Epigenetic Reprogramming by Decitabine in Triple-Negative Breast Cancer: Mechanisms, Immune Modulation, and Therapeutic Synergy
Simple Summary
Abstract
1. Introduction
- The mechanistic rationale for its use based on epigenetic alterations in TNBC;
- Evidence of its efficacy and synergistic potential in preclinical models;
- Clinical outcomes, safety profiles, and combinatorial strategies being tested;
- Existing challenges and gaps in translating epigenetic therapy into clinical benefit;
- By consolidating this knowledge, we aim to inform future translational research and clinical trial design, with the goal of expanding therapeutic options for patients with this formidable breast cancer subtype.
2. Materials and Methods
2.1. Literature Search
2.2. Inclusion and Exclusion Criteria
2.3. Literature Screening
2.4. Data Extraction
2.5. Risk of Bias Assessment
3. Results
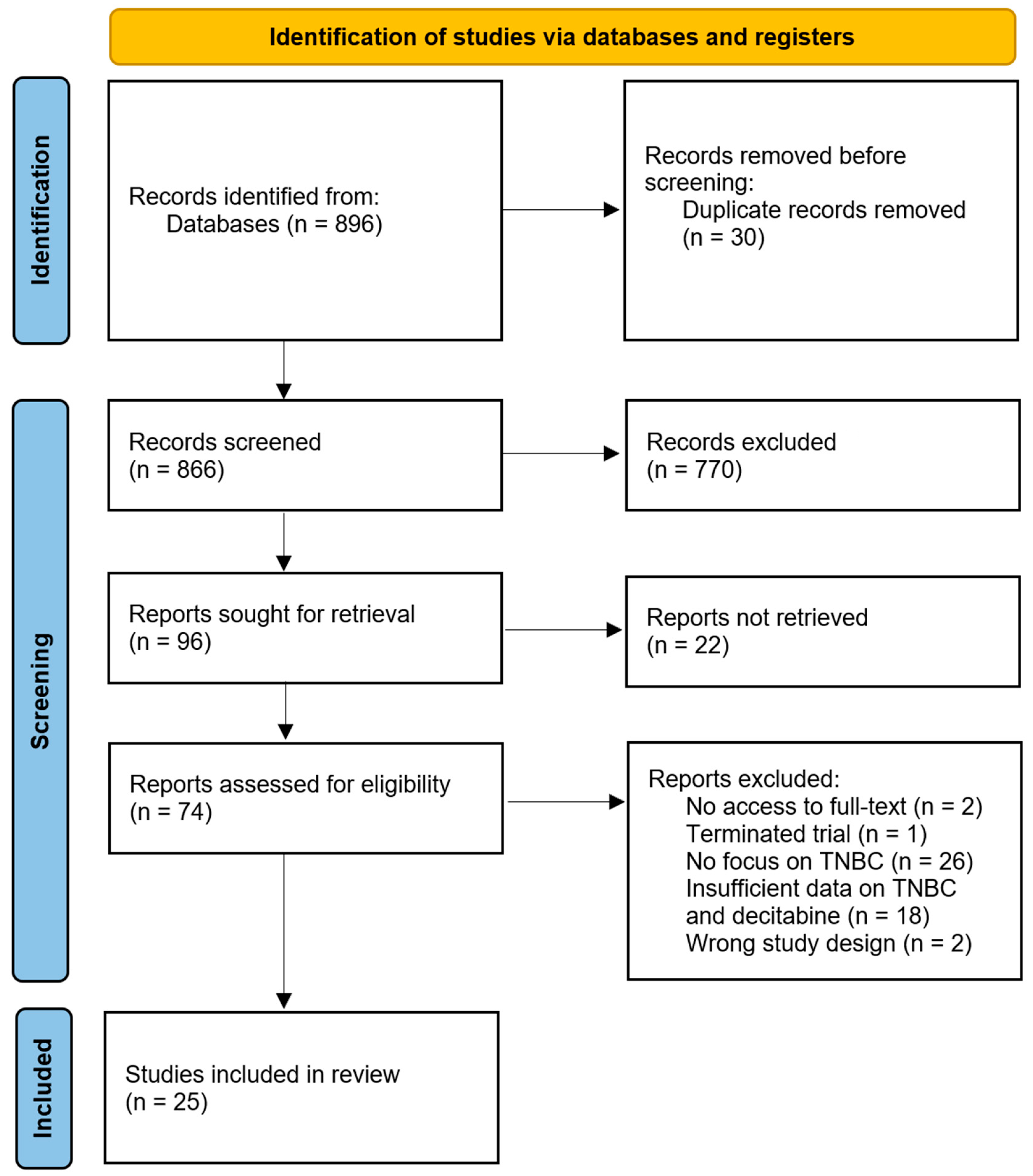
3.1. Study Selection
3.2. Risk of Bias Assessment
3.3. Study Characteristics
3.4. Anti-Tumor Efficacy of Decitabine
3.4.1. In Vitro Evidence of Cytotoxicity and Clonogenic Suppression
3.4.2. Tumor Volume Reduction in Murine Models
3.4.3. Patient-Derived Xenograft (PDX) Models and Clinical Translation
- Stromal TILs (sTILs): increased by 6.1% (p = 0.008);
- PD-L1 H-score: increased by 51.1% (p = 0.012);
- Monocytic MDSCs: decreased by 59% in blood (p < 0.01).
