Revealing the Angiogenic Signature of FH-Deficient Breast Cancer: Genomic Profiling and Clinical Implications
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
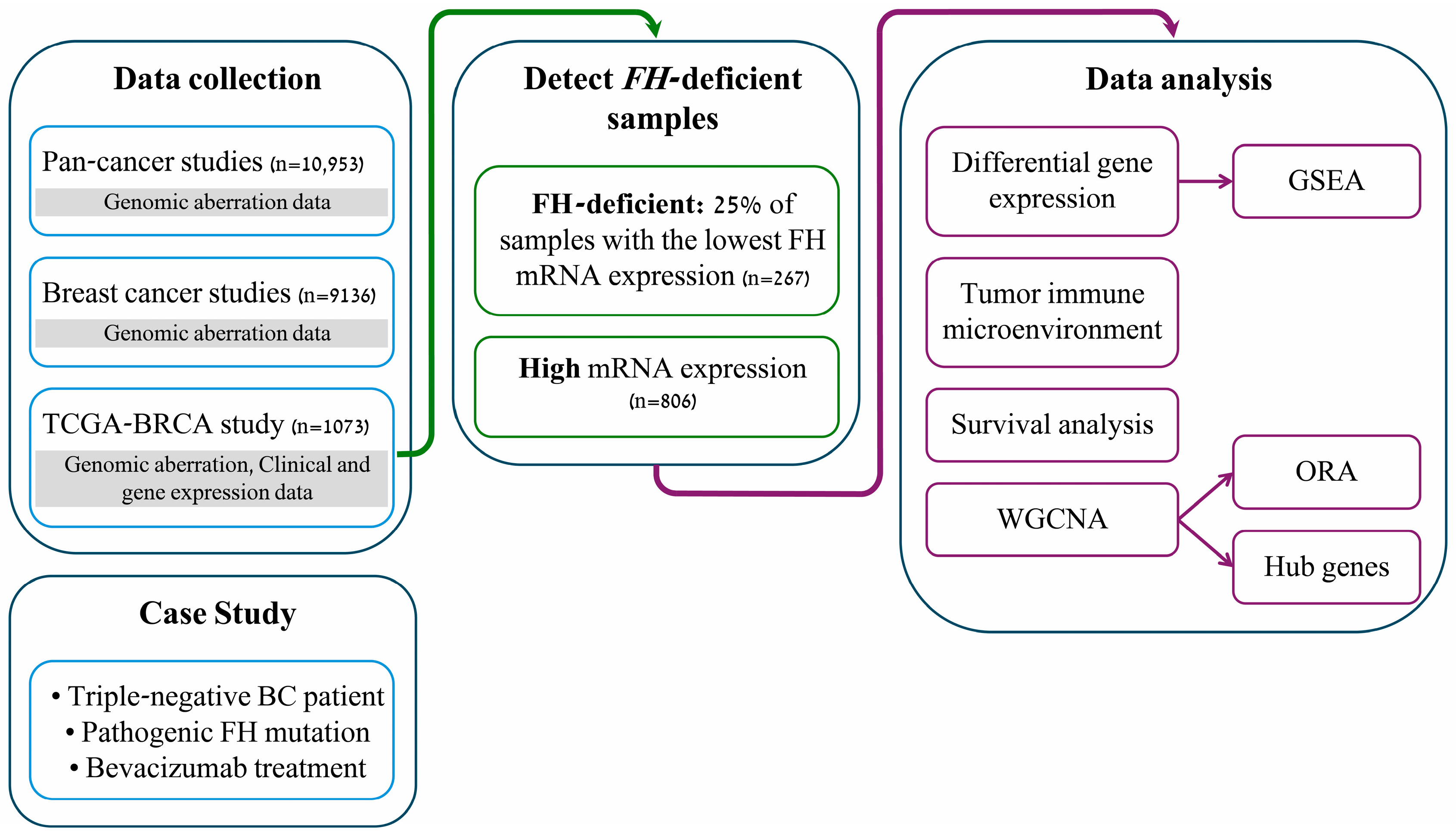
2.1. Data Resource and Study Design
2.2. Calculation of Angiogenesis Score
2.3. Differentially Expressed Gene (DEG) Identification
2.4. Estimation of Tumor-Infiltrating Immune Cells
2.5. Weighted Correlation Network Analysis (WGCNA)
2.6. Identifying Hub Genes
2.7. Pathways Enrichment Analysis
2.8. Statistical Analysis
3. Results
3.1. Prevalence of FH Alterations in Cancer Patients
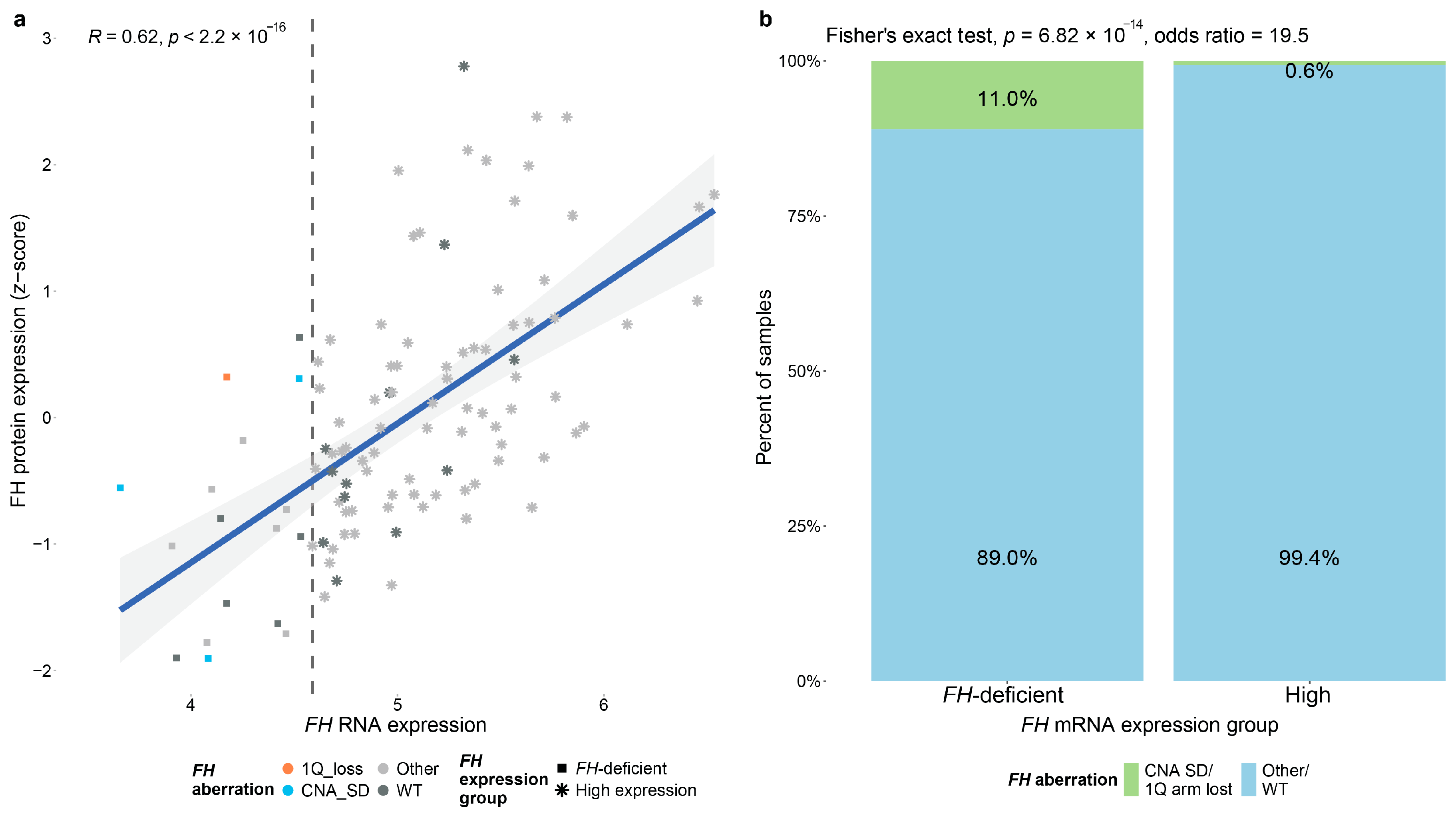
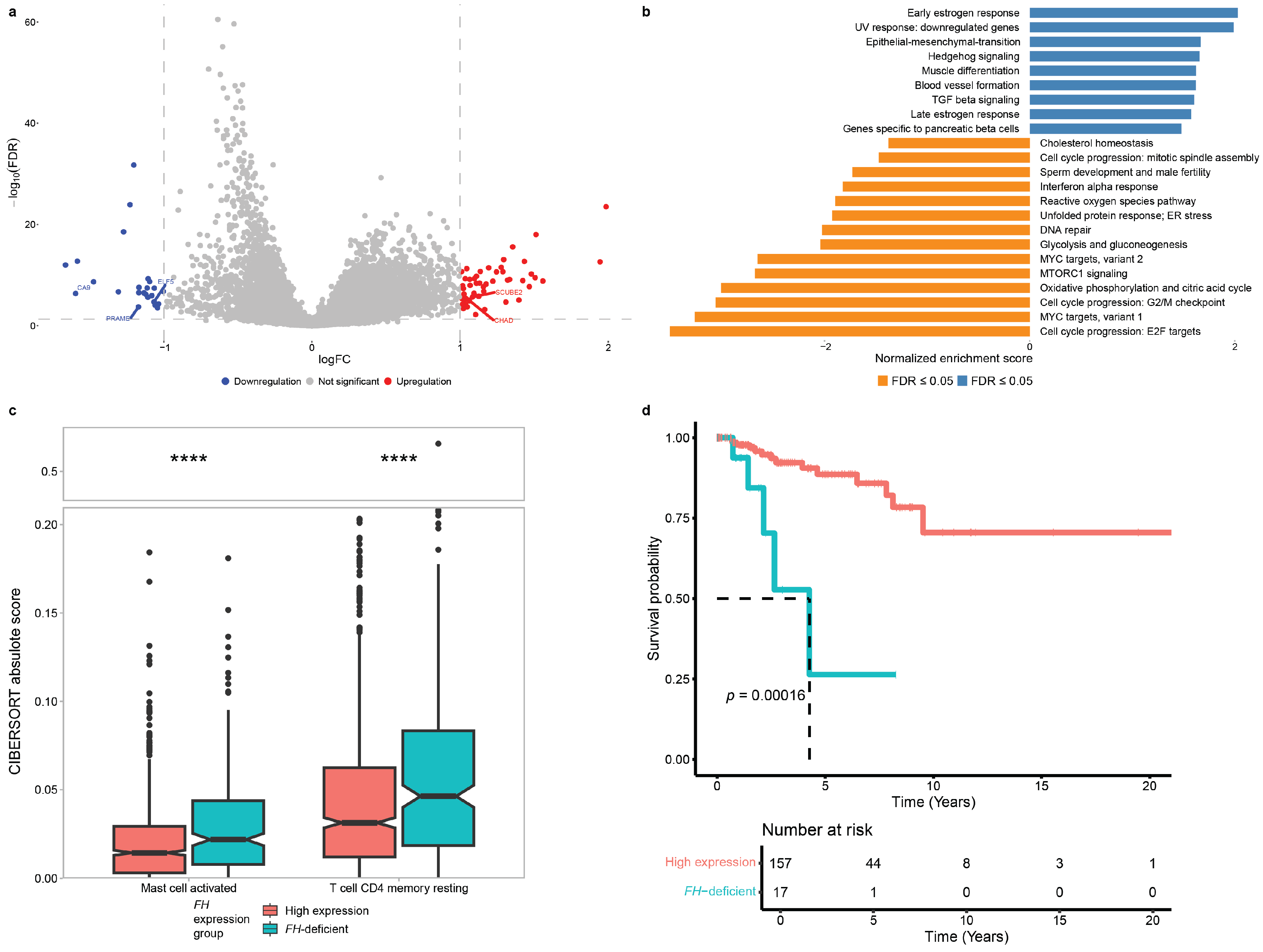
3.2. Effect of FH Deficiency on Primary Breast Cancer Tumors
3.3. Molecular and Clinical Consequences of FH Deficiency in Breast Cancer
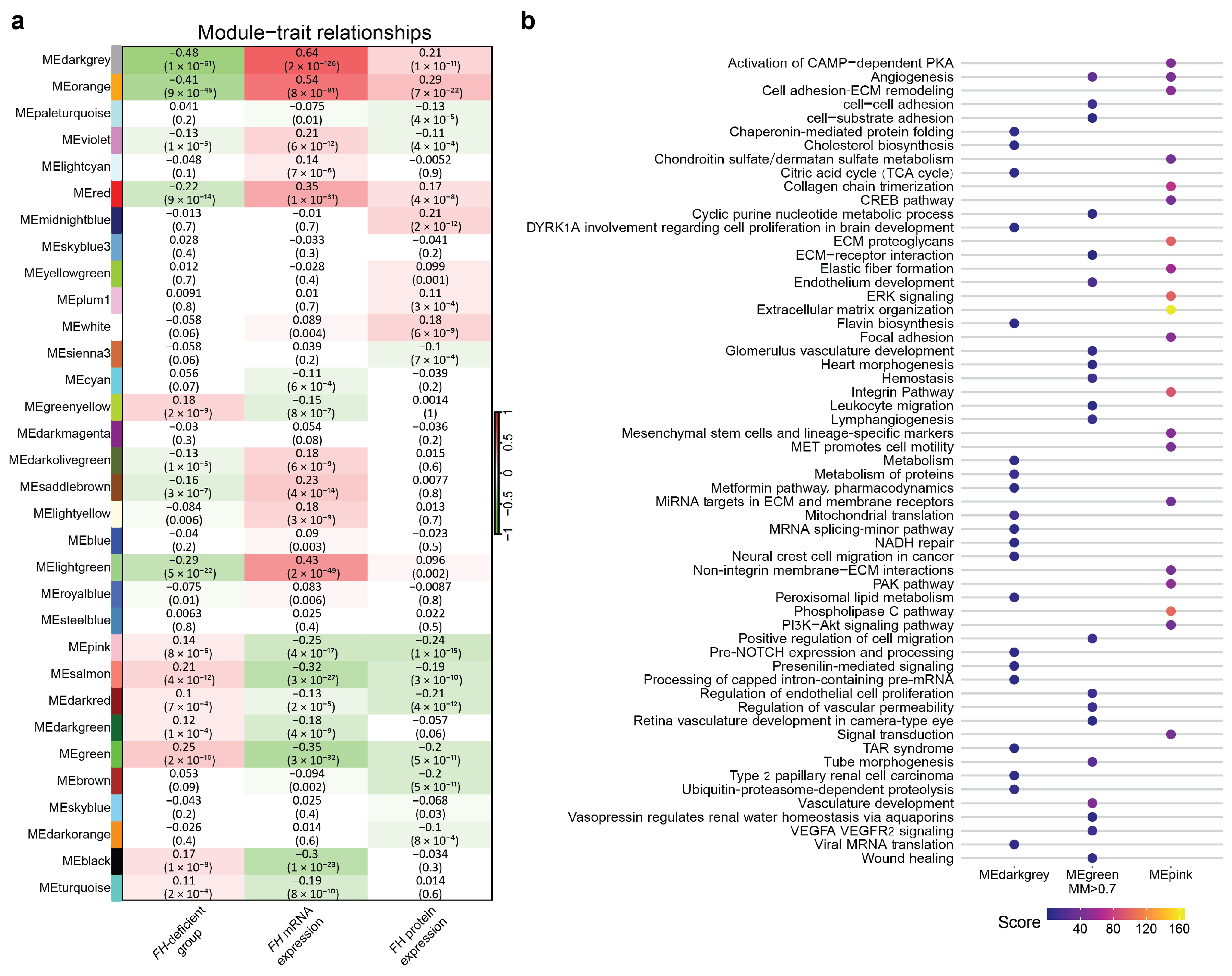
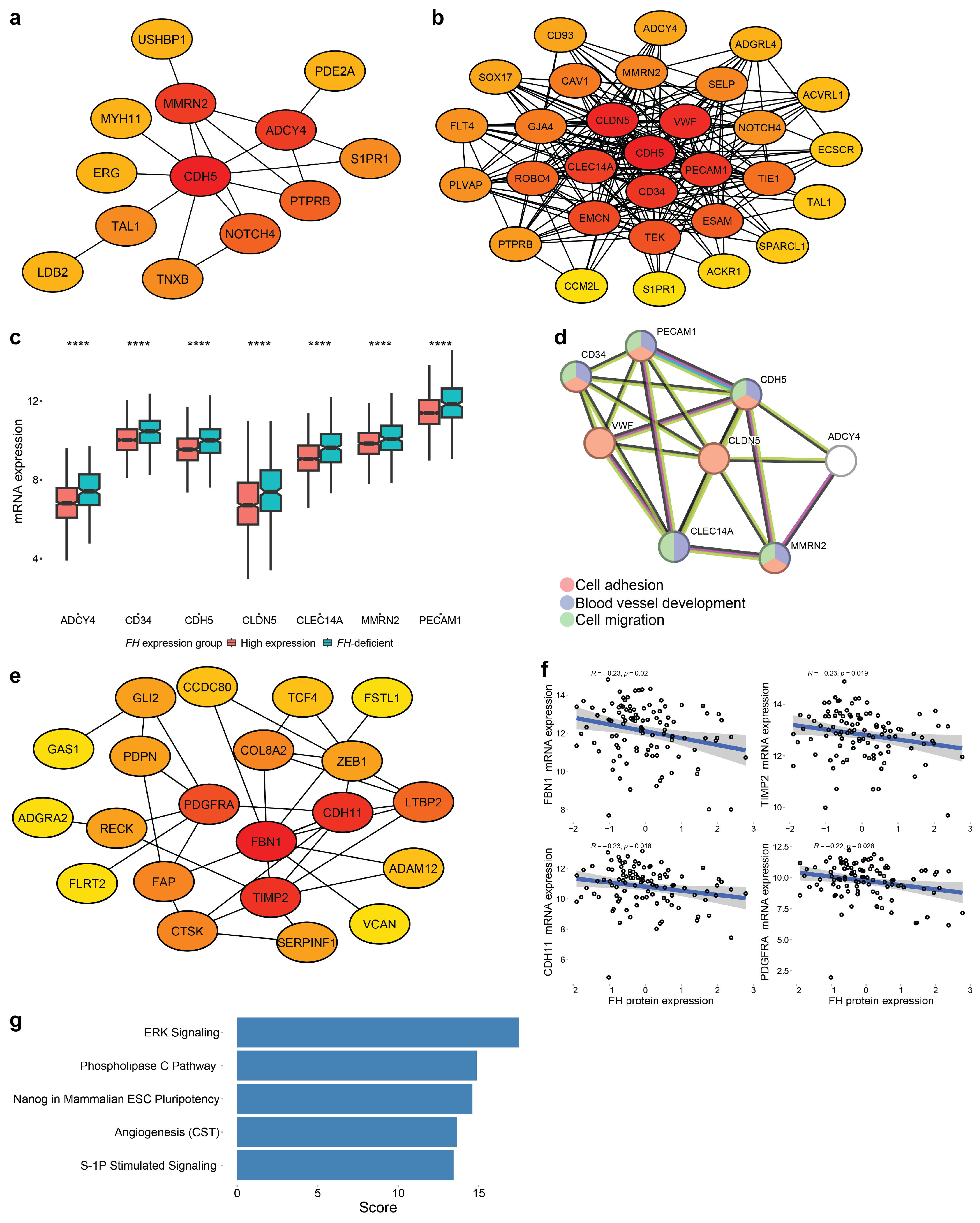
3.4. Weighted Correlation Network Analysis
3.5. Identifying Key Hub Genes
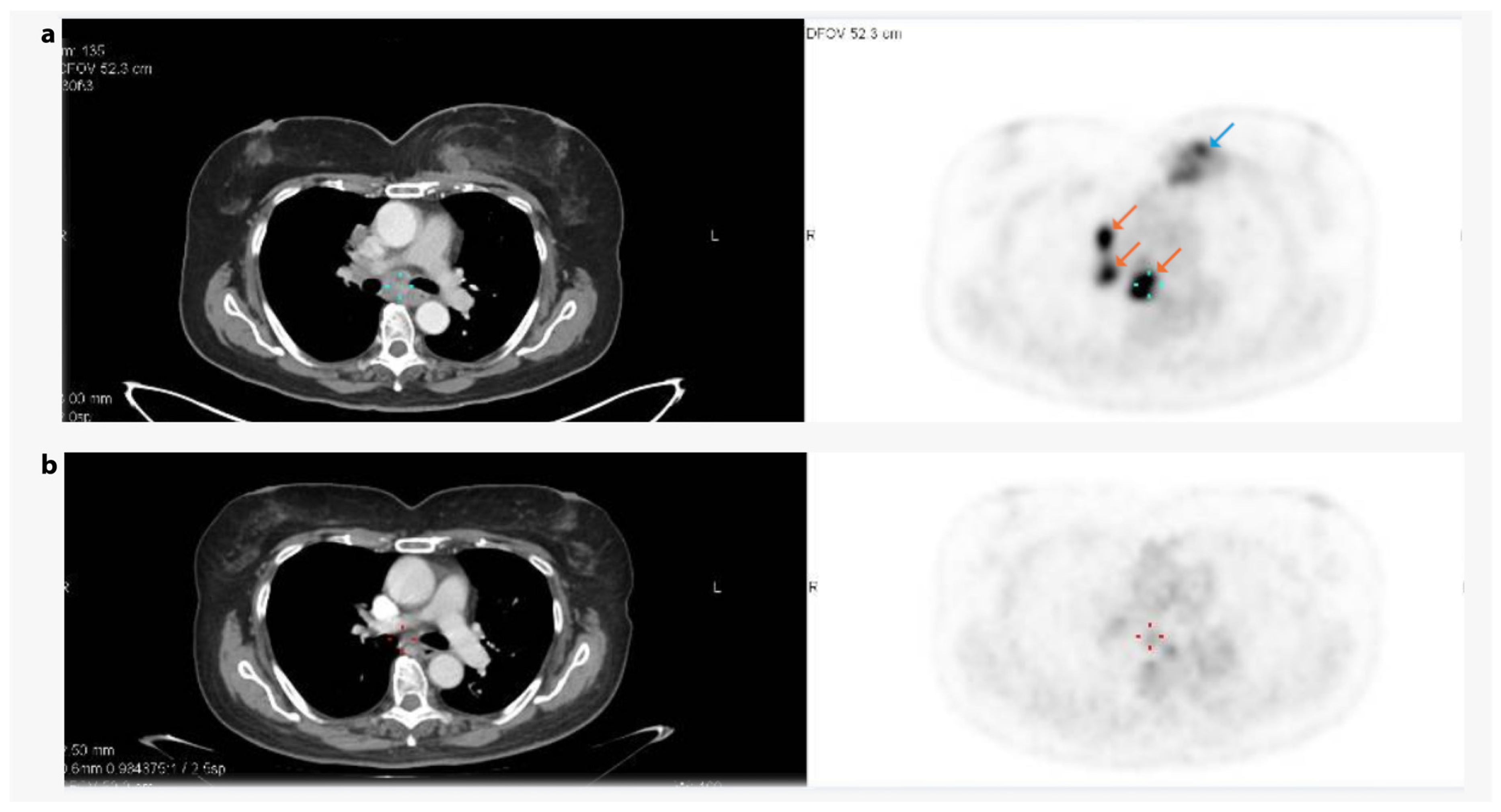
3.6. Case Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| CNA | Copy number alterations |
| DD | Deep deletion |
| DEG | Differentially expressed genes |
| EMT | Epithelial–mesenchymal transition |
| FH | Fumarate hydratase |
| GS | Gene significance |
| GSEA | Gene set enrichment analysis |
| HLRCC | Hereditary leiomyomatosis and renal cell carcinoma |
| IDC | Invasive ductal carcinoma |
