Amplification-Free Testing of microRNA Biomarkers in Cancer
Simple Summary
Abstract
1. Introduction
2. Conventional Analytical Methods
2.1. Quantitative and Digital PCR-Based Technologies
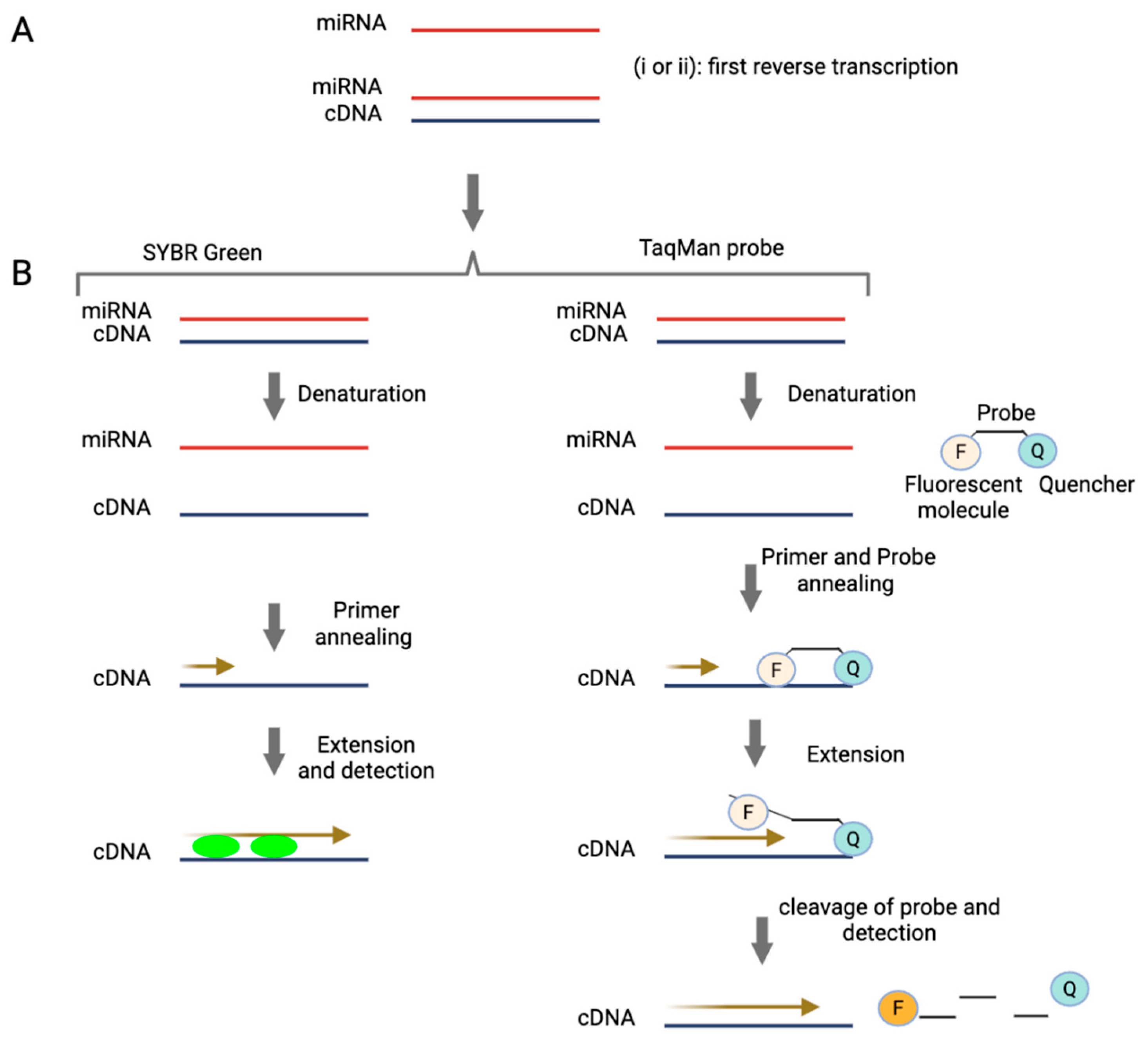
2.2. miRNA Microarrays
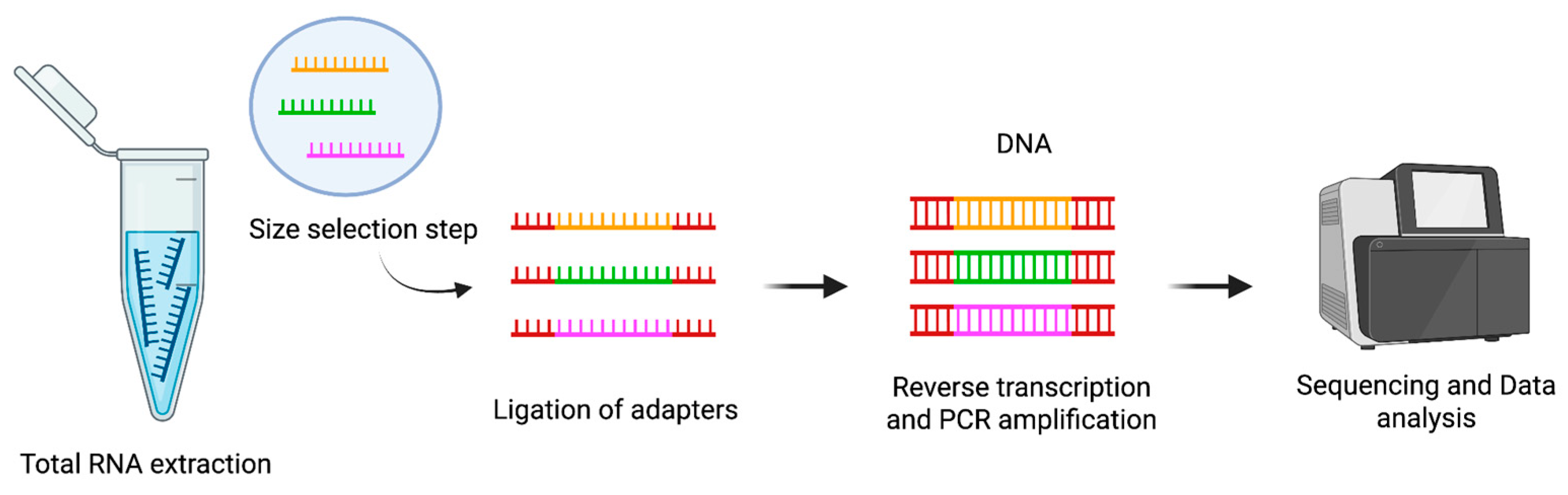
2.3. Next-Generation Sequencing for miRNA Profiling
3. Aim and Methodology
- A fluorescent spherical nucleic acid (FSNA)-assisted microfluidic chip developed for miRNA detection [66].