3.5. Immune Landscape Modulation
3.5.1. Antigen Presentation and T-Cell Recruitment
3.5.2. Combination with Immune Checkpoint Inhibitors
3.6. Epigenetic Reactivation and Molecular Pathways
3.6.1. Tumor Suppressor Reactivation
3.6.2. Post-Transcriptional Regulation
3.7. Other Combination Strategies to Enhance Therapeutic Efficacy
3.7.1. Hormonal Therapies
3.7.2. Chemotherapy
3.8. Mechanisms of Resistance to Decitabine
3.9. Safety and Tolerability
3.10. Histological and Phenotypical Changes
4. Discussion
4.1. Summary and Interpretation of Findings
4.2. Context Within Current Literature
4.3. Preclinical and Translational Insights on Resistance
4.4. Clinical Implications and Future Research Directions
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TNBC | Triple-Negative Breast Cancer |
| AKT | Protein Kinase B |
| BRCA | Breast Cancer Genes |
| CDH-1 | E-Cadherin |
| CREB1 | cAMP Response Element Binding Protein 1 |
| DAB2IP | Disabled Homolog 2 Interacting Protein |
| DNMT/DNMT1 | DNA Methyltransferase |
| DNMTi | DNA Methyltransferase Inhibitor |
| DPN | Diarylpropionitrile |
| ERα | Estrogen Receptor Alpha |
| ERβ | Estrogen Receptor Beta |
| ERBB2 | Erb-B2 Receptor Tyrosine Kinase 2 |
| ESSRA | Estrogen-Related Receptor Alpha |
| FOXM1 | Forkhead Box Protein M1 |
| FTH1 | Ferritin Heavy Chain 1 |
| GSK | Glycogen Synthase Kinase-3 |
| HMGB1 | High-Mobility Group Box 1 |
| HRR | Homologous Recombination Repair |
| hTERT | Human Telomerase Reverse Transcriptase |
| IGF-1 | Insulin-like Growth Factor 1 |
| IKKb | Inhibitor of Nuclear Factor Kappa-B Kinase Subunit Beta |
| IRF4 | Interferon Regulatory Factor 4 |
| LPAR1 | Lysophosphatidic Acid Receptor 1 |
| LRRC26 | Leucine-Rich Repeat-Containing Protein 26 |
| MYC | MYC Proto-Oncogene, bHLH Transcription Factor |
| NOXA | Phorbol-12-myristate-13-acetate-Induced Protein 1 |
| NUPR1 | Nuclear Protein 1 |
| PARP | Poly (ADP-ribose) Polymerase |
| PDT | Photodynamic Therapy |
| PIK3CB | Phosphoinositide 3-Kinase Catalytic Subunit Beta |
| POLD3 | Polymerase Delta 3 |
| Rap1B | Ras-Related Protein 1b |
| RUX3 | Runt-Related Transcription Factor 3 |
| sFRP1 | Secreted Frizzled-Related Protein 1 |
| STING | Stimulator of Interferon Genes |
| TP53 | Tumor Protein P53 |
| TPX2 | Targeting Protein for Xklp2 |
| TRAF6 | Tumor Necrosis Factor Receptor-Associated Factor 6 |
| TSPAN5 | Tetraspanin 5 |
| VEGF | Vascular Endothelial Growth Factor |
| AZA | Azacitidine |
| DAC | Decitabine |
| ADC | 5-aza-2′-deoxycytidine |
| DEX | Dexamethasone |
| DOC | Docetaxel |
| PAN | Panobinostat |
| PTX | Paclitaxel |
| ATP | Adenosine Triphosphate |
| CRT | Chemoradiotherapy |
| CSCs | Cancer Stem Cells |
| DNA | Deoxyribonucleic Acid |
| DSBs | Double Strand Breaks |
| EMT | Epithelial–Mesenchymal Transition |
| EPR | Enhanced Permeability and Retention |
| γ-H2AX | Phosphorylated Form of Histone H2AX |
| miR | microRNA |
| lncRNA | Long Non-Coding RNA |
| shRNA | Short Hairpin RNA |
| CTLs | Cytotoxic T Lymphocytes |
| ICD | Immunogenic Cell Death |
| IFN-γ | Interferon Gamma |
| IFN-β | Interferon Beta |
| IFN-I | Interferon Type I |
| ISGs | Interferon-Stimulated Genes |
| MHC-I | Major Histocompatibility Complex Class I |
| M-MDSCs | Monocytic Myeloid-Derived Suppressor Cells |
| PD-1 | Programmed Cell Death Protein 1 |
| PD-L1 | Programmed Cell Death Protein Ligand 1 |
| Treg | Regulatory T Cells |
| TEM | Effector Memory T Cells |
| ALT | Alternative Lengthening of Telomeres |
| DFS | Disease-Free Survival |
| LNEs | Lipid Nanoemulsions |
| mMSNs | Macrophage-Membrane-Camouflaged Mesoporous Silica Nanoparticles |
| NCT | Neoadjuvant Chemotherapy |
| OS | Overall Survival |
| pCR | Pathological Complete Response |
| sTILs | Stromal Tumor-Infiltrating Lymphocytes |
| iTILs | Intratumoral Tumor-Infiltrating Lymphocytes |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DMEM | Dulbecco’s Modified Eagle Medium |
References
- Penault-Llorca, F.; Viale, G. Pathological and Molecular Diagnosis of Triple-Negative Breast Cancer: A Clinical Perspective. Ann. Oncol. 2012, 23, vi19–vi22. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Oh, M.H.; Go, J.-H.; Han, K.; Choi, S.-Y. Molecular Subtypes of Triple-Negative Breast Cancer: Understanding of Subtype Categories and Clinical Implication. Genes Genom. 2020, 42, 1381–1387. [Google Scholar] [CrossRef]
- Uscanga-Perales, G.I.; Santuario-Facio, S.K.; Ortiz-López, R. Triple Negative Breast Cancer: Deciphering the Biology and Heterogeneity. Available online: https://www.elsevier.es/en-revista-medicina-universitaria-304-resumen-triple-negative-breast-cancer-deciphering-S1665579616300667 (accessed on 17 July 2025).