| MM | Module membership |
| OR | Odd ratio |
| ORA | Overrepresentation analysis |
| OS | Overall survival |
| PFS | Progression-free survival |
| PPI | Protein–protein interaction |
| SD | Shallow deletion |
| SV | Structural variants |
| TCA | Tricarboxylic acid |
| TIL | Tumor-infiltrating lymphocytes |
| TMB | Tumor mutation burden |
| TME | Tumor microenvironment |
| VEGF | Vascular endothelial growth factor |
| WGCNA | Weighted correlation network analysis |
References
- Chitoran, E.; Rotaru, V.; Stefan, D.-C.; Gullo, G.; Simion, L. Blocking Tumoral Angiogenesis VEGF/VEGFR Pathway: Bevacizumab—20 Years of Therapeutic Success and Controversy. Cancers 2025, 17, 1126. [Google Scholar] [CrossRef]
- Kristensen, T.B.; Knutsson, M.L.T.; Wehland, M.; Laursen, B.E.; Grimm, D.; Warnke, E.; Magnusson, N.E. Anti-Vascular Endothelial Growth Factor Therapy in Breast Cancer. Int. J. Mol. Sci. 2014, 15, 23024–23041. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Wang, M.; Gralow, J.; Dickler, M.; Cobleigh, M.; Perez, E.A.; Shenkier, T.; Cella, D.; Davidson, N.E. Paclitaxel plus Bevacizumab versus Paclitaxel Alone for Metastatic Breast Cancer. N. Engl. J. Med. 2007, 357, 2666–2676. [Google Scholar] [CrossRef]
- Hey, S.P.; Gyawali, B.; D’Andrea, E.; Kanagaraj, M.; Franklin, J.M.; Kesselheim, A.S. A Systematic Review and Meta-Analysis of Bevacizumab in First-Line Metastatic Breast Cancer: Lessons for Research and Regulatory Enterprises. JNCI J. Natl. Cancer Inst. 2020, 112, 335–342. [Google Scholar] [CrossRef]