- A dual-aptamer modified gold nanoparticle (AuNP) system enabling universal miRNA detection via dye-fading readout [67].
- A cascade CRISPR/Cas-based method for amplification-free miRNA sensing [68].
- An ultra-fast, clickable, fluorescence-based detection strategy [69].
4. Emerging Amplification-Free Methods for Circulating miRNA Detection in Cancer
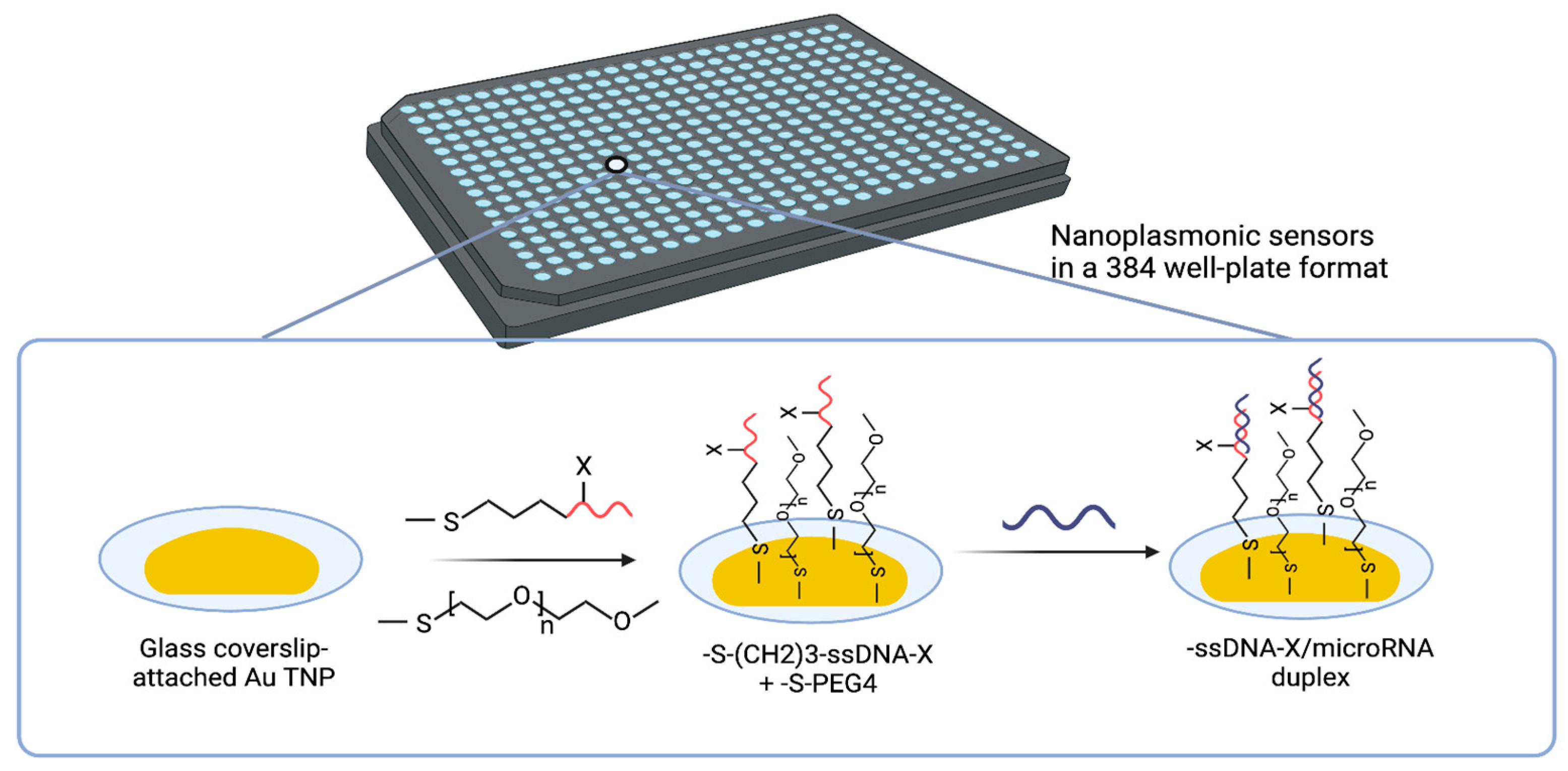
4.1. Solid-State Nanoplasmonic Sensor
4.2. Electro-Optical Nanopore Sensing
4.3. Singlet Oxygen-Based Photoelectrochemical
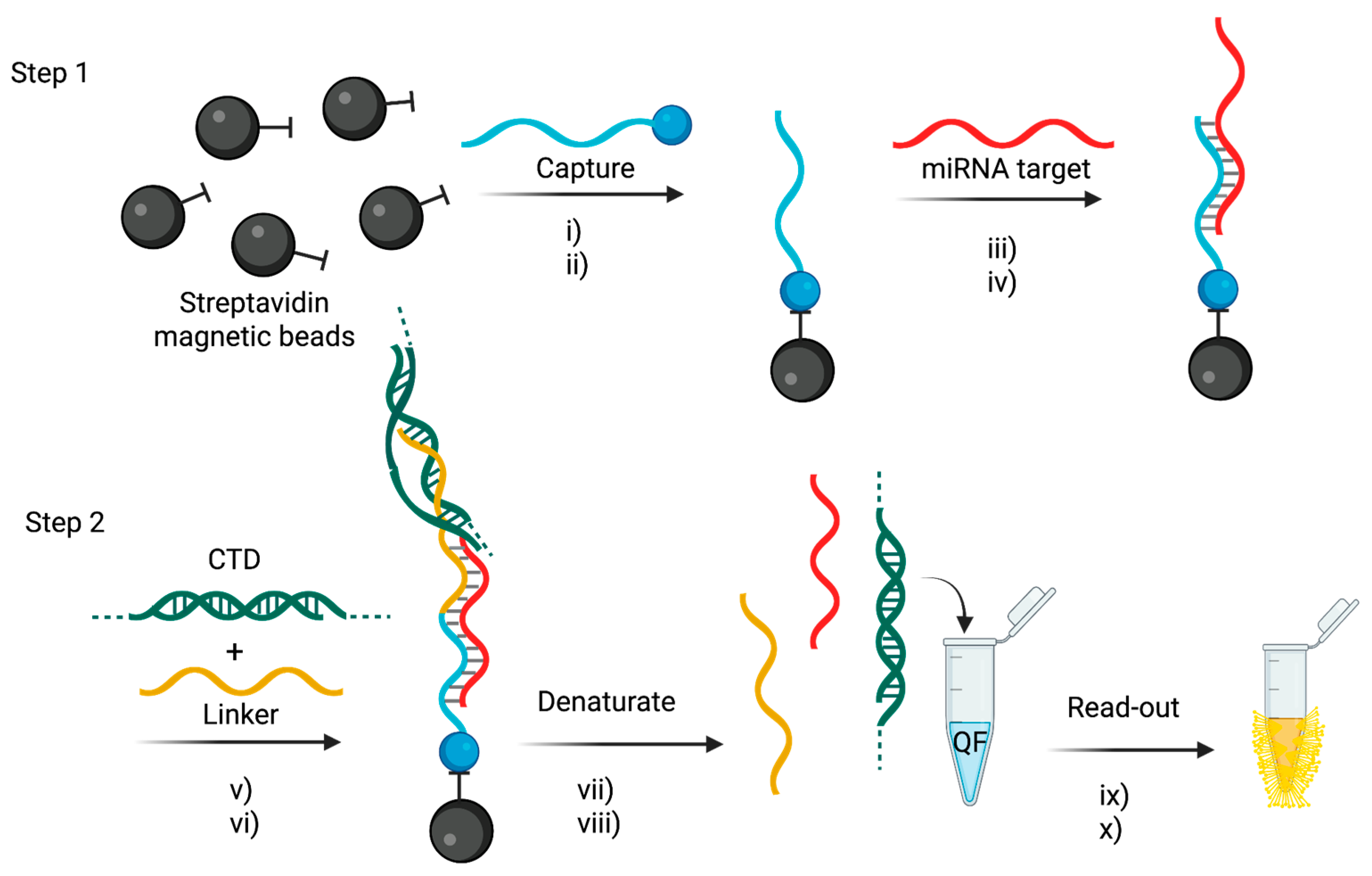
4.4. Tandem Bead-Based Hybridization Assay
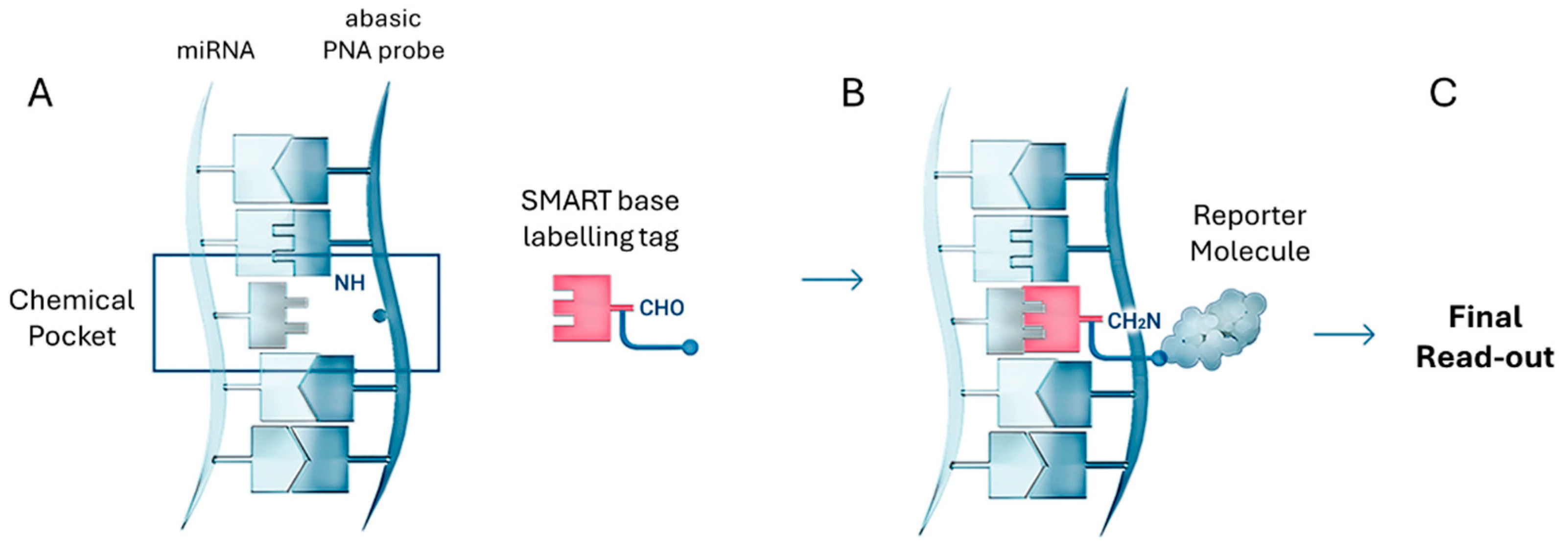
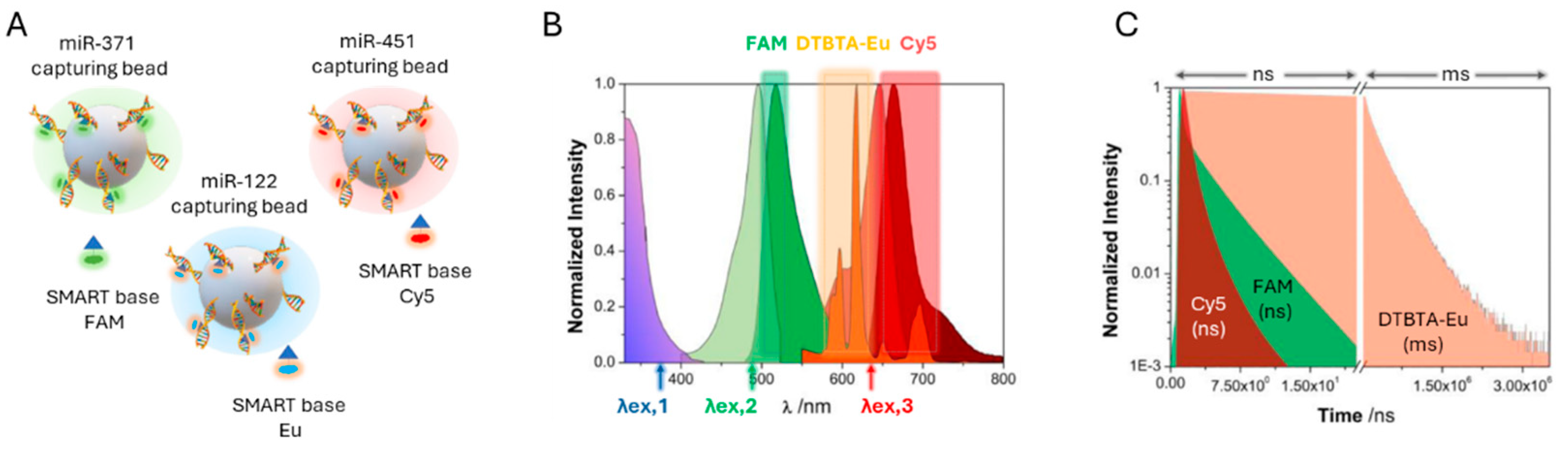
4.5. Dynamic Chemical Labeling
5. Conclusions and Future Direction
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [PubMed]
- Dizon, D.S.; Kamal, A.H. Cancer statistics 2024: All hands on deck. CA Cancer J. Clin. 2024, 74, 8–9. [Google Scholar] [CrossRef]
- American Cancer Society. American Cancer Society Annual Cancer Statistics 2024 Shows Drop in Cancer Mortality but Increasing Incidence for Six of the Top Ten Cancers. Del. J. Public Health 2024, 10, 6–7. [Google Scholar]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer current: Status, challenges and prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, S.; Matsuzaki, J.; Ochiya, T. Circulating microRNAs: Challenges with their use as liquid biopsy biomarkers. Cancer Biomark. 2022, 35, 1–9. [Google Scholar] [CrossRef]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating tumour DNA—looking beyond the blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef]
- Sallam, R.M. Proteomics in cancer biomarkers discovery: Challenges and applications. Dis. Markers 2015, 2015, 321370. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Fan, Y.; Liu, Y.; Yang, M. Recent advances in droplet-based microfluidics in liquid biopsy for cancer diagnosis. Droplet 2024, 3, e92. [Google Scholar] [CrossRef]
- Primorac, D.; Ciechanover, A. Personalized medicine: The future is here. Croat. Med. J. 2024, 65, 169–173. [Google Scholar] [CrossRef]
- Bartel, D.P. Review MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. (Lond.) 2018, 15, 68. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N. Micro RNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Ha, T.Y. MicroRNAs in Human Diseases: From Cancer to Cardiovascular Disease. Immune Netw. 2011, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, G.A.D. MicroRNAs: Circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene 2024, 43, 2135–2142. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. 2024 Nobel Prize awarded for work on microRNAs. Lancet 2024, 404, 1507–1508. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Papaneophytou, C. The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins. Int. J. Mol. Sci. 2024, 25, 3403. [Google Scholar] [CrossRef]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef]
- Cicatiello, A.G.; Musone, M.; Imperatore, S.; Giulioni, C.; La Rocca, R.; Cafarelli, A.; Del Giudice, F.; Dentice, M.; Crocetto, F. Circulating miRNAs in genitourinary cancer: Pioneering advances in early detection and diagnosis. J. Liq. Biopsy 2025, 8, 100296. [Google Scholar] [CrossRef]
- Berger, F.; Reiser, M.F. Micro-RNAs as potential new molecular biomarkers in oncology: Have they reached relevance for the clinical imaging sciences? Theranostics 2013, 3, 943–952. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. BioMed Res. Int. 2015, 2015, 125094. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2024, 24, 37. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef]
- Norouzi, S.; Soltani, S.; Alipour, E. Recent advancements in biosensor designs toward the detection of intestine cancer miRNA biomarkers. Int. J. Biol. Macromol. 2023, 245, 125509. [Google Scholar] [CrossRef]
