Targeting NRF2 and FSP1 to Overcome Ferroptosis Resistance in TSC2-Deficient and Cancer Cells
Simple Summary
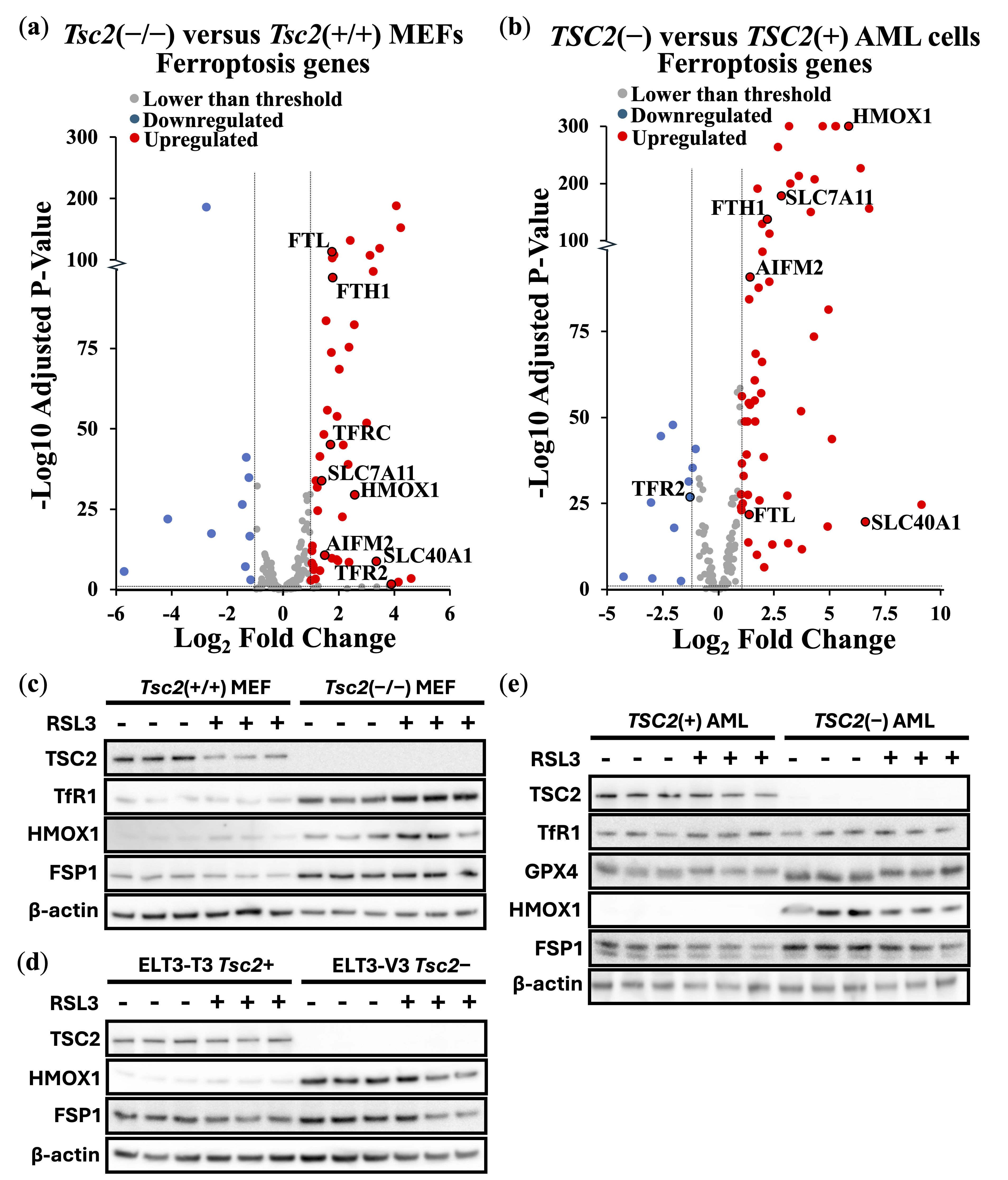
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Drugs and Reagents
2.2. Flow Cytometry
2.3. siRNA Transfections
2.4. Cell Viability Assays Using Crystal Violet
2.5. RNA Sequencing and Analysis
2.6. Western Blotting
2.7. Immunofluorescence
2.8. Statistical Analysis
3. Results
3.1. TSC2-Deficiency Protects Cells from Ferroptosis Inducers
3.2. NRF2 Protects TSC2-Deficient Cells from Ferroptosis
3.3. FSP1 and NRF2 Mediate Ferroptosis Resistance in Cancer Through Independent Mechanisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIFM2 | AIF family member 2 |
| AML | Angiomyolipoma |
| BCUHB | Betsi Cadwaladr University Health Board |
| CoQ10 | Ubiquinone |
| CoQ10H2 | Ubiquinol |
| DEG | Differential expressed gene |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| FSP1 | Ferroptosis suppressor protein 1 |
| GO | Gene ontology |
| GPX4 | Glutathione peroxidase 4 |
| GSH | Glutathione |
| HMOX1 | Heme oxygenase 1 |
| HIF-1α | Hypoxia-inducible factor 1α |
| KEAP1 | Kelch-like ECH-associated protein 1 |
| LIP | Labile iron pool |
| LPO | Lipid peroxidation |
| mTOR | Mechanistic target of rapamycin |
| MEF | Mouse embryonic fibroblasts |
| NFE2L2 | Nuclear factor E2-related factor 2 |
| PANX2 | Pannexin 2 |
| PBS | Phosphate-buffered saline |
| PUFAs | Polyunsaturated fatty acids |
| REF-1 | Redox factor-1 |
| ROS | Reactive oxygen species |
| RSL3 | RAS-selective lethal |
| RT-PCR | Real-time PCR |
| SLC7A11 | SLC7A11 Solute carrier family 7 member 11 |
| STAT3 | Signal transducer and activator of transcription 3 |
| TfR1 | Transferrin receptor 1 |
| TSC2 | Tuberous Sclerosis Complex |
References
- Lee, E.; Song, C.H.; Bae, S.J.; Ha, K.T.; Karki, R. Regulated cell death pathways and their roles in homeostasis, infection, inflammation, and tumorigenesis. Exp. Mol. Med. 2023, 55, 1632–1643. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1672. [Google Scholar] [CrossRef] [PubMed]
- Averill-Bates, D.A. The antioxidant glutathione. Vitam. Horm. 2023, 121, 109–141. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Kuang, F.; Liu, J.; Tang, D.; Kang, R. Oxidative Damage and Antioxidant Defense in Ferroptosis. Front. Cell Dev. Biol. 2020, 8, 586578. [Google Scholar] [CrossRef]
- Shakya, A.; McKee, N.W.; Dodson, M.; Chapman, E.; Zhang, D.D. Anti-Ferroptotic Effects of Nrf2: Beyond the Antioxidant Response. Mol. Cells 2023, 46, 165–175. [Google Scholar] [CrossRef]