- Linehan, W.M.; Rouault, T.A. Molecular Pathways: Fumarate Hydratase-Deficient Kidney Cancer—Targeting the Warburg Effect in Cancer. Clin. Cancer Res. 2013, 19, 3345–3352. [Google Scholar] [CrossRef]
- Sudarshan, S.; Linehan, W.M.; Neckers, L. HIF and Fumarate Hydratase in Renal Cancer. Br. J. Cancer 2007, 96, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Kancherla, P.; Daneshvar, M.; Sager, R.A.; Mollapour, M.; Bratslavsky, G. Fumarate Hydratase as a Therapeutic Target in Renal Cancer. Expert Opin. Ther. Targets 2020, 24, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Ferlicot, S.; Guillaud-Bataille, M.; Le Teuff, G.; Genestie, C.; Deveaux, S.; Slama, A.; Poulalhon, N.; Escudier, B.; Albiges, L.; et al. Reassessing the Clinical Spectrum Associated with Hereditary Leiomyomatosis and Renal Cell Carcinoma Syndrome in FrenchFHMutation Carriers. Clin. Genet. 2017, 92, 606–615. [Google Scholar] [CrossRef]
- Schultz, K.A.P.; Rednam, S.P.; Kamihara, J.; Doros, L.; Achatz, M.I.; Wasserman, J.D.; Diller, L.R.; Brugières, L.; Druker, H.; Schneider, K.A.; et al. PTEN, DICER1, FH, and Their Associated Tumor Susceptibility Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin. Cancer Res. 2017, 23, e76–e82. [Google Scholar] [CrossRef]