- Quang, M.T.; Nguyen, M.N. The potential of microRNAs in cancer diagnostic and therapeutic strategies: A narrative review. J. Basic Appl. Zool. 2024, 85, 7. [Google Scholar] [CrossRef]
- Mou, G.; Wang, K.; Xu, D.; Zhou, G. Evaluation of three RT-qPCR-based miRNA detection methods using seven rice miRNAs. Biosci. Biotechnol. Biochem. 2013, 77, 1349–1353. [Google Scholar] [CrossRef]
- Falak, S.; O’Sullivan, D.M.; Cleveland, M.H.; Cowen, S.; Busby, E.J.; Devonshire, A.S.; Valiente, E.; Jones, G.M.; Kammel, M.; Milavec, M.; et al. The Application of Digital PCR as a Reference Measurement Procedure to Support the Accuracy of Quality Assurance for Infectious Disease Molecular Diagnostic Testing. Clin. Chem. 2025, 71, 378–386. [Google Scholar] [CrossRef]
- Strutt, R.; Xiong, B.; Abegg, V.F.; Dittrich, P.S. Open microfluidics: Droplet microarrays as next generation multiwell plates for high throughput screening. Lab a Chip 2024, 24, 1064–1075. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 977. [Google Scholar] [CrossRef]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Gines, G.; Menezes, R.; Xiao, W.; Rondelez, Y.; Taly, V. Emerging isothermal amplification technologies for microRNA biosensing: Applications to liquid biopsies. Mol. Asp. Med. 2020, 72, 100832. [Google Scholar] [CrossRef]
- Redshaw, N.; Wilkes, T.; Whale, A.; Cowen, S.; Huggett, J.; Foy, C.A. A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. Biotechniques 2013, 54, 155–164. [Google Scholar] [CrossRef]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Considerations for accurate gene expression measurement by reverse transcription quantitative PCR when analysing clinical samples. Anal. Bioanal. Chem. 2014, 406, 6471–6483. [Google Scholar] [CrossRef]
- Choi, C.; Yoon, S.; Moon, H.; Bae, Y.U.; Kim, C.B.; Diskul-Na-Ayudthaya, P.; Ngu, T.V.; Munir, J.; Han, J.; Park, S.B.; et al. mirRICH, a simple method to enrich the small RNA fraction from over-dried RNA pellets. RNA Biol. 2018, 15, 763–772. [Google Scholar] [CrossRef]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Chen, C.; Tan, R.; Wong, L.; Fekete, R.; Halsey, J. Quantitation of MicroRNAs by Real-Time RT-qPCR. In PCR Protocols; Humana Press: Totowa, NJ, USA, 2011; pp. 43–55. [Google Scholar]
- Tajadini, M.; Panjehpour, M.; Javanmard, S. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Sauzade, M.; Brouzes, E. DPCR: A technology review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Binderup, H.G.; Madsen, J.S.; Helweg Heegaard, N.H.; Houlind, K.; Andersen, R.F.; Brasen, C.L. Quantification of microRNA levels in plasma—Impact of preanalytical and analytical conditions. PLoS ONE 2018, 13, e0201069. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Beaulieu, Y.B.; Kleinman, C.L.; Landry-Voyer, A.M.; Majewski, J.; Bachand, F. Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1. PLoS Genet. 2012, 8, e1003078. [Google Scholar] [CrossRef]
- Nejad, C.; Pépin, G.; Behlke, M.A.; Gantier, M.P. Modified polyadenylation-based RT-qPCR increases selectivity of amplification of 3’-MicroRNA isoforms. Front. Genet. 2018, 9, 11. [Google Scholar] [CrossRef]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef]
- Love, C.; Dave, S. MicroRNA Expression Profiling Using Microarrays. In Hematological Malignancies Methods and Protocols; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef]
- Jet, T.; Gines, G.; Rondelez, Y.; Taly, V. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem. Soc. Rev. 2021, 50, 4141–4161. [Google Scholar] [CrossRef]
- Li, W.; Ruan, K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 2009, 394, 1117–1124. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systemati c comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef]