- Chiang, S.K.; Chen, S.E.; Chang, L.C. A Dual Role of Heme Oxygenase-1 in Cancer Cells. Int. J. Mol. Sci. 2018, 20, 39. [Google Scholar] [CrossRef]
- Torti, S.V.; Torti, F.M. Iron and Cancer: 2020 Vision. Cancer Res. 2020, 80, 5435–5448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.; Xia, R.; Zhang, H.S. Ferroptosis resistance in cancer: Recent advances and future perspectives. Biochem. Pharmacol. 2024, 219, 115933. [Google Scholar] [CrossRef]
- Henske, E.P.; Jóźwiak, S.; Kingswood, J.C.; Sampson, J.R.; Thiele, E.A. Tuberous sclerosis complex. Nat. Rev. Dis. Prim. 2016, 2, 16035. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.R.; Fingar, D.C.; Manning, B.D.; Kwiatkowski, D.J.; Cantley, L.C.; Blenis, J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 13571–13576. [Google Scholar] [CrossRef]
- Tee, A.R.; Manning, B.D.; Roux, P.P.; Cantley, L.C.; Blenis, J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003, 13, 1259–1268. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Liu, J.; Kang, R.; Tang, D. Interplay between MTOR and GPX4 signaling modulates autophagy-dependent ferroptotic cancer cell death. Cancer Gene Ther. 2021, 28, 55–63. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 2009, 37, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Swanda, R.V.; Nie, L.; Liu, X.; Wang, C.; Lee, H.; Lei, G.; Mao, C.; Koppula, P.; Cheng, W.; et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat. Commun. 2021, 12, 1589. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Zhu, J.; Wu, J.; Thompson, C.B.; Jiang, X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 31189–31197. [Google Scholar] [CrossRef]
- Armenta, D.A.; Laqtom, N.N.; Alchemy, G.; Dong, W.; Morrow, D.; Poltorack, C.D.; Nathanson, D.A.; Abu-Remalieh, M.; Dixon, S.J. Ferroptosis inhibition by lysosome-dependent catabolism of extracellular protein. Cell Chem. Biol. 2022, 29, 1588–1600.e7. [Google Scholar] [CrossRef]
- Zhang, H.; Cicchetti, G.; Onda, H.; Koon, H.B.; Asrican, K.; Bajraszewski, N.; Vazquez, F.; Carpenter, C.L.; Kwiatkowski, D.J. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 2003, 112, 1223–1233. [Google Scholar] [CrossRef]
- Astrinidis, A.; Cash, T.P.; Hunter, D.S.; Walker, C.L.; Chernoff, J.; Henske, E.P. Tuberin, the tuberous sclerosis complex 2 tumor suppressor gene product, regulates Rho activation, cell adhesion and migration. Oncogene 2002, 21, 8470–8476. [Google Scholar] [CrossRef]