- Adam, J.; Hatipoglu, E.; O’Flaherty, L.; Ternette, N.; Sahgal, N.; Lockstone, H.; Baban, D.; Nye, E.; Stamp, G.W.; Wolhuter, K.; et al. Renal Cyst Formation in Fh1-Deficient Mice Is Independent of the Hif/Phd Pathway: Roles for Fumarate in KEAP1 Succination and Nrf2 Signaling. Cancer Cell 2011, 20, 524–537. [Google Scholar] [CrossRef]
- O’Flaherty, L.; Adam, J.; Heather, L.C.; Zhdanov, A.V.; Chung, Y.-L.; Miranda, M.X.; Croft, J.; Olpin, S.; Clarke, K.; Pugh, C.W.; et al. Dysregulation of Hypoxia Pathways in Fumarate Hydratase-Deficient Cells Is Independent of Defective Mitochondrial Metabolism. Hum. Mol. Genet. 2010, 19, 3844–3851. [Google Scholar] [CrossRef]
- Valcarcel-Jimenez, L.; Frezza, C. Fumarate Hydratase (FH) and Cancer: A Paradigm of Oncometabolism. Br. J. Cancer 2023, 129, 1546–1557. [Google Scholar] [CrossRef]
- Shuch, B.; Li, S.; Risch, H.; Bindra, R.S.; McGillivray, P.D.; Gerstein, M. Estimation of the Carrier Frequency of Fumarate Hydratase Alterations and Implications for Kidney Cancer Risk in Hereditary Leiomyomatosis and Renal Cancer. Cancer 2020, 126, 3657–3666. [Google Scholar] [CrossRef]
- Lu, E.; Hatchell, K.E.; Nielsen, S.M.; Esplin, E.D.; Ouyang, K.; Nykamp, K.; Zavoshi, S.; Li, S.; Zhang, L.; Wilde, B.R.; et al. Fumarate Hydratase Variant Prevalence and Manifestations Among Individuals Receiving Germline Testing. Cancer 2022, 128, 675–684. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438.e6. [Google Scholar] [CrossRef]