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review about Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef] [PubMed]
- Siddharth, S.; Sharma, D. Racial Disparity and Triple-Negative Breast Cancer in African-American Women: A Multifaceted Affair between Obesity, Biology, and Socioeconomic Determinants. Cancers 2018, 10, 514. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, X.; Liu, Y.; Wang, C. Prognostic Disparities in Young Patients Based on Breast Cancer Subtype: A Population-Based Study from the SEER Database. Medicine 2023, 102, e33416. [Google Scholar] [CrossRef]
- Zevallos, A.; Bravo, L.; Bretel, D.; Paez, K.J.; Infante, U.; Cardenas, N.K.; Alvarado, H.; Posada, A.; Pinto, J.A. The Hispanic Landscape of Triple Negative Breast Cancer. Crit. Rev. Oncol. Hematol. 2020, 155, 103094. [Google Scholar] [CrossRef]
- O’Reilly, D.; Sendi, M.A.; Kelly, C.M. Overview of Recent Advances in Metastatic Triple Negative Breast Cancer. World J. Clin. Oncol. 2021, 12, 164–182. [Google Scholar] [CrossRef]
- Lv, Y.; Ma, X.; Du, Y.; Feng, J. Understanding Patterns of Brain Metastasis in Triple-Negative Breast Cancer and Exploring Potential Therapeutic Targets. OncoTargets Ther. 2021, 14, 589–607. [Google Scholar] [CrossRef]
- Yao, H.; He, G.; Yan, S.; Chen, C.; Song, L.; Rosol, T.J.; Deng, X. Triple-Negative Breast Cancer: Is There a Treatment on the Horizon? Oncotarget 2016, 8, 1913–1924. [Google Scholar] [CrossRef]
- Newton, E.E.; Mueller, L.E.; Treadwell, S.M.; Morris, C.A.; Machado, H.L. Molecular Targets of Triple-Negative Breast Cancer: Where Do We Stand? Cancers 2022, 14, 482. [Google Scholar] [CrossRef]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-Negative Breast Cancer: Challenges and Opportunities of a Heterogeneous Disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Dogra, A.K.; Prakash, A.; Gupta, S.; Gupta, M. Prognostic Significance and Molecular Classification of Triple Negative Breast Cancer: A Systematic Review. Eur. J. Breast Health 2025, 21, 101–114. [Google Scholar] [CrossRef]
- Xiong, N.; Wu, H.; Yu, Z. Advancements and Challenges in Triple-Negative Breast Cancer: A Comprehensive Review of Therapeutic and Diagnostic Strategies. Front. Oncol. 2024, 14, 1405491. [Google Scholar] [CrossRef]
- Obidiro, O.; Battogtokh, G.; Akala, E.O. Triple Negative Breast Cancer Treatment Options and Limitations: Future Outlook. Pharmaceutics 2023, 15, 1796. [Google Scholar] [CrossRef]
- Kim, E.-S. Molecular Targets and Therapies Associated with Poor Prognosis of Triple-Negative Breast Cancer (Review). Int. J. Oncol. 2025, 66, 52. [Google Scholar] [CrossRef]
- Liu, Q.; Song, X.; Liu, Z.; Yu, Z. Investigation of Candidate Genes and Pathways in Basal/TNBC Patients by Integrated Analysis. Technol. Cancer Res. Treat. 2021, 20, 153303382110195. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.Y.; Zhang, X.J.; Wang, L.; Hu, L.N.; Zhang, X.D.; Li, L.; Gao, J.N. A Six-Epithelial–Mesenchymal Transition Gene Signature May Predict Metastasis of Triple-Negative Breast Cancer. OncoTargets Ther. 2020, 13, 6497–6509. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Xu, X.; Li, X.; Guan, W.; Meng, T.; Xu, G. Transcriptome-Based Network Analysis Unveils Eight Immune-Related Genes as Molecular Signatures in the Immunomodulatory Subtype of Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 1787. [Google Scholar] [CrossRef]
- Thompson, K.J.; Leon-Ferre, R.A.; Sinnwell, J.P.; Zahrieh, D.; Suman, V.; Metzger, F.; Asad, S.; Stover, D.; Carey, L.; Sikov, W.M.; et al. Luminal Androgen Receptor Breast Cancer Subtype and Investigation of the Microenvironment and Neoadjuvant Chemotherapy Response. NAR Cancer 2022, 4, zcac018. [Google Scholar] [CrossRef] [PubMed]
- Błaszczak, E.; Miziak, P.; Odrzywolski, A.; Baran, M.; Gumbarewicz, E.; Stepulak, A. Triple-Negative Breast Cancer Progression and Drug Resistance in the Context of Epithelial–Mesenchymal Transition. Cancers 2025, 17, 228. [Google Scholar] [CrossRef] [PubMed]
- Fultang, N.; Chakraborty, M.; Peethambaran, B. Regulation of Cancer Stem Cells in Triple Negative Breast Cancer. Cancer Drug Resist. 2021, 4, 321–342. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, H.; Yang, B.B.; Qadir, J.; Yuan, H.; Ye, T. Tumor-Infiltrating Immune Cells State-Implications for Various Breast Cancer Subtypes. Front. Immunol. 2025, 16, 1550003. [Google Scholar] [CrossRef]
- Cai, W.; Cai, X.; Fei, Y.; Ye, R.; Song, D.; Hu, D.; Zhang, W.; Xia, M.; Yang, X. DNA Methylation and Immune Evasion in Triple-Negative Breast Cancer: Challenges and Therapeutic Opportunities. Front. Oncol. 2025, 15, 1534055. [Google Scholar] [CrossRef] [PubMed]
- Nedeljković, M.; Damjanović, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer—How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yu, C.-W. Epigenetic Modulations in Triple-Negative Breast Cancer: Therapeutic Implications for Tumor Microenvironment. Pharmacol. Res. 2024, 204, 107205. [Google Scholar] [CrossRef]
- Temian, D.C.; Pop, L.A.; Irimie, A.I.; Berindan-Neagoe, I. The Epigenetics of Triple-Negative and Basal-like Breast Cancer: Current Knowledge. J. Breast Cancer 2018, 21, 233. [Google Scholar] [CrossRef]
- Al Aboud, N.M.; Simpson, B.; Jialal, I. Genetics, Epigenetic Mechanism. Available online: https://www.ncbi.nlm.nih.gov/books/NBK532999/ (accessed on 17 July 2025).
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic Modifications. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Aine, M.; Nacer, D.F.; Arbajian, E.; Veerla, S.; Karlsson, A.; Häkkinen, J.; Johansson, H.J.; Rosengren, F.; Vallon-Christersson, J.; Borg, Å.; et al. The DNA Methylation Landscape of Primary Triple-Negative Breast Cancer. Nat. Commun. 2025, 16, 3041. [Google Scholar] [CrossRef] [PubMed]
- Tarhonska, K.; Wichtowski, M.; Wow, T.; Kołacińska-Wow, A.; Płoszka, K.; Fendler, W.; Zawlik, I.; Paszek, S.; Zuchowska, A.; Jabłońska, E. DNA Methylation and Demethylation in Triple-Negative Breast Cancer: Associations with Clinicopathological Characteristics and the Chemotherapy Response. Biomedicines 2025, 13, 585. [Google Scholar] [CrossRef]