- Del Vescovo, V.; Meier, T.; Inga, A.; Denti, M.A.; Borlak, J. A cross-platform comparison of Affymetrix and Agilent microarrays reveals discordant miRNA expression in lung tumors of c-Raf transgenic mice. PLoS ONE 2013, 8, e78870. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative mirnA expression platforms in the micrornA quality control (mirQC) study. Nat. Methods 2014, 11, 809–815. [Google Scholar] [CrossRef]
- Willenbrock, H.; Salomon, J.; Søkilde, R.; Barken, K.B.; Hansen, T.N.; Nielsen, F.C.; Møller, S.; Litman, T. Quantitative miRNA expression analysis: Comparing microarrays with next-generation sequencing. RNA 2009, 15, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ma, X.; Li, J.; Shao, C. Creating and maintaining a high-confidence microRNA repository for crop research: A brief review and re-examination of the current crop microRNA registries. J. Plant Physiol. 2022, 270, 153636. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bletsa, M.; Boonen, I.; Lemey, P. NGS Library Preparation Using NEXTFLEX Rapid Directional RNAseq Kit (NOVA-5138-08) for Animal Tissue Samples. Version v1. 2022. Available online: https://www.protocols.io/ (accessed on 18 August 2025).
- Fuchs, R.T.; Sun, Z.; Zhuang, F.; Robb, G.B. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS ONE 2015, 10, e0126049. [Google Scholar] [CrossRef]
- Wang, B.; Sun, F.; Luan, Y. Comparison of the effectiveness of different normalization methods for metagenomic cross-study phenotype prediction under heterogeneity. Sci. Rep. 2024, 14, 7024. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef]
- Cheng, C.; Fei, Z.; Xiao, P. Methods to improve the accuracy of next-generation sequencing. Front. Bioeng. Biotechnol. 2023, 11, 982111. [Google Scholar] [CrossRef]
- Bustin, S.; Dhillon, H.S.; Kirvell, S.; Greenwood, C.; Parker, M.; Shipley, G.L.; Nolan, T. Variability of the reverse transcription step: Practical implications. Clin. Chem. 2015, 61, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tang, Q.; Zhang, R.; Chen, H.; Khoo, B.L.; Zhang, X.; Chen, Y.; Yan, H.; Li, J.; Shao, H.; et al. Sensitive detection of microRNAs using polyadenine-mediated fluorescent spherical nucleic acids and a microfluidic electrokinetic signal amplification chip. J. Pharm. Anal. 2022, 12, 808–813. [Google Scholar] [CrossRef]
- Zhang, R.; Shao, H.; Hu, X.; Liang, Y.; Chen, H.; Zheng, S.; Liu, L. Universal, sensitive, and visual sandwich-type biosensor based on nanogold-catalyzed reduction and its application for detecting C-reactive protein in serum by a portable colorimeter. Sens. Actuators B Chem. 2025, 431, 137408. [Google Scholar] [CrossRef]
- Sha, Y.; Huang, R.; Huang, M.; Yue, H.; Shan, Y.; Huab, J.; Xing, D. Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity. Chem. Commun. 2021, 57, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Uhd, J.; Miotke, L.; Ji, H.P.; Dunaeva, M.; Pruijn, G.J.M.; Jørgensen, C.D.; Kristoffersen, E.L.; Birkedal, V.; Yde, C.W.; Nielsen, F.C.; et al. Ultra-fast detection and quantification of nucleic acids by amplification-free fluorescence assay. Analyst 2020, 17, 5836–5844. [Google Scholar] [CrossRef]
- Masterson, A.N.; Chowdhury, N.N.; Fang, Y.; Yip-Schneider, M.T.; Hati, S.; Gupta, P.; Cao, S.; Wu, H.; Schmidt, C.M.; Fishel, M.L.; et al. Amplification-Free, High-Throughput Nanoplasmonic Quantification of Circulating MicroRNAs in Unprocessed Plasma Microsamples for Earlier Pancreatic Cancer Detection. ACS Sens. 2023, 8, 1085–1100. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C.L.; Ladame, S.; Ivanov, A.P.; Edel, J.B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515. [Google Scholar] [CrossRef]
- Shanmugam, S.T.; Campos, R.; Trashin, S.; Daems, E.; Carneiro, D.; Fraga, A.; Ribeiro, R.; De Wael, K. Singlet oxygen-based photoelectrochemical detection of miRNAs in prostate cancer patients’ plasma: A novel diagnostic tool for liquid biopsy. Bioelectrochemistry 2024, 158, 108698. [Google Scholar] [CrossRef]
