prev. Med. 2023, 177, 107775. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Llovet, J.M.; Yarchoan, M.; Mehta, N.; Heimbach, J.K.; Dawson, L.A.; Jou, J.H.; Kulik, L.M.; Agopian, V.G.; Marrero, J.A.; et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology 2023, 78, 1922–1965. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.Y.; Danpanichkul, P.; Yong, J.N.; Yu, Z.; Tan, D.J.H.; Lim, W.H.; Koh, B.; Lim, R.Y.Z.; Tham, E.K.J.; Mitra, K.; et al. Liver cancer in 2021: Global Burden of Disease study. J. Hepatol. 2025, 82, 851–860. [Google Scholar] [CrossRef]
- Weisel, J.W.; Litvinov, R.I. Mechanisms of fibrin polymerization and clinical implications. Blood 2013, 121, 1712–1719. [Google Scholar] [CrossRef]
- Cesarman-Maus, G.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar] [CrossRef]
- Binder, B.R.; Mihaly, J.; Prager, G.W. uPAR-uPA-PAI-1 interactions and signaling: A vascular biologist’s view. Thromb. Haemost. 2007, 97, 336–342. [Google Scholar]
- Lo, L.; Valentine, H.; Harrison, J.; Hayes, S.; Welch, I.; Pritchard, S.; West, C.; Ang, Y. Tissue factor expression in the metaplasia-adenoma-carcinoma sequence of gastric cancer in a European population. Br. J. Cancer 2012, 107, 1125–1130. [Google Scholar] [CrossRef]
- Yamashita, H.; Kitayama, J.; Ishikawa, M.; Nagawa, H. Tissue factor expression is a clinical indicator of lymphatic metastasis and poor prognosis in gastric cancer with intestinal phenotype. J. Surg. Oncol. 2007, 95, 324–331. [Google Scholar] [CrossRef]
- Ebert, M.P.A.; Lamer, S.; Meuer, J.; Malfertheiner, P.; Reymond, M.; Buschmann, T.; Röcken, C.; Seibert, V. Identification of the thrombin light chain a as the single best mass for differentiation of gastric cancer patients from individuals with dyspepsia by proteome analysis. J. Proteome Res. 2005, 4, 586–590. [Google Scholar] [CrossRef]
- Arigami, T.; Uenosono, Y.; Ishigami, S.; Okubo, K.; Kijima, T.; Yanagita, S.; Okumura, H.; Uchikado, Y.; Kijima, Y.; Nakajo, A.; et al. A Novel Scoring System Based on Fibrinogen and the Neutrophil-Lymphocyte Ratio as a Predictor of Chemotherapy Response and Prognosis in Patients with Advanced Gastric Cancer. Oncology 2016, 90, 186–192. [Google Scholar] [CrossRef]
- Yu, W.; Wang, Y.; Shen, B. An elevated preoperative plasma fibrinogen level is associated with poor overall survival in Chinese gastric cancer patients. Cancer Epidemiol. 2016, 42, 39–45. [Google Scholar] [CrossRef]
- Suzuki, T.; Shimada, H.; Nanami, T.; Oshima, Y.; Yajima, S.; Ito, M.; Washizawa, N.; Kaneko, H. Hyperfibrinogenemia is associated with inflammatory mediators and poor prognosis in patients with gastric cancer. Surg. Today 2016, 46, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Luo, Z.; Tang, D.; Liu, L.; Yao, D.; Zhu, L.; Wang, Z. Identification of carboxyl terminal peptide of Fibrinogen as a potential serum biomarker for gastric cancer. Tumor Biol. 2016, 37, 6963–6970. [Google Scholar] [CrossRef] [PubMed]
- Abramowicz, A.; Wojakowska, A.; Gdowicz-Klosok, A.; Polanska, J.; Rodziewicz, P.; Polanowski, P.; Namysl-Kaletka, A.; Pietrowska, M.; Wydmanski, J.; Widlak, P. Identification of serum proteome signatures of locally advanced and metastatic gastric cancer: A pilot study. J. Transl. Med. 2015, 13, 304. [Google Scholar] [CrossRef] [PubMed]
- Abbasciano, V.; Tassinari, D.; Sartori, S.; Trevisani, L.; Arcudi, D.; Bianchi, M.P.; Liboni, A. Usefulness of coagulation markers in staging of gastric cancer. Cancer Detect. Prev. 1995, 19, 331–336. [Google Scholar]
- Ay, C.; Dunkler, D.; Pirker, R.; Thaler, J.; Quehenberger, P.; Wagner, O.; Zielinski, C.; Pabinger, I. High D-dimer levels are associated with poor prognosis in cancer patients. Haematologica 2012, 97, 1158–1164. [Google Scholar] [CrossRef]
- Diao, D.; Wang, Z.; Cheng, Y.; Zhang, H.; Guo, Q.; Song, Y.; Zhu, K.; Li, K.; Liu, D.; Dang, C. D-dimer: Not just an indicator of venous thrombosis but a predictor of asymptomatic hematogenous metastasis in gastric cancer patients. PLoS ONE 2014, 9, e101125. [Google Scholar] [CrossRef]
- Kwaan, H.C.; Mazar, A.P.; McMahon, B.J. The apparent uPA/PAI-1 paradox in cancer: More than meets the eye. Semin. Thromb. Hemost. 2013, 39, 382–391. [Google Scholar] [CrossRef]
- Liu, D.; Aguirre Ghiso, J.; Estrada, Y.; Ossowski, L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell 2002, 1, 445–457. [Google Scholar] [CrossRef]
- Aguirre-Ghiso, J.A.; Estrada, Y.; Liu, D.; Ossowski, L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res. 2003, 63, 1684–1695. [Google Scholar] [CrossRef]