- Esteller, M. CpG Island Hypermethylation and Tumor Suppressor Genes: A Booming Present, a Brighter Future. Oncogene 2002, 21, 5427–5440. [Google Scholar] [CrossRef]
- Oubaddou, Y.; Oukabli, M.; Fenniche, S.; Elktaibi, A.; Elochi, M.R.; Al Bouzidi, A.; Qmichou, Z.; Dakka, N.; Diorio, C.; Richter, A.; et al. BRCA1 Promoter Hypermethylation in Malignant Breast Tumors and in the Histologically Normal Adjacent Tissues to the Tumors: Exploring Its Potential as a Biomarker and Its Clinical Significance in a Translational Approach. Genes 2023, 14, 1680. [Google Scholar] [CrossRef] [PubMed]
- Sheaffer, K.L.; Elliott, E.N.; Kaestner, K.H. DNA Hypomethylation Contributes to Genomic Instability and Intestinal Cancer Initiation. Cancer Prev. Res. 2016, 9, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Besselink, N.; Keijer, J.; Vermeulen, C.; Boymans, S.; de Ridder, J.; van Hoeck, A.; Cuppen, E.; Kuijk, E. The Genome-Wide Mutational Consequences of DNA Hypomethylation. Sci. Rep. 2023, 13, 6874. [Google Scholar] [CrossRef]
- Choi, J.Y.; James, S.R.; Link, P.A.; McCann, S.E.; Hong, C.C.; Davis, W.; Nesline, M.; Ambrosone, C.B.; Karpf, A.R. Association between Global DNA Hypomethylation in Leukocytes and Risk of Breast Cancer. Carcinogenesis 2009, 30, 1889–1897. [Google Scholar] [CrossRef]
- Gu, M.; Ren, B.; Fang, Y.; Ren, J.; Liu, X.; Wang, X.; Zhou, F.; Xiao, R.; Luo, X.; You, L.; et al. Epigenetic Regulation in Cancer. MedComm (2020) 2024, 5, e495. [Google Scholar] [CrossRef]
- Karahoca, M.; Momparler, R.L. Pharmacokinetic and Pharmacodynamic Analysis of 5-Aza-2′-Deoxycytidine (Decitabine) in the Design of Its Dose-Schedule for Cancer Therapy. Clin. Epigenetics 2013, 5, 3. [Google Scholar] [CrossRef]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA Methylation Inhibitor 5-Aza-2′-Deoxycytidine Induces Reversible Genome-Wide DNA Damage That Is Distinctly Influenced by DNA Methyltransferases 1 and 3B. Mol. Cell. Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef]
- Patel, K.; Dickson, J.; Din, S.; Macleod, K.; Jodrell, D.; Ramsahoye, B. Targeting of 5-Aza-2′-Deoxycytidine Residues by Chromatin-Associated DNMT1 Induces Proteasomal Degradation of the Free Enzyme. Nucleic Acids Res. 2010, 38, 4313–4324. [Google Scholar] [CrossRef]
- Kikuchi, A.; Onoda, H.; Yamaguchi, K.; Kori, S.; Matsuzawa, S.; Chiba, Y.; Tanimoto, S.; Yoshimi, S.; Sato, H.; Yamagata, A.; et al. Structural Basis for Activation of DNMT1. Nat. Commun. 2022, 13, 7130. [Google Scholar] [CrossRef] [PubMed]
- Hattori, N.; Sako, M.; Kimura, K.; Iida, N.; Takeshima, H.; Nakata, Y.; Kono, Y.; Ushijima, T. Novel Prodrugs of Decitabine with Greater Metabolic Stability and Less Toxicity. Clin. Epigenet. 2019, 11, 111. [Google Scholar] [CrossRef]
- Noto, F.; Mancini, J.; Gambardella, A.R.; Curcio, C.; Ninno, A.D.; Andreone, S.; Buccione, C.; D’Urso, M.T.; Macchia, D.; Pacca, A.M.; et al. Decitabine Co-Operates with the IL-33/ST2 Axis Modifying the Tumor Microenvironment and Improving the Response to PD-1 Blockade in Melanoma. J. Exp. Clin. Cancer Res. 2025, 44, 137. [Google Scholar] [CrossRef]
- Wang, L.; Amoozgar, Z.; Huang, J.; Saleh, M.H.; Xing, D.; Orsulic, S.; Goldberg, M.S. Decitabine Enhances Lymphocyte Migration and Function and Synergizes with CTLA-4 Blockade in a Murine Ovarian Cancer Model. Cancer Immunol. Res. 2015, 3, 1030–1041. [Google Scholar] [CrossRef]
- Yu, J.; Qin, B.; Moyer, A.M.; Nowsheen, S.; Liu, T.; Qin, S.; Zhuang, Y.; Liu, D.; Lu, S.W.; Kalari, K.R.; et al. DNA Methyltransferase Expression in Triple-Negative Breast Cancer Predicts Sensitivity to Decitabine. J. Clin. Investig. 2018, 128, 2376–2388. [Google Scholar] [CrossRef]
- Yu, J.; Zayas, J.; Qin, B.; Wang, L. Targeting DNA Methylation for Treating Triple-Negative Breast Cancer. Pharmacogenomics 2019, 20, 1151–1157. [Google Scholar] [CrossRef]
- Dahn, M.L.; Cruickshank, B.M.; Jackson, A.J.; Dean, C.; Holloway, R.W.; Hall, S.R.; Coyle, K.M.; Maillet, H.; Waisman, D.M.; Goralski, K.B.; et al. Decitabine Response in Breast Cancer Requires Efficient Drug Processing and Is Not Limited by Multidrug Resistance. Mol. Cancer Ther. 2020, 19, 1110–1122. [Google Scholar] [CrossRef]
- Nik Amirah Auni, N.M.A.; Mohd Redzwan, N.; Fauzi, A.N.; Yahya, M.M.; Wong, K.K. Hypomethylating Agents as Emerging Therapeutics for Triple-Negative Breast Cancer. Life Sci. 2025, 363, 123403. [Google Scholar] [CrossRef]
- Zhu, S.; Wu, Y.; Song, B.; Yi, M.; Yan, Y.; Mei, Q.; Wu, K. Recent Advances in Targeted Strategies for Triple-Negative Breast Cancer. J. Hematol. Oncol. 2023, 16, 100. [Google Scholar] [CrossRef]
- Shen, K.; Lu, M. Decitabine Plus Carboplatin in the Treatment of Metastatic TNBC (DETECT). Available online: https://clinicaltrials.gov/study/NCT03295552?cond=TNBC (accessed on 17 July 2025).
- Wang, H.; Wang, Z.; Wang, Z.; Li, X.; Li, Y.; Yan, N.; Wu, L.; Liang, Y.; Wu, J.; Song, H.; et al. Decitabine Induces IRF7-Mediated Immune Responses in P53-Mutated Triple-Negative Breast Cancer: A Clinical and Translational Study. Front. Med. 2024, 18, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Bear, H.D.; Deng, X.; Bandyopadhyay, D.; Idowu, M.; Jenkins, T.M.; Kmieciak, M.; Williams, M.; Archer, G.; Gwaltney, L.; Dillon, P.; et al. T-Cell Immune Checkpoint Inhibition plus Hypomethylation for Locally Advanced HER2-Negative Breast Cancer: A Phase 2 Neoadjuvant Window Trial of Decitabine and Pembrolizumab Followed by Standard Neoadjuvant Chemotherapy. J. Immunother. Cancer 2025, 13, e010294. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Br. Med. J. 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Shea, B.J.; Reeves, B.C.; Wells, G.; Thuku, M.; Hamel, C.; Moran, J.; Moher, D.; Tugwell, P.; Welch, V.; Kristjansson, E.; et al. AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ 2017, 358, j4008. [Google Scholar] [CrossRef]
- Amir-Behghadami, M.; Janati, A. Population, Intervention, Comparison, Outcomes and Study (PICOS) Design as a Framework to Formulate Eligibility Criteria in Systematic Reviews. Emerg. Med. J. 2020, 37, 387. [Google Scholar] [CrossRef] [PubMed]
- Rayyan AI. Available online: https://rayyan.ai/reviews (accessed on 4 April 2025).