- Ferreira, I.; Slott, S.; Schi, C. Mutations in microRNA-128-2-3p identified with amplification-free hybridization assay. PLoS ONE 2023, 18, e0289556. [Google Scholar]
- Padial-Jaudenes, M.; Tabraue-Chávez, M.; Detassis, S.; Ruedas-Rama, M.J.; Gonzalez-Garcia, M.C.; Fara, M.A.; López-Delgado, F.J.; Gonzàlez-Vera, J.A.; Guardia-Monteagudo, J.J.; Diaz-Mochon, J.J.; et al. Multiplexed MicroRNA biomarker detection by bridging lifetime filtering imaging and dynamic chemical labeling. Sens. Actuators B Chem. 2024, 417, 136136. [Google Scholar] [CrossRef]
- Sarantis, P.; Koustas, E.; Papadimitropoulou, A.; Papavassiliou, A.G.; Karamouzis, M.V. Pancreatic ductal adenocarcinoma: Treatment hurdles, tumor microenvironment and immunotherapy. World J. Gastrointest. Oncol. 2020, 12, 173–181. [Google Scholar] [CrossRef]
- Beatty, G.L.; Werba, G.; Lyssiotis, C.A.; Simeone, D.M. The biological underpinnings of therapeutic resistance in pancreatic cancer. Genes Dev. 2021, 35, 940–962. [Google Scholar] [CrossRef]
- Willets, K.A.; Van Duyne, R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef]
- Mayer, K.M.; Hafner, J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011, 111, 3828–3857. [Google Scholar] [CrossRef]
- Kelly, K.L.; Coronado, E.; Zhao, L.L.; Schatz, G.C. The optical properties of metal nanoparticles: The influence of size, shape, and dielectric environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Joshi, G.K.; Deitz-McElyea, S.; Liyanage, T.; Lawrence, K.; Mali, S.; Sardar, R.; Korc, M. Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing at Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS Nano 2015, 9, 11075–11089. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.K.; Deitz-Mcelyea, S.; Johnson, M.; Mali, S.; Korc, M.; Sardar, R. Highly specific plasmonic biosensors for ultrasensitive MicroRNA detection in plasma from pancreatic cancer patients. Nano Lett. 2014, 14, 6955–6963. [Google Scholar] [CrossRef]
- Couture, M.; Ray, K.K.; Poirier-Richard, H.P.; Crofton, A.; Masson, J.F. 96-Well Plasmonic Sensing with Nanohole Arrays. ACS Sens. 2016, 1, 287–294. [Google Scholar] [CrossRef]
- Salleh, S.; Thyagarajan, A.; Sahu, R.P. Exploiting the relevance of CA 19-9 in pancreatic cancer. J. Cancer Metastasis Treat. 2020, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.J.; Matsumoto, Y.; Dombek, G.E.; Stackhouse, K.A.; Ore, A.S.; Glickman, J.N.; Heimburg-Molinaro, J.; Cummings, R.D. Differential expression of CD175 and CA19-9 in pancreatic adenocarcinoma. Sci. Rep. 2025, 15, 4177. [Google Scholar] [CrossRef]
- Quirico, L.; Orso, F. The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist. 2020, 3, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Zedan, A.H.; Osther, P.J.S.; Assenholt, J.; Madsen, J.S.; Hansen, T.F. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci. Rep. 2020, 10, 227. [Google Scholar] [CrossRef]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y.; et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Fälth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sültmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef]
- Trashin, S.; Rahemi, V.; Ramji, K.; Neven, L.; Gorun, S.M.; De Wael, K. Singlet oxygen-based electrosensing by molecular photosensitizers. Nat. Commun. 2017, 8, 16108. [Google Scholar] [CrossRef]
- Daems, E.; Bassini, S.; Mariën, L.; Op de Beeck, H.; Stratulat, A.; Zwaenepoel, K.; Vandamme, T.; Op de Beeck, K.; Koljenović, S.; Peeters, M.; et al. Singlet oxygen-based photoelectrochemical detection of single-point mutations in the KRAS oncogene. Biosens. Bioelectron. 2024, 249, 115957. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, S.T.; Trashin, S.; De Wael, K. Singlet oxygen-based photoelectrochemical detection of DNA. Biosens. Bioelectron. 2022, 195, 113652. [Google Scholar] [CrossRef]