- Razavi, P.; Dickler, M.N.; Shah, P.D.; Toy, W.; Brown, D.N.; Won, H.H.; Li, B.T.; Shen, R.; Vasan, N.; Modi, S.; et al. Alterations in PTEN and ESR1 Promote Clinical Resistance to Alpelisib plus Aromatase Inhibitors. Nat. Cancer 2020, 1, 382–393. [Google Scholar] [CrossRef]
- Smith, A.E.; Ferraro, E.; Safonov, A.; Morales, C.B.; Lahuerta, E.J.A.; Li, Q.; Kulick, A.; Ross, D.; Solit, D.B.; de Stanchina, E.; et al. HER2 + Breast Cancers Evade Anti-HER2 Therapy via a Switch in Driver Pathway. Nat. Commun. 2021, 12, 6667. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, B.; Guo, J.; Shao, H.; Del Priore, I.S.; Chang, Q.; Kudo, R.; Li, Z.; Razavi, P.; Liu, B.; et al. INK4 Tumor Suppressor Proteins Mediate Resistance to CDK4/6 Kinase Inhibitors. Cancer Discov. 2022, 12, 356–371. [Google Scholar] [CrossRef]
- Nguyen, B.; Fong, C.; Luthra, A.; Smith, S.A.; DiNatale, R.G.; Nandakumar, S.; Walch, H.; Chatila, W.K.; Madupuri, R.; Kundra, R.; et al. Genomic Characterization of Metastatic Patterns from Prospective Clinical Sequencing of 25,000 Patients. Cell 2022, 185, 563–575.e11. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- cBioPortal for Cancer Genomics. Available online: https://www.cbioportal.org/study/summary?id=ilc_msk_2023 (accessed on 5 September 2025).