- Grooten, W.J.A.; Tseli, E.; Äng, B.O.; Boersma, K.; Stålnacke, B.-M.; Gerdle, B.; Enthoven, P. Elaborating on the Assessment of the Risk of Bias in Prognostic Studies in Pain Rehabilitation Using QUIPS—Aspects of Interrater Agreement. Diagn. Progn. Res. 2019, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Cochrane Methods Risk of Bias 2 (RoB 2) Tool. Available online: https://methods.cochrane.org/risk-bias-2 (accessed on 4 September 2025).
- Russo, S.; Feola, S.; Feodoroff, M.; Chiaro, J.; Antignani, G.; Fusciello, M.; D’Alessio, F.; Hamdan, F.; Pellinen, T.; Mölsä, R.; et al. Low-Dose Decitabine Enhances the Efficacy of Viral Cancer Vaccines for Immunotherapy. Deleted J. 2024, 32, 200766. [Google Scholar] [CrossRef]
- Chu, Y.-H.; Huang, Y.-C.; Chiu, P.-Y.; Kuo, W.-H.; Pan, Y.-R.; Kuo, Y.-T.; Wang, R.-H.; Kao, Y.-C.; Wang, Y.-H.; Lin, Y.-F.; et al. Combating Breast Cancer Progression through Combination Therapy with Hypomethylating Agent and Glucocorticoid. iScience 2023, 26, 106597. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-W.; Wu, X.-J.; Liang, Y.; Ye, G.-Q.; Che, Y.-C.; Wu, X.-Z.; Zhu, X.-J.; Fan, H.-L.; Fan, X.-P.; Xu, J.-F. MiR-155 Increases Stemness and Decitabine Resistance in Triple-Negative Breast Cancer Cells by Inhibiting TSPAN5. Mol. Carcinog. 2020, 59, 447–461. [Google Scholar] [CrossRef]
- Kim, B.; Pena, C.D.; Auguste, D.T. Targeted Lipid Nanoemulsions Encapsulating Epigenetic Drugs Exhibit Selective Cytotoxicity on CDH1−/FOXM1+ Triple Negative Breast Cancer Cells. Mol. Pharm. 2019, 16, 1813–1826. [Google Scholar] [CrossRef]
- Cooper, S.; Von, C.A.; Kang, K.-T.; Marlow, L.A.; Grebe, S.K.; Menefee, M.E.; Tun, H.W.; Colón-Otero, G.; Perez, E.A.; Copland, J.A. Reexpression of Tumor Suppressor, SFRP1, Leads to Antitumor Synergy of Combined HDAC and Methyltransferase Inhibitors in Chemoresistant Cancers. Mol. Cancer Ther. 2012, 11, 2105–2115. [Google Scholar] [CrossRef]
- Fan, W.; Li, W.; Li, L.; Qin, M.; Mao, C.; Yuan, Z.; Wang, P.; Chu, B.; Jiang, Y. Bifunctional HDAC and DNMT Inhibitor Induces Viral Mimicry Activates the Innate Immune Response in Triple-Negative Breast Cancer. Eur. J. Pharm. Sci. 2024, 197, 106767. [Google Scholar] [CrossRef]
- Gao, T.; Sang, X.; Huang, X.; Gu, P.; Liu, J.; Liu, Y.; Zhang, N. Macrophage-Camouflaged Epigenetic Nanoinducers Enhance Chemoimmunotherapy in Triple Negative Breast Cancer. Acta Pharm. Sin. B 2022, 13, 4305–4317. [Google Scholar] [CrossRef]
- Nakajima, W.; Miyazaki, K.; Sakaguchi, M.; Asano, Y.; Ishibashi, M.; Kurita, T.; Yamaguchi, H.; Takei, H.; Tanaka, N. Epigenetic Priming with Decitabine Augments the Therapeutic Effect of Cisplatin on Triple-Negative Breast Cancer Cells through Induction of Proapoptotic Factor NOXA. Cancers 2022, 14, 248. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.; Lv, Z.-D.; Wang, Y.; Jin, L.-Y.; Ding, L.; Yang, Z.-C. Down-Regulation of BRMS1 by DNA Hypermethylation and Its Association with Metastatic Progression in Triple-Negative Breast Cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11076–11083. [Google Scholar]
- He, Y.; Hu, Q.; Wang, L.; Chen, C. Decitabine/Paclitaxel Co-Delivery Systems Modified with Anti-PD-L1 Antibodies Mediate Chemoimmunotherapy for Triple Negative Breast Cancer. Mater. Des. 2023, 237, 112562. [Google Scholar] [CrossRef]
- Xiong, Z.; Yang, L.; Li, N.; Fu, J.; Liu, P.; Sun, P.; Wei, W.; Xie, X. DAB2IP Attenuates Chemoresistance of Triple-Negative Breast Cancer through Sequestration of RAC1 to Prevent β-Catenin Nuclear Accumulation. Clin. Transl. Med. 2022, 12, e1133. [Google Scholar] [CrossRef]
- Elango, R.; Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Transcriptional Alterations of Protein Coding and Noncoding RNAs in Triple Negative Breast Cancer in Response to DNA Methyltransferases Inhibition. Cancer Cell Int. 2021, 21, 515. [Google Scholar] [CrossRef]
- Vernier, M.; McGuirk, S.; Dufour, C.R.; Wan, L.; Audet-Walsh, E.; St-Pierre, J.; Giguère, V. Inhibition of DNMT1 and ERRα Crosstalk Suppresses Breast Cancer via Derepression of IRF4. Oncogene 2020, 39, 6406–6420. [Google Scholar] [CrossRef]
- Pacaud, R.; Thomas, S.; Chaudhuri, S.; Lazar, A.; Timmerman, L.A.; Munster, P.N. Low Dose DNA Methyltransferase Inhibitors Potentiate PARP Inhibitors in Homologous Recombination Repair Deficient Tumors. Breast Cancer Res. 2025, 27, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-Y.; Xiao, Y.; Wei, J.-L.; Xu, X.-E.; Jin, X.; Hu, X.; Li, D.-Q.; Jiang, Y.-Z. MYC Suppresses STING-Dependent Innate Immunity by Transcriptionally Upregulating DNMT1 in Triple-Negative Breast Cancer. J. Immunother. Cancer 2021, 9, e002528. [Google Scholar] [CrossRef]
- Salahuddin, A.; Ghanem, H.; Omran, G.A.; Helmy, M.W. Epigenetic Restoration and Activation of ERβ: An Inspiring Approach for Treatment of Triple-Negative Breast Cancer. Med. Oncol. 2022, 39, 150. [Google Scholar] [CrossRef]
- Butler, C.; Sprowls, S.; Szalai, G.; Arsiwala, T.; Saralkar, P.; Straight, B.; Hatcher, S. Hypomethylating Agent Azacitidine Is Effective in Treating Brain Metastasis Triple-Negative Breast Cancer through Regulation of DNA Methylation of Keratin 18 Gene. Transl. Oncol. 2020, 13, 100775. [Google Scholar] [CrossRef]
- Banerjee, S.; Acedo, P.; Sheikh, S.E.; Harati, R.; Meecham, A.; Williams, N.; Gerard, G.F.; Keshtgar, M.; MacRobert, A.J. Rifat Hamoudi Combination of Verteporfin-Photodynamic Therapy with 5-Aza-2′-Deoxycytidine Enhances the Anti-Tumour Immune Response in Triple Negative Breast Cancer. Front. Immunol. 2023, 14, 1188087. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, Y.; Matsushita, Y.; Suzuki, H.; Komatsu, M.; Yoshimaru, T.; Kimura, R.; Yanai, A.; Honda, J.; Tangoku, A.; Sasa, M.; et al. Frequent Downregulation of LRRC26 by Epigenetic Alterations Is Involved in the Malignant Progression of Triple-Negative Breast Cancer. Int. J. Oncol. 2018, 52, 1539–1558. [Google Scholar] [CrossRef]