- Verrucchi, M.; Giacomazzo, G.E.; Sfragano, P.S.; Laschi, S.; Conti, L.; Pagliai, M.; Gellini, C.; Ricci, M.; Ravera, E.; Valtancoli, B.; et al. Characterization of a Ruthenium(II) Complex in Singlet Oxygen-Mediated Photoelectrochemical Sensing. Langmuir 2023, 39, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Sita-Lumsden, A.; Fletcher, C.E.; Dart, D.A.; Brooke, G.N.; Waxman, J.; Bevan, C.L. Circulating nucleic acids as biomarkers of prostate cancer. Biomark. Med. 2013, 7, 867–877. [Google Scholar] [CrossRef]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Sachdeva, A.; Peeters, M.; McClements, J. Point-of-Care Prostate Specific Antigen Testing: Examining Translational Progress toward Clinical Implementation. ACS Sens. 2023, 8, 3643–3658. [Google Scholar] [CrossRef]
- Fajardo, P.; Taskova, M.; Martín-Serrano, M.A.; Hansen, J.; Slott, S.; Jakobsen, A.K.; Wibom, M.L.; Salegi, B.; Muñoz, A.; Barbachano, A.; et al. p38γ and p38δ as biomarkers in the interplay of colon cancer and inflammatory bowel diseases. Cancer Commun. 2022, 42, 897–901. [Google Scholar] [CrossRef]
- Fonseca, A.; Ramalhete, S.V.; Mestre, A.; das Neves, R.P.; Marreiros, A.; Castelo-Branco, P.; Roberto, V.P. Identification of colorectal cancer associated biomarkers: An integrated analysis of miRNA expression. Aging (Albany N. Y.) 2021, 13, 21991–22029. [Google Scholar] [CrossRef]
- Domljanovic, I.; Taskova, M.; Miranda, P.; Weber, G.; Astakhova, K. Optical and theoretical study of strand recognition by nucleic acid probes. Commun. Chem. 2020, 3, 111. [Google Scholar] [CrossRef]
- Kalies, S.; Kuetemeyer, K.; Heisterkamp, A.; Denk, W.; Strickler, J.H.; Webb, W.W. Mechanisms of high-order photobleaching and its relationship to intracellular ablation. Biomed. Opt. Express 2011, 2, 805–816. [Google Scholar] [CrossRef]
- Bowler, F.R.; Diaz-Mochon, J.J.; Swift, M.D.; Bradley, M. DNA analysis by dynamic chemistry. Angew. Chem. Int. Ed. 2010, 49, 1809–1812. [Google Scholar] [CrossRef]
- Venkateswaran, S.; Luque-González, M.A.; Tabraue-Chávez, M.; Fara, M.A.; López-Longarela, B.; Cano-Cortes, V.; López-Delgado, F.J.; Sánchez-Martín, R.M.; Ilyine, H.; Bradley, M.; et al. Novel bead-based platform for direct detection of unlabelled nucleic acids through Single Nucleobase Labelling. Talanta 2016, 161, 489–496. [Google Scholar] [CrossRef]
- Rissin, D.M.; López-Longarela, B.; Pernagallo, S.; Ilyine, H.; Vliegenthart, A.D.B.; Dear, J.W.; Díaz-Mochón, J.J. Polymerase-free measurement of microRNA-122 with single base specificity using single molecule arrays: Detection of drug-induced liver injury. PLoS ONE 2017, 12, e0179669. [Google Scholar] [CrossRef]
- Garcia-Fernandez, E.; Gonzalez-Garcia, M.C.; Pernagallo, S.; Ruedas-Rama, M.J.; Fara, M.A.; López-Delgado, F.J.; Dear, J.W.; Ilyine, H.; Ress, C.; Díaz-Mochón, J.J.; et al. MiR-122 direct detection in human serum by time-gated fluorescence imaging. Chem. Commun. 2019, 55, 14958–14961. [Google Scholar] [CrossRef]
- Marín-Romero, A.; Robles-Remacho, A.; Tabraue-Chávez, M.; López-Longarela, B.; Sánchez-Martín, R.M.; Guardia-Monteagudo, J.J.; Fara, M.A.; López-Delgado, F.J.; Pernagallo, S.; Díaz-Mochón, J.J. A PCR-free technology to detect and quantify microRNAs directly from human plasma. Analyst 2018, 143, 5676–5682. [Google Scholar] [CrossRef]
- López-Longarela, B.; Morrison, E.E.; Tranter, J.D.; Chahman-Vos, L.; Léonard, J.F.; Gautier, J.C.; Laurent, S.; Lartigau, A.; Boitier, E.; Sautier, L.; et al. Direct Detection of miR-122 in Hepatotoxicity Using Dynamic Chemical Labeling Overcomes Stability and isomiR Challenges. Anal. Chem. 2020, 92, 3388–3395. [Google Scholar] [CrossRef]
- Marín-Romero, A.; Tabraue-Chávez, M.; Dear, J.W.; Sánchez-Martín, R.M.; Ilyine, H.; Guardia-Monteagudo, J.J.; Fara, M.A.; López-Delgado, F.J.; Díaz-Mochón, J.J.; Pernagallo, S. Amplification-free profiling of microRNA-122 biomarker in DILI patient serums, using the luminex MAGPIX system. Talanta 2020, 219, 121265. [Google Scholar] [CrossRef]