- Wu, L.; Yao, H.; Chen, H.; Wang, A.; Guo, K.; Gou, W.; Yu, Y.; Li, X.; Yao, M.; Yuan, S.; et al. Landscape of Somatic Alterations in Large-Scale Solid Tumors from an Asian Population. Nat. Commun. 2022, 13, 4264. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef]
- cBioPortal for Cancer Genomics. Available online: https://www.cbioportal.org/study/summary?id=brca_mbcproject_2022 (accessed on 5 September 2025).
- Jee, J.; Fong, C.; Pichotta, K.; Tran, T.N.; Luthra, A.; Waters, M.; Fu, C.; Altoe, M.; Liu, S.-Y.; Maron, S.B.; et al. Automated Real-World Data Integration Improves Cancer Outcome Prediction. Nature 2024, 636, 728–736. [Google Scholar] [CrossRef]
- cBioPortal for Cancer Genomics. Available online: https://www.cbioportal.org/study/summary?id=breast_msk_2025 (accessed on 5 September 2025).
- The Cancer Genome Atlas Network. Cancer Genome Atlas Network Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Zon, M.; Gendoo, D.M.A.; Haibe-Kains, B. MetaGxBreast: Transcriptomic Breast Cancer Datasets. 2025. Available online: https://www.bioconductor.org/packages/devel/data/experiment/html/MetaGxBreast.html (accessed on 31 July 2025).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Dolgalev, I. msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format; Zenodo: Geneva, Switzerland, 2025. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Cui, H.; Zhao, G.; Lu, Y.; Zuo, S.; Duan, D.; Luo, X.; Zhao, H.; Li, J.; Zeng, Z.; Chen, Q.; et al. TIMER3: An Enhanced Resource for Tumor Immune Analysis. Nucleic Acids Res. 2025, 53, W534–W541. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 2012, 46, 1–17. [Google Scholar] [CrossRef]
- Horvath, S. Weighted Network Analysis: Applications in Genomics and Systems Biology; Springer: New York, NY, 2011; ISBN 978-1-4419-8818-8. [Google Scholar]
- Sinberger, L.A.; Zahavi, T.; Sonnenblick, A.; Salmon-Divon, M. Coexistent ARID1A-PIK3CA Mutations Are Associated with Immune-Related Pathways in Luminal Breast Cancer. Sci. Rep. 2023, 13, 20911. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, H.-Y.; Zhu, J.; Niu, Y.-M.; Zhang, C.; Guo, G.-L. Identification of Hub Genes and Key Pathways Associated With Bipolar Disorder Based on Weighted Gene Co-Expression Network Analysis. Front. Physiol. 2019, 10, 1081. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster Gene Set Analysis and New Support for Metabolomics and Multi-Omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Ben-Ari Fuchs, S.; Lieder, I.; Stelzer, G.; Mazor, Y.; Buzhor, E.; Kaplan, S.; Bogoch, Y.; Plaschkes, I.; Shitrit, A.; Rappaport, N.; et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. OMICS A J. Integr. Biol. 2016, 20, 139–151. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using ’ggplot2’, R package version 0.4.9; 2021. Available online: https://CRAN.R-project.org/package=survminer (accessed on 31 July 2025).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia—A Key Regulatory Factor in Tumour Growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef]
- Chappell, J.C.; Payne, L.B.; Rathmell, W.K. Hypoxia, Angiogenesis, and Metabolism in the Hereditary Kidney Cancers. J. Clin. Investig. 2019, 129, 442–451. [Google Scholar] [CrossRef]
- van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Hollern, D.P.; Swiatnicki, M.R.; Andrechek, E.R. Histological Subtypes of Mouse Mammary Tumors Reveal Conserved Relationships to Human Cancers. PLoS Genet. 2018, 14, e1007135. [Google Scholar] [CrossRef]
- KEGG PATHWAY: Focal Adhesion—Homo Sapiens (Human). Available online: https://www.kegg.jp/pathway/hsa04510 (accessed on 24 February 2025).