- Wang, H.; Wu, D.; Cai, L.; Li, X.; Zhang, Z.; Chen, S. Aberrant Methylation of WD-Repeat Protein 41 Contributes to Tumour Progression in Triple-Negative Breast Cancer. J. Cell. Mol. Med. 2020, 24, 6869–6882. [Google Scholar] [CrossRef] [PubMed]
- Umeh-Garcia, M.; O’Geen, H.; Simion, C.; Gephart, M.H.; Segal, D.J.; Sweeney, C.A. Aberrant Promoter Methylation Contributes to LRIG1 Silencing in Basal/Triple-Negative Breast Cancer. Br. J. Cancer 2022, 127, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Al-dulaimi, S.; Matta, S.; Slijepcevic, P.; Roberts, T. 5-Aza-2′-Deoxycytidine Induces Telomere Dysfunction in Breast Cancer Cells. Biomed. Pharmacother. 2024, 178, 117173. [Google Scholar] [CrossRef]
- Song, P.; Gao, Z.; Bao, Y.; Chen, L.; Huang, Y.; Liu, Y.; Dong, Q.; Wei, X. Wnt/β-Catenin Signaling Pathway in Carcinogenesis and Cancer Therapy. J. Hematol. Oncol. 2024, 17, 46. [Google Scholar] [CrossRef]
- Al-Yozbaki, M.; Jabre, I.; Syed, N.H.; Wilson, C.M. Targeting DNA Methyltransferases in Non-Small-Cell Lung Cancer. Semin. Cancer Biol. 2022, 83, 77–87. [Google Scholar] [CrossRef]
- Shafa, A.; Kailasam, A.; Prokop, L.; Hou, X.; Weroha, S.J. The Role of DNA Methyltransferase Inhibitors (DNMTi) in the Treatment of Patients with Recurrent Epithelial Ovarian Cancer: A Systematic Review and Meta-Analysis. Gynecol. Oncol. Rep. 2022, 44, S24. [Google Scholar] [CrossRef]
- Pawlak, A.; Hybicka, K.C.; Zioło, E.; Strządała, L.; Kałas, W. The Contrasting Delayed Effects of Transient Exposure of Colorectal Cancer Cells to Decitabine or Azacitidine. Cancers 2022, 14, 1530. [Google Scholar] [CrossRef]
- Li, H.-T.; Jang, H.J.; Rohena-Rivera, K.; Liu, M.; Gujar, H.; Kulchycki, J.; Zhao, S.; Billet, S.; Zhou, X.; Weisenberger, D.J.; et al. RNA Mis-Splicing Drives Viral Mimicry Response after DNMTi Therapy in SETD2-Mutant Kidney Cancer. Cell Rep. 2023, 42, 112016. [Google Scholar] [CrossRef]
- Wang, R.; Dong, X.; Zhang, X.; Liao, J.; Cui, W.; Li, W. Exploring Viral Mimicry Combined with Epigenetics and Tumor Immunity: New Perspectives in Cancer Therapy. Int. J. Biol. Sci. 2025, 21, 958–973. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.E.; Rawling, D.C.; Vela, A.; Pyle, A.M. An Evolving Arsenal: Viral RNA Detection by RIG-I-like Receptors. Curr. Opin. Microbiol. 2014, 20, 76–81. [Google Scholar] [CrossRef]
- Gaudy, A.; Laille, E.; Bailey, R.; Zhou, S.; Skikne, B.; Beach, C.L. Population Pharmacokinetics of Oral Azacitidine, and Exposure–Response Analysis in Acute Myeloid Leukemia. Clin. Pharmacol. Ther. 2023, 114, 845–852. [Google Scholar] [CrossRef]
- Fan, H.; Lu, X.; Wang, X.; Liu, Y.; Guo, B.; Zhang, Y.; Zhang, W.; Nie, J.; Feng, K.; Chen, M.; et al. Low-Dose Decitabine-Based Chemoimmunotherapy for Patients with Refractory Advanced Solid Tumors: A Phase I/II Report. J. Immunol. Res. 2014, 2014, 371087. [Google Scholar] [CrossRef]
- Lv, M.; Wang, Y.; Yuan, Z.; Zhai, L.; Iqbal, H.; Ur-Rehman, U.; Ning, X.; Wei, H.; Xin, J.; Jin, Z.; et al. Decitabine Promotes the Differentiation of Poorly Differentiated Gastric Cancer Cells and Enhances the Sensitivity of NK Cell Cytotoxicity via TNF-α. Sci. Rep. 2025, 15, 13119. [Google Scholar] [CrossRef]
- Kastl, L.; Brown, I.; Schofield, A.C. Effects of Decitabine on the Expression of Selected Endogenous Control Genes in Human Breast Cancer Cells. Mol. Cell. Probes 2010, 24, 87–92. [Google Scholar] [CrossRef]
- Datta, J.; Ghoshal, K.; Motiwala, T.; Jacob, S.T. Novel Insights into the Molecular Mechanism of Action of DNA Hypomethylating Agents: Role of Protein Kinase C δ in Decitabine-Induced Degradation of DNA Methyltransferase 1. Genes Cancer 2012, 3, 71–81. [Google Scholar] [CrossRef]
- Buocikova, V.; Tyciakova, S.; Pilalis, E.; Mastrokalou, C.; Urbanova, M.; Matuskova, M.; Demkova, L.; Medova, V.; Longhin, E.M.; Rundén-Pran, E.; et al. Decitabine-Induced DNA Methylation-Mediated Transcriptomic Reprogramming in Human Breast Cancer Cell Lines; the Impact of DCK Overexpression. Front. Pharmacol. 2022, 13, 991751. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Castoro, R.; El Ahdab, S.; Jelinek, J.; Wang, X.; Si, J.; Shu, J.; He, R.; Zhang, N.; Chung, W.; et al. Mechanisms of Resistance to Decitabine in the Myelodysplastic Syndrome. PLoS ONE 2011, 6, e23372. [Google Scholar] [CrossRef]
- Welch, J.S.; Petti, A.A.; Miller, C.A.; Fronick, C.C.; O’Laughlin, M.; Fulton, R.S.; Wilson, R.K.; Baty, J.D.; Duncavage, E.J.; Tandon, B.; et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N. Engl. J. Med. 2016, 375, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Derissen, E.J.B.; Beijnen, J.H.; Schellens, J.H.M. Concise Drug Review: Azacitidine and Decitabine. Oncologist 2013, 18, 619–624. [Google Scholar] [CrossRef]
- Manchado, E.; Huang, C.-H.; Tasdemir, N.; Tschaharganeh, D.F.; Wilkinson, J.E.; Lowe, S.W. A Pipeline for Drug Target Identification and Validation. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Leon-Ferre, R.A.; Farber, D. Testing the Addition of an Anti-Cancer Drug, ASTX727 (Cedazuridine, Decitabine), to Chemotherapy (Paclitaxel) and Immunotherapy (Pembrolizumab) for Metastatic Triple-Negative Breast Cancer. Available online: https://clinicaltrials.gov/study/NCT05673200?cond=TNBC (accessed on 17 July 2025).