- Marín-Romero, A.; Di Zeo-Sánchez, D.E.; Tabraue-Chávez, M.; Villanueva-Paz, M.; Pinazo-Bandera, J.M.; Sanabria-Cabrera, J.; García-Cortés, M.; Díaz-Mochón, J.J.; Lucena, M.I.; Andrade, R.J.; et al. Short communication: miRNA122 interrogation via PCR-Free method to track liver recovery. PLoS ONE 2025, 20, e0324858. [Google Scholar] [CrossRef]
- Robles-Remacho, A.; Martos-Jamai, I.; Tabraue-Chávez, M.; Aguilar-González, A.; Laz-Ruiz, J.A.; Cano-Cortés, M.V.; López-Delgado, F.J.; Guardia-Monteagudo, J.J.; Pernagallo, S.; Diaz-Mochon, J.J.; et al. Click chemistry-based dual nanosystem for microRNA-122 detection with single-base specificity from tumour cells. J. Nanobiotechnol. 2024, 22, 791. [Google Scholar] [CrossRef] [PubMed]
- Robles-Remacho, A.; Luque-Gonzalez, M.A.; López-Delgado, F.J.; Guardia-Monteagudo, J.J.; Fara, M.A.; Pernagallo, S.; Sanchez-Martin, R.M.; Diaz-Mochon, J.J. Direct detection of alpha satellite DNA with single-base resolution by using abasic Peptide Nucleic Acids and Fluorescent in situ Hybridization. Biosens. Bioelectron. 2023, 219, 114770. [Google Scholar] [CrossRef] [PubMed]
- Pernagallo, S.; Ventimiglia, G.; Cavalluzzo, C.; Alessi, E.; Ilyine, H.; Bradley, M.; Diaz-Mochon, J.J. Novel biochip platform for nucleic acid analysis. Sensors 2012, 12, 8100–8111. [Google Scholar] [CrossRef] [PubMed]
- Marín-Romero, A.; Pernagallo, S. A comprehensive review of Dynamic Chemical Labelling on Luminex xMAP technology: A journey towards Drug-Induced Liver Injury testing. Anal. Methods 2023, 15, 6139–6149. [Google Scholar] [CrossRef]
- Marín-Romero, A.; Tabraue-Chávez, M.; Dear, J.W.; Díaz-Mochón, J.J.; Pernagallo, S. Open a new window in the world of circulating microRNAs by merging ChemiRNA Tech with a Luminex platform. Sens. Diagn. 2022, 1, 1243–1251. [Google Scholar] [CrossRef]
- Detassis, S.; Precazzini, F.; Brentari, I.; Ruffilli, R.; Ress, C.; Maglione, A.; Pernagallo, S.; Denti, M.A. SA-ODG platform: A semi-automated and PCR-free method to analyse microRNAs in solid tissues. Analyst 2024, 149, 3891–3899. [Google Scholar] [CrossRef]
- Marín-Romero, A.; Regele, V.; Kolanovic, D.; Hofner, M.; Díaz-Mochón, J.J.; Nöhammer, C.; Pernagallo, S. MAGPIX and FLEXMAP 3D Luminex platforms for direct detection of miR-122-5p through dynamic chemical labelling. Analyst 2023, 148, 5658–5666. [Google Scholar] [CrossRef] [PubMed]
- Tabraue-Chávez, M.; Luque-González, M.A.; Marín-Romero, A.; Sánchez-Martín, R.M.; Escobedo-Araque, P.; Pernagallo, S.; Díaz-Mochón, J.J. A colorimetric strategy based on dynamic chemistry for direct detection of Trypanosomatid species. Sci. Rep. 2019, 9, 3696. [Google Scholar] [CrossRef]
- Luque-González, M.A.; Tabraue-Chávez, M.; López-Longarela, B.; Sánchez-Martín, R.M.; Ortiz-González, M.; Soriano-Rodríguez, M.; García-Salcedo, J.A.; Pernagallo, S.; Díaz-Mochón, J.J. Identification of Trypanosomatids by detecting Single Nucleotide Fingerprints using DNA analysis by dynamic chemistry with MALDI-ToF. Talanta 2018, 176, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Martín-Sierra, C.; Chavez, M.T.; Escobedo, P.; García-Cabrera, V.; López-Delgado, F.J.; Guardia-Monteagudo, J.J.; Ruiz-García, I.; Erenas, M.M.; Sanchez-Martin, R.M.; Capitán-Vallvey, L.F.; et al. SARS-CoV-2 viral RNA detection using the novel CoVradar device associated with the CoVreader smartphone app Biosensors and Bioelectronics SARS-CoV-2 viral RNA detection using the novel CoVradar device associated with the CoVreader smartphone app Luis Ferm. Biosens. Bioelectron. 2023, 230, 115268. [Google Scholar] [CrossRef] [PubMed]