- Aden, D.; Sureka, N.; Zaheer, S.; Chaurasia, J.K.; Zaheer, S. Metabolic Reprogramming in Cancer: Implications for Immunosuppressive Microenvironment. Immunology 2025, 174, 30–72. [Google Scholar] [CrossRef]
- Baran, J.; Sobiepanek, A.; Mazurkiewicz-Pisarek, A.; Rogalska, M.; Gryciuk, A.; Kuryk, L.; Abraham, S.N.; Staniszewska, M. Mast Cells as a Target—A Comprehensive Review of Recent Therapeutic Approaches. Cells 2023, 12, 1187. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, H.; Li, Z.; Li, D.; Liu, S.; Huang, H.; Li, M. Application of Weighted Gene Co-Expression Network Analysis to Identify Key Modules and Hub Genes in Oral Squamous Cell Carcinoma Tumorigenesis. OncoTargets Ther. 2018, 11, 6001–6021. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, J.; Huang, G.; Yan, J.; Xu, C.; Dou, Z.; Sun, C.; Zhang, H. The Crosstalk between HIFs and Mitochondrial Dysfunctions in Cancer Development. Cell Death Dis. 2021, 12, 215. [Google Scholar] [CrossRef]
- Zyla, R.E.; Hodgson, A. Gene of the Month: FH. J. Clin. Pathol. 2021, 74, 615–619. [Google Scholar] [CrossRef]
- Kupiec-Weglinski, J.W. NRF2: New Mechanistic Insights and Therapeutic Perspectives. Antioxid. Redox Signal. 2024, 40, 632–635. [Google Scholar] [CrossRef]
- Sudarshan, S.; Sourbier, C.; Kong, H.-S.; Block, K.; Romero, V.A.V.; Yang, Y.; Galindo, C.; Mollapour, M.; Scroggins, B.; Goode, N.; et al. Fumarate Hydratase Deficiency in Renal Cancer Induces Glycolytic Addiction and Hypoxia-Inducible Transcription Factor 1α Stabilization by Glucose-Dependent Generation of Reactive Oxygen Species. Mol. Cell Biol. 2009, 29, 4080–4090. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Gonçalves, E.; Isaac Johnson, T.; Roberto Zecchini, V.; da Costa, A.S.H.; Gaude, E.; Vercauteren Drubbel, A.; Julian Theobald, S.; Abbo, S.; Tran, M.; et al. Fumarate Is an Epigenetic Modifier That Elicits Epithelial-to-Mesenchymal Transition. Nature 2016, 537, 544–547. [Google Scholar] [CrossRef]
- Delgado-Bellido, D.; Oliver, F.J.; Vargas Padilla, M.V.; Lobo-Selma, L.; Chacón-Barrado, A.; Díaz-Martin, J.; de Álava, E. VE-Cadherin in Cancer-Associated Angiogenesis: A Deceptive Strategy of Blood Vessel Formation. Int. J. Mol. Sci. 2023, 24, 9343. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, L.; Wu, B.; Guo, X.; Shi, Y.; Lv, C.; Yu, Y.; Zhang, Y.; Liang, Z.; Zhong, C.; et al. Angiogenesis Modulated by CD93 and Its Natural Ligands IGFBP7 and MMRN2: A New Target to Facilitate Solid Tumor Therapy by Vasculature Normalization. Cancer Cell Int. 2023, 23, 189. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, T.; Chen, F.; Yu, Z. The Impact of Hsa-miR-1972 on the Expression of von Willebrand Factor in Breast Cancer Progression Regulation. PeerJ 2024, 12, e18476. [Google Scholar] [CrossRef] [PubMed]
- Grizzi, F.; Hegazi, M.A.A.A.; Zanoni, M.; Vota, P.; Toia, G.; Clementi, M.C.; Mazzieri, C.; Chiriva-Internati, M.; Taverna, G. Prostate Cancer Microvascular Routes: Exploration and Measurement Strategies. Life 2023, 13, 2034. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, W.; Zuo, L.; Xu, M.; Wu, Y.; Huang, J.; Zhang, X.; Li, Y.; Wang, J.; Chen, J.; et al. The Fibrillin-1/VEGFR2/STAT2 Signaling Axis Promotes Chemoresistance via Modulating Glycolysis and Angiogenesis in Ovarian Cancer Organoids and Cells. Cancer Commun. 2022, 42, 245–265. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Z.; Shen, J.; Ma, X.; Zheng, S.; Zheng, Y.; Cao, K.; Dong, N. Identifying and Validating Angiogenesis-Related Genes Remodeling Tumor Microenvironment and Suppressing Immunotherapy Response in Gastric Cancer. Gene 2024, 928, 148796. [Google Scholar] [CrossRef]
- Ling, Y.; Kang, X.; Yi, Y.; Feng, S.; Ma, G.; Qu, H. CLDN5: From Structure and Regulation to Roles in Tumors and Other Diseases beyond CNS Disorders. Pharmacol. Res. 2024, 200, 107075. [Google Scholar] [CrossRef] [PubMed]
| Study | Sample Type | Total (N) | FH Mutation | FH Structural Variance (SV) | FH Copy Number Alterations (CNAs) | 1Q Arm Status | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Mutated Samples | Total Number of Samples | Number of Samples with SV | Total Number of Samples | Amp. * | Gain | SD * | DD * | Diploid | Total Number of Samples | Gain | Loss | Total Number of Samples | |||
| Breast Cancer (MSK, Cancer Cell 2018 [17]) | Metastasis | 1000 | 2 (0.2%) | 1000 | 1 (0.1%) | 1000 | 20 | 0 | 0 | 1 (0.1%) | 979 | 1000 | NA | NA | NA |
| Primary | 918 | 4 (0.44%) | 918 | 1 (0.11%) | 918 | 1 | 0 | 0 | 0 | 917 | 918 | ||||
| Breast Cancer (MSK, Nature Cancer 2020) [18] | Metastasis | 30 | 0 | 30 | 0 | 30 | 1 | 1 | 0 | 0 | 28 | 30 | NA | NA | NA |
| Primary | 8 | 0 | 8 | 0 | 8 | 0 | 0 | 0 | 0 | 8 | 8 | ||||
| MAPK on Resistance to Anti-HER2 Therapy for Breast Cancer (MSK, Nat Commun. 2022) [19] | Metastasis | 91 | 0 | 91 | 0 | 91 | 1 | 0 | 0 | 0 | 90 | 91 | NA | NA | NA |
| Primary | 54 | 0 | 54 | 0 | 54 | 0 | 0 | 0 | 0 | 54 | 54 | ||||