- Miller, K. Study of ASTX727 Plus Talazoparib in Patients with Triple Negative or Hormone Resistant/HER2-Negative Metastatic Breast Cancer. Available online: https://clinicaltrials.gov/study/NCT04134884?cond=TNBC (accessed on 17 July 2025).
- Jan, N.; Sofi, S.; Qayoom, H.; Shabir, A.; Haq, B.U.; Macha, M.A.; Almilaibary, A.; Mir, M.A. Metronomic Chemotherapy and Drug Repurposing: A Paradigm Shift in Oncology. Heliyon 2024, 10, e24670. [Google Scholar] [CrossRef] [PubMed]



| Framework | Description |
|---|---|
| P (Participants) | Patients with triple-negative breast cancer (TNBC); preclinical in vivo and in vitro models involving TNBC cell lines or xenografts. |
| I (Intervention) | Administration of decitabine (5-aza-2′-deoxycytidine), either as monotherapy or in combination with other therapeutic agents. |
| C (Comparison) | Standard chemotherapeutic agents, placebo, or untreated control groups. |
| O (Outcomes) | Tumor progression metrics (overall response rate, progression-free survival, overall survival, disease-free survival), biomarker responses, immune response, epigenetic modulation, histopathological changes, safety profile. |
| S (Study Design) | Preclinical in vivo/in vitro studies and clinical studies (including randomized trials, observational studies, and case series) evaluating the role of decitabine in TNBC. |
| Study | Cell Lines Used | Intervention | Key Findings |
|---|---|---|---|
| Dahn et al., 2020 [47] | MDA-MB-468, MDA-MB-231, SUM159 | 0.5 mg/kg DAC for 3/5-day cycles over 3–4 weeks | Dose-dependent cytotoxicity; significant reduction in clonogenic potential; apoptosis induction; observed dependence on DCK expression levels for drug activation. |
| Chu et al., 2023 [60] | MDA-MB-231, HCC-1395, Hs578T, MCF-10A (non-tumorigenic), MCF-7 | 50 mg/kg DAC 5 mg/kg DEX | Upregulation of miR-708; suppression of CD44 and Rap1B expression; inhibited migration, invasion, and clonogenicity. |
| Salahuddin et al., 2022 [74] | MDA-MB-231 | 4 µM DAC 0.26 µM Vorinostat 0.093 µM DPN (ERβ agonist) Combinations: Decitabine + Vorinostat, DPN + Vorinostat, DPN + Decitabine, and all three together | Hormonal resensitization through ERβ reactivation; increased apoptotic signaling via caspase-3 elevation; VEGF downregulation. |
| Yang et al., 2020 [61] | MDA-MB-231, BT-549 | DAC (concentrations ranged from 2 μmol/L to 256 μmol/L) + Modulatory agents (miR-155 mimics, miR-155 inhibitors, siTSPAN5, pcDNA3.1-TSPAN5 plasmids) | DAC increased miR-155, promoting resistance via TSPAN5 suppression; co-treatment with miR-155 inhibitor restored TSPAN5 and reduced tumorsphere formation. |
| Kim et al., 2019 [62] | MDA-MB-231, MDA-MB-436, MDA-MB-468, Hs578T, HCC1806, HCC1569, DU4475 | 5 μM DAC + 120 nM PAN for 24–48 h | TNBC-specific cytotoxicity; CDH1 upregulation and FOXM1 suppression; induced cell cycle arrest at G2/M phase. |
| Cooper et al., 2012 [63] | MDA-MB-231, BT20 | 1 μM DAC for 72 h + 5 nM Romidepsin during the last 24 h | Reduced colony-forming ability; synergistic cytotoxicity with HDAC inhibitor; reactivation of Wnt antagonist sFRP1. |
| Vernier et al., 2020 [71] | MDA-MB-231, MDA-MB-436, MDA-MB-468 | 5 μM C29 + 3–5 μM AZA | Reduced proliferation; de-repression of IRF4 via disruption of DNMT1–ERα loop; induction of type I interferon-related genes. |
| Study | Murine Model | Intervention | Key Findings |
|---|---|---|---|
| Dahn et al., 2020 [47] | MDA-MB-468, MDA-MB-231, SUM159 | 0.5 mg/kg DAC for 3/5-day cycles over 3–4 weeks | Significant tumor volume reduction (~60%); effect reversed in DCK-deficient tumors. |
| Russo et al., 2024 [59] | 4T1 | 0.5 mg/kg DAC + PeptiCRAd (1 × 109 viral particles per tumor + 20 µg peptide) | Significant tumor shrinkage in DAC + PeptiCRAd group compared to monotherapies. |
| Wu et al., 2021 [73] | 66cl4, 4T1 | 0.8 mg/kg DAC daily + 100 µ Anti-PD-1 antibody on days 3, 7, 10, 14 | Combination significantly reduced tumor burden compared to monotherapy. |
| Gao et al., 2022 [65] | 4T1 | 10 mg/kg PTX + 2.5 mg/kg DAC + 5 mg/kg aPD-1 | DAC + PTX + aPD-1 resulted in tumor rejection in 75% of mice. |
| Banerjee et al., 2023 [76] | 4T1 | 6.25 mg/kg 5-ADC + 15 mg/kg Verteporfin | Combo group showed smallest tumor volumes. |
| He et al., 2024 [68] | 4T1 and PTX-resistant 4T1/PTX | 30 µM DAC + 87.69 µM PTX + 0.3 mg/kg αPD-L1 | Greatest tumor volume reduction in triple combo group. |
| Vernier et al., 2020 [71] | NIC-5231, NIC-5257 | 5 μM C29 + 3–5 μM 5-azadC | DAC + C29 group showed maximal tumor suppression. |