- Marín-Romero, A.; Tabraue-Chávez, M.; López-Longarela, B.; Fara, M.A.; Sánchez-Martín, R.M.; Dear, J.W.; Ilyine, H.; Díaz-Mochón, J.J.; Pernagallo, S. Simultaneous Detection of Drug-Induced Liver Injury Protein and microRNA Biomarkers Using Dynamic Chemical Labelling on a Luminex MAGPIX System. Analytica 2021, 2, 130–139. [Google Scholar] [CrossRef]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Detassis, S.; Grasso, M.; Tabraue-Chávez, M.; Marín-Romero, A.; López-Longarela, B.; Ilyine, H.; Ress, C.; Ceriani, S.; Erspan, M.; Maglione, A.; et al. New Platform for the Direct Profiling of microRNAs in Biofluids. Anal. Chem. 2019, 91, 5874–5880. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Gonzalez, A.; Robles-Remacho, A.; Marin-Romero, A.; Detassis, S.; Lopez-Longarela, B.; Lopez-Delgado, F.J.; de Miguel-Perez, D.; Guardia-Monteagudo, J.J.; Fara, M.A.; Tabraue-Chavez, M.; et al. PCR-free and chemistry-based technology for miR-21 rapid detection directly from tumour cells. Talanta 2019, 200, 51–56. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, L.; Ding, R.; Deng, X.; Chen, Z. Role of miRNA-122 in cancer (Review). Int. J. Oncol. 2024, 65, 83. [Google Scholar] [CrossRef]
- He, D.; Miao, H.; Xu, Y.; Xiong, L.; Wang, Y.; Xiang, H.; Zhang, H.; Zhang, Z. MiR-371-5p facilitates pancreatic cancer cell proliferation and decreases patient survival. PLoS ONE 2014, 9, e112930. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Fu, J.; Li, N.; Shen, L. Clinical utility of microRNA-451 as diagnostic biomarker for human cancers. Biosci. Rep. 2019, 39, BSR20180653. [Google Scholar] [CrossRef]
- Lu, Y.; Lu, J.; Zhao, J.; Cusido, J.; Raymo, F.M.; Yuan, J.; Yang, S.; Leif, R.C.; Huo, Y.; Piper, J.A.; et al. On-the-fly decoding luminescence lifetimes in the microsecond region for lanthanide-encoded suspension arrays. Nat. Commun. 2014, 5, 3741. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Zhou, T.; Gu, W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell 2021, 12, 911–946. [Google Scholar] [CrossRef]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer: Long non-coding RNAs and cancer. Noncoding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef] [PubMed]



| Technology | Type of Detection | Target miRNA | Limit of Detection (LoD) | Type of Clinical Sample | Types of Cancer | Multiplexing Capability | POCT Suitability | Reference |
|---|---|---|---|---|---|---|---|---|
| Solid-state nanoplasmonic sensor | UV-Vis | miR-10b-5p miR-let7a-5p | 637.7 aM | Plasma | Pancreatic ductal adenocarcinoma | Yes (can be converted to multiplexed by adding receptors) | Partial (not fully POCT-ready) | [70] |
| Electro-optical nanopore sensing | Fluorescence | miR-141-3p miR-375-3p | 5–8 fM | Serum | Prostate cancer | Yes (up to ~10 targets with potential for more) | Yes | [71] |
| Singlet oxygen-based photoelectrochemical | Photoelectrochemical | miR-145-5p miR-141-3p | 3.5–8.3 pM | Plasma | Prostate cancer | Partial (multiplexing possible with multi-array platforms) | Partial | [72] |
| Tandem bead-based hybridization assay | Fluorescence | miR-128-2-3p | 2.2 pM | Plasma | Colorectal cancer | No (single target focused) | Partial | [73] |
| Dynamic chemical labeling (DCL) | Fluorescence | miR-21-5p miR-122-5p miR-371a-3p miR-451a-5p | pM range | Serum | Various cancers | Yes | Yes | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soleimanpour, B.; Diaz Mochon, J.J.; Pernagallo, S. Amplification-Free Testing of microRNA Biomarkers in Cancer. Cancers 2025, 17, 2715. https://doi.org/10.3390/cancers17162715
Soleimanpour B, Diaz Mochon JJ, Pernagallo S. Amplification-Free Testing of microRNA Biomarkers in Cancer. Cancers. 2025; 17(16):2715. https://doi.org/10.3390/cancers17162715
Chicago/Turabian StyleSoleimanpour, Bahareh, Juan Jose Diaz Mochon, and Salvatore Pernagallo. 2025. "Amplification-Free Testing of microRNA Biomarkers in Cancer" Cancers 17, no. 16: 2715. https://doi.org/10.3390/cancers17162715
APA StyleSoleimanpour, B., Diaz Mochon, J. J., & Pernagallo, S. (2025). Amplification-Free Testing of microRNA Biomarkers in Cancer. Cancers, 17(16), 2715. https://doi.org/10.3390/cancers17162715