| Metastatic Breast Cancer (MSK, Cancer Discovery 2022) [20] | Metastasis | 1365 | 6 (0.44%) | 1365 | 1 (0.07%) | 1365 | 25 | 0 | 0 | 0 | 1340 | 1365 | NA | NA | NA |
| MSK MetTropism (MSK, Cell 2021) [21] | Metastasis | 1048 | 4 (0.38%) | 1048 | 1 (0.1%) | 1048 | 19 | 0 | 0 | 1 (0.1%) | 1028 | 1048 | NA | NA | NA |
| Primary | 1561 | 6 (0.38%) | 1561 | 1 (0.06%) | 1561 | 6 | 0 | 0 | 0 | 1555 | 1561 | ||||
| MSK-IMPACT Clinical Sequencing Cohort (MSK, Nat Med 2017) [22] | Metastasis | 837 | 1 (0.12%) | 837 | 1 (0.12%) | 837 | 14 | 0 | 0 | 1 (0.12%) | 822 | 837 | NA | NA | NA |
| Primary | 500 | 2 (0.4%) | 500 | 1 (0.2%) | 500 | 1 | 0 | 0 | 0 | 499 | 500 | ||||
| Non-CDH1 Invasive Lobular Carcinoma (MSK, 2023) [23] | Primary | 25 | 0 | 25 | 0 | 25 | 0 | 0 | 0 | 0 | 25 | 25 | NA | NA | NA |
| China Pan-cancer (OrigiMed, Nature 2022) [24] | Metastasis | 25 | 0 | 25 | 0 | 25 | 0 | 0 | 0 | 0 | 25 | 25 | NA | NA | NA |
| Primary | 71 | 0 | 71 | 0 | 71 | 0 | 0 | 0 | 0 | 71 | 71 | ||||
| Breast Invasive Carcinoma (TCGA, PanCancer Atlas) [25] | Primary | 1052 | 4 (0.38%) | 1052 | 0 | 1052 | 101 | 667 | 37 (3.5%) | 0 | 247 | 1052 | 625 | 10 (1.13%) | 888 |
| The Metastatic Breast Cancer Project (provisional, December 2021) [26] | NA | 334 | 1 (0.3%) | 334 | 0 | 156 | 49 | 115 | 96 (28.7%) | 6 (1.8%) | 68 | 334 | NA | NA | NA |
| MSK-CHORD (MSK, Nature 2024) [27] | Metastasis | 2564 | 9 (0.35%) | 2564 | 1 (0.04%) | 2464 | 42 | 0 | 0 | 1 (0.04%) | 2421 | 2464 | NA | NA | NA |
| Primary | 2859 | 13 (0.45%) | 2859 | 1 (0.034%) | 2859 | 14 | 0 | 0 | 0 | 2845 | 2859 | ||||
| Breast Cancer (MSK, 2025) [28] | Metastasis | 2048 | 12 (0.58%) | 2048 | 2 (0.1%) | 2048 | 38 | 0 | 0 | 1 (0.05%) | 2009 | 2048 | NA | NA | NA |
| Primary | 1812 | 5 (0.27%) | 1812 | 1 (0.06) | 1812 | 8 | 0 | 0 | 0 | 1804 | 1812 | ||||
| Patient ID | Sample ID | Study | Sample Type | TMB | Protein Change | Mutation Type | Variant Type | Mutation Status | Chr. | Start Pos | End Pos | Ref | Var | Fraction Genome Altered |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-0022525 | P-0022525-T01-IM6 | MSK, Cell 2021 [21] | P | 3.46 | L208Vfs*9 | Frameshift Insertion | INS | Somatic | 1 | 241,672,019 | 241,672,020 | - | C | 0.222 |
| P-0005712 | P-0005712-T01-IM5 | MSK, Cell 2021 [21] | P | 3.91 | FH intragenic | Fusion | Dup | Somatic | 1 | 241,665,729 | NA | 0.4247 | ||
| P-0005712 | P-0005712-T01-IM5 | MSK, Cancer Cell 2018 [17] | P | 0.133 | FH intragenic | Fusion | Dup | Somatic | 1 | 241,665,729 | NA | 0.5076 | ||
| P-0045182 | P-0045182-T01-IM6 | MSK, Nature 2024 [27] | P | 1.73 | Q237* | Nonsense | SNP | Somatic | 1 | 241,671,932 | 241,671,932 | G | A | 0.4 |
| Patient0707 | P-0707 | OrigiMed, Nature 2022 [24] | P | 0.166 | X186_splice | Splice Region | SNP | NA | 1 | 241,672,089 | 241,672,089 | T | C | NA |
| P-0017116 | P-0017116-T01-IM6 | MSK, Cancer Discovery 2022 [20] | M | 8.65 | P63Ifs*9 | Frameshift Deletion | DEL | Somatic | 1 | 241,680,556 | 241,680,562 | CATTTGG | - | 0.2691 |
| P-0004918 | P-0004918-T02-IM6 | MSK, Cell 2021 [21] | M | 6.92 | RGS7-FH Fusion | Fusion | Dup | Somatic | 1 | 241,357,653 | NA | 0.2868 | ||
| P-0000532 | P-0000532-T02-IM5 | MSK, Cancer Discovery 2022 [20] | M | 8.81 | PDE1C-FH Fusion | Fusion | Trans | Somatic | 7 | 31,926,696 | NA | 0.6164 | ||
| P-0048392 | P-0048392-T01-IM6 | MSK, Nature 2024 [27] | M | 8.65 | MIR-1273E/1273E-FH Fusion | Fusion | Inversion | Somatic | 1 | 240,716,629 | NA | 0.136 | ||
| P-0000532 | P-0000532-T02-IM5 | MSK, Cancer Cell 2018 [17] | M | 0.3 | PDE1C-FH Fusion | Fusion | Trans | Somatic | 7 | 31,926,696 | NA | 0.5412 | ||
| P-0008574 | P-0008574-T03-IM6 | MSK, Cell 2021 [21] | M | 7.78 | M336K | Missense | SNP | Somatic | 1 | 241,667,443 | 241,667,443 | A | T | 0.2097 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinberger, L.A.; Keren-Khadmy, N.; Goldberg, A.; Peretz-Yablonski, T.; Sonnenblick, A.; Salmon-Divon, M. Revealing the Angiogenic Signature of FH-Deficient Breast Cancer: Genomic Profiling and Clinical Implications. Cancers 2025, 17, 2942. https://doi.org/10.3390/cancers17182942
Sinberger LA, Keren-Khadmy N, Goldberg A, Peretz-Yablonski T, Sonnenblick A, Salmon-Divon M. Revealing the Angiogenic Signature of FH-Deficient Breast Cancer: Genomic Profiling and Clinical Implications. Cancers. 2025; 17(18):2942. https://doi.org/10.3390/cancers17182942
Chicago/Turabian StyleSinberger, Liat Anabel, Noa Keren-Khadmy, Assaf Goldberg, Tamar Peretz-Yablonski, Amir Sonnenblick, and Mali Salmon-Divon. 2025. "Revealing the Angiogenic Signature of FH-Deficient Breast Cancer: Genomic Profiling and Clinical Implications" Cancers 17, no. 18: 2942. https://doi.org/10.3390/cancers17182942
APA StyleSinberger, L. A., Keren-Khadmy, N., Goldberg, A., Peretz-Yablonski, T., Sonnenblick, A., & Salmon-Divon, M. (2025). Revealing the Angiogenic Signature of FH-Deficient Breast Cancer: Genomic Profiling and Clinical Implications. Cancers, 17(18), 2942. https://doi.org/10.3390/cancers17182942