| Study | Model | Intervention | Immune Markers Affected | Other Notes |
|---|---|---|---|---|
| Bear et al., 2020 [52] | Primary breast tumor tissue (biopsy-derived) | 15 mg/m2 DAC × 4 days 200 mg Pembrolizumab on days 8 and 22 | ↑ sTILs, ↑ PD-L1 (H-score, CPS), ↓ M-MDSCs | Correlated with pCR; effective immune recruitment. |
| Russo et al., 2024 [59] | 4T1, MDA-MB-436 | 0.5 mg/kg DAC + PeptiCRAd (1 × 109 viral particles per tumor + 20 µg peptide) | ↑ MHC-I, PD-L1, CD8+ T-cell infiltration; ↓ Tregs | Enhanced spatial relocalization of CD8+ T cells; tumor growth control. |
| Wu et al., 2021 [73] | 66cl4, 4T1 | 0.8 mg/kg DAC daily + 100 µ Anti-PD-1 antibody on days 3, 7, 10, 14 | ↑ IFN-β, ISGs (CXCL10); ↑ CD8+ T cells | Strong STING pathway activation; immune inflamed phenotype. |
| Gao et al., 2022 [65] | 4T1 | 10 mg/kg PTX + 2.5 mg/kg DAC + 5 mg/kg aPD-1 | ↑ IFN-γ, CD8+, Granzyme B+ cells | Induced ICD and tumor clearance in 75% of mice. |
| Study | Model | Intervention | Synergistic Effects | Other Notes |
|---|---|---|---|---|
| Bear et al., 2020 [52] | Primary breast tumor tissue (biopsy-derived) | 15 mg/m2 DAC × 4 days 200 mg Pembrolizumab on days 8 and 22 | 40.7% pCR rate, ↑ sTILs, ↓ M-MDSCs | Correlated with pCR; effective immune recruitment. |
| Wu et al., 2021 [73] | 66cl4, 4T1 | 0.8 mg/kg DAC daily + 100 µ Anti-PD-1 antibody on days 3, 7, 10, 14 | Enhanced IFN-I and CD8+ response; greater tumor volume reduction vs. monotherapy. | MYC-DNMT1-STING axis central to synergy. |
| Gao et al., 2022 [65] | 4T1 | 10 mg/kg PTX + 2.5 mg/kg DAC + 5 mg/kg aPD-1 | Tumor rejection in 75% of mice. | Synergized with chemo and immune priming effect. |
| He et al., 2024 [68] | 4T1 and PTX-resistant 4T1/PTX | 30 µM DAC + 87.69 µM PTX + 0.3 mg/kg αPD-L1 (in imaging studies) | Triple combo showed best survival and tumor suppression. | EMT reversal enhanced checkpoint efficacy. |
| Study | Model | Tumor Suppressor Activated | Mechanisms | Other Notes |
|---|---|---|---|---|
| Yu et al., 2019 [46] | Hs 578T, BT-549, MDA-MB-231 | Global gene reprogramming including MYC downregulation | TRAF6-mediated DNMT degradation. | Broad reactivation of suppressed pathways; resensitization to chemotherapy. |
| Dahn et al., 2020 [47] | MDA-MB-468, MDA-MB-231, SUM159 | BRCA1, CDH1, RUNX3 | Partial promoter demethylation; viral mimicry. | Linked to DAC activation via DCK; reduced tumor volume. |
| Cooper et al., 2012 [63] | MDA-MB-231, BT20 | sFRP1 | Histone and DNA demethylation with DAC + Romidepsin. | Wnt pathway inhibition; induced apoptosis. |
| Fan et al., 2024 [64] | BT-549, HCC1937, MDA-MB-231 | EMT-related markers (such as E-cadherin) | Dual HDAC and DNMT inhibition. | Reduced migration and invasiveness. |
| Nakajima et al., 2022 [66] | D-type: MDA-MB-468, HCC38 G-type: MDA-MB-453, MDA-MB-157, MDA-MB-231, HCC1143 R-type: Hs578T, HCC1187, HCC1937 | NOXA (pro-apoptotic gene) | Epigenetic class profiling; unsilenced in R-type. | Correlated with resistance; failure to silence pro-apoptotic gene in resistant class. |
| Vernier et al., 2020 [71] | MDA-MB-231, MDA-MB-436, MDA-MB-468 | IRF4 | Disruption of ERα-DNMT1 complex. | Enhanced interferon signaling and growth suppression. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riyas Mohamed, F.R.; Aldubaisi, S.; Akbar, A.; Khan, M.I.; Yaqinuddin, A. Epigenetic Reprogramming by Decitabine in Triple-Negative Breast Cancer: Mechanisms, Immune Modulation, and Therapeutic Synergy. Cancers 2025, 17, 2953. https://doi.org/10.3390/cancers17182953
Riyas Mohamed FR, Aldubaisi S, Akbar A, Khan MI, Yaqinuddin A. Epigenetic Reprogramming by Decitabine in Triple-Negative Breast Cancer: Mechanisms, Immune Modulation, and Therapeutic Synergy. Cancers. 2025; 17(18):2953. https://doi.org/10.3390/cancers17182953
Chicago/Turabian StyleRiyas Mohamed, Fathima Raahima, Safiah Aldubaisi, Arshiya Akbar, Mohammad Imran Khan, and Ahmed Yaqinuddin. 2025. "Epigenetic Reprogramming by Decitabine in Triple-Negative Breast Cancer: Mechanisms, Immune Modulation, and Therapeutic Synergy" Cancers 17, no. 18: 2953. https://doi.org/10.3390/cancers17182953
APA StyleRiyas Mohamed, F. R., Aldubaisi, S., Akbar, A., Khan, M. I., & Yaqinuddin, A. (2025). Epigenetic Reprogramming by Decitabine in Triple-Negative Breast Cancer: Mechanisms, Immune Modulation, and Therapeutic Synergy. Cancers, 17(18), 2953. https://doi.org/10.3390/cancers17182953