- Villa, E.; Critelli, R.; Lei, B.; Marzocchi, G.; Cammà, C.; Giannelli, G.; Pontisso, P.; Cabibbo, G.; Enea, M.; Colopi, S.; et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut 2016, 65, 861–869. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e15. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, H.; Zhao, Q.; Li, J.-H.; Peng, S.-F.; Zhang, Z.; Yang, J.-H.; Fu, Y. Immune-related cell death index and its application for hepatocellular carcinoma. NPJ Precis. Oncol. 2024, 8, 194. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Ye, Z.; Shen, R.; Yu, G.; Wu, J.; Xiong, Y.; Zhou, R.; Qiu, W.; Huang, N.; Sun, L.; et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front. Immunol. 2021, 12, 687975. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gao, J.; Ren, L.; Chen, P.; Yang, Y.; She, S.; Xie, Y.; Liao, W.; Chen, H. A signature based on NKG2D ligands to predict the recurrence of hepatocellular carcinoma after radical resection. Cancer Med. 2023, 12, 6337–6347. [Google Scholar] [CrossRef]
- Lee, A.Y.Y.; Levine, M.N.; Baker, R.I.; Bowden, C.; Kakkar, A.K.; Prins, M.; Rickles, F.R.; Julian, J.A.; Haley, S.; Kovacs, M.J.; et al. Low-molecular-weight heparin versus a coumarin for the prevention of recurrent venous thromboembolism in patients with cancer. N. Engl. J. Med. 2003, 349, 146–153. [Google Scholar] [CrossRef]
- Quan, Y.; He, J.; Zou, Q.; Zhang, L.; Sun, Q.; Huang, H.; Li, W.; Xie, K.; Wei, F. Low molecular weight heparin synergistically enhances the efficacy of adoptive and anti-PD-1-based immunotherapy by increasing lymphocyte infiltration in colorectal cancer. J. Immunother. Cancer 2023, 11, e007080. [Google Scholar] [CrossRef]
- Tolosano, E.; Fagoonee, S.; Morello, N.; Vinchi, F.; Fiorito, V. Heme scavenging and the other facets of hemopexin. Antioxid. Redox Signal 2010, 12, 305–320. [Google Scholar] [CrossRef]
- Ascenzi, P.; Fasano, M. Heme-hemopexin: A ‘chronosteric’ heme-protein. IUBMB Life 2007, 59, 700–708. [Google Scholar] [CrossRef]
- Fiorito, V.; Tolosano, E. Hemopexin and Cancer. Int. J. Mol. Sci. 2022, 23, 997. [Google Scholar] [CrossRef]
- Suzuki, Y.; Takadate, T.; Mizuma, M.; Shima, H.; Suzuki, T.; Tachibana, T.; Shimura, M.; Hata, T.; Iseki, M.; Kawaguchi, K.; et al. Stromal expression of hemopexin is associated with lymph-node metastasis in pancreatic ductal adenocarcinoma. PLoS ONE 2020, 15, e0235904. [Google Scholar] [CrossRef]
- Comunale, M.A.; Lowman, M.; Long, R.E.; Krakover, J.; Philip, R.; Seeholzer, S.; Evans, A.A.; Hann, H.-W.L.; Block, T.M.; Mehta, A.S. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J. Proteome Res. 2006, 5, 308–315. [Google Scholar] [CrossRef]
- Morota, K.; Nakagawa, M.; Sekiya, R.; Hemken, P.M.; Sokoll, L.J.; Elliott, D.; Chan, D.W.; Dowell, B.L. A comparative evaluation of Golgi protein-73, fucosylated hemopexin, α-fetoprotein, and PIVKA-II in the serum of patients with chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Clin. Chem. Lab. Med. 2011, 49, 711–718. [Google Scholar] [CrossRef]
- Canesin, G.; Di Ruscio, A.; Li, M.; Ummarino, S.; Hedblom, A.; Choudhury, R.; Krzyzanowska, A.; Csizmadia, E.; Palominos, M.; Stiehm, A.; et al. Scavenging of Labile Heme by Hemopexin Is a Key Checkpoint in Cancer Growth and Metastases. Cell Rep. 2020, 32, 108181. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Man, Y.; Zhang, X.; Guo, Q.; She, S.; Yang, Y.; Fei, R.; Cong, X.; Chen, D.; Wei, W.; et al. Hemopexin Suppresses Hepatocellular Carcinoma via TNF-α-Mediated Mitochondrial Apoptosis. Cancers 2025, 17, 2969. https://doi.org/10.3390/cancers17182969
Ren L, Man Y, Zhang X, Guo Q, She S, Yang Y, Fei R, Cong X, Chen D, Wei W, et al. Hemopexin Suppresses Hepatocellular Carcinoma via TNF-α-Mediated Mitochondrial Apoptosis. Cancers. 2025; 17(18):2969. https://doi.org/10.3390/cancers17182969
Chicago/Turabian StyleRen, Liying, Yuxin Man, Xue Zhang, Qian Guo, Shaoping She, Yao Yang, Ran Fei, Xu Cong, Dongbo Chen, Wen Wei, and et al. 2025. "Hemopexin Suppresses Hepatocellular Carcinoma via TNF-α-Mediated Mitochondrial Apoptosis" Cancers 17, no. 18: 2969. https://doi.org/10.3390/cancers17182969
APA StyleRen, L., Man, Y., Zhang, X., Guo, Q., She, S., Yang, Y., Fei, R., Cong, X., Chen, D., Wei, W., & Chen, H. (2025). Hemopexin Suppresses Hepatocellular Carcinoma via TNF-α-Mediated Mitochondrial Apoptosis. Cancers, 17(18), 2969. https://doi.org/10.3390/cancers17182969