- Yu, J.; Astrinidis, A.; Howard, S.; Henske, E.P. Estradiol and tamoxifen stimulate LAM-associated angiomyolipoma cell growth and activate both genomic and nongenomic signaling pathways. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 286, L694–L700. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.R.; Zhou, W.; Bowman, M.J.; Shih, J.; Au, K.S.; Dittenhafer-Reed, K.E.; Sisson, K.A.; Koeman, J.; Weisenberger, D.J.; Cottingham, S.L.; et al. The genomic landscape of tuberous sclerosis complex. Nat. Commun. 2017, 8, 15816. [Google Scholar] [CrossRef]
- Xiang, T.; Jia, Y.; Sherris, D.; Li, S.; Wang, H.; Lu, D.; Yang, Q. Targeting the Akt/mTOR pathway in Brca1-deficient cancers. Oncogene 2011, 30, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, E.J.; Jin, C.; Kim, D.H. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol. Oncol. 2005, 97, 26–34. [Google Scholar] [CrossRef]
- Ricoult, S.J.; Yecies, J.L.; Ben-Sahra, I.; Manning, B.D. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 2016, 35, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinform. 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinform. 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Johnson, C.E.; Dunlop, E.A.; Seifan, S.; McCann, H.D.; Hay, T.; Parfitt, G.J.; Jones, A.T.; Giles, P.J.; Shen, M.H.; Sampson, J.R.; et al. Loss of tuberous sclerosis complex 2 sensitizes tumors to nelfinavir-bortezomib therapy to intensify endoplasmic reticulum stress-induced cell death. Oncogene 2018, 37, 5913–5925. [Google Scholar] [CrossRef]
- Dunlop, E.A.; Hunt, D.K.; Acosta-Jaquez, H.A.; Fingar, D.C.; Tee, A.R. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 2011, 7, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.J.; Huang, X.; Zhu, B.T. Mechanism of RSL3-induced ferroptotic cell death in HT22 cells: Crucial role of protein disulfide isomerase. Acta Biochim. Biophys. Sin. 2024, 57, 616–632. [Google Scholar] [CrossRef]
- Sun, S.; Shen, J.; Jiang, J.; Wang, F.; Min, J. Targeting ferroptosis opens new avenues for the development of novel therapeutics. Signal Transduct. Target Ther. 2023, 8, 372. [Google Scholar] [CrossRef]
- Yuk, H.; Abdullah, M.; Kim, D.H.; Lee, H.; Lee, S.J. Necrostatin-1 Prevents Ferroptosis in a RIPK1- and IDO-Independent Manner in Hepatocellular Carcinoma. Antioxidants 2021, 10, 1347. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Fakhari, D.; Saffari, A.; Wahlster, L.; Di Nardo, A.; Turner, D.; Lewis, T.L., Jr.; Conrad, C.; Rothberg, J.M.; Lipton, J.O.; Kölker, S.; et al. Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell Rep. 2016, 17, 1053–1070. [Google Scholar] [CrossRef]
- Smiley, S.T.; Reers, M.; Mottola-Hartshorn, C.; Lin, M.; Chen, A.; Smith, T.W.; Steele, G.D., Jr.; Chen, L.B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. USA 1991, 88, 3671–3675. [Google Scholar] [CrossRef]
- Endale, H.T.; Tesfaye, W.; Mengstie, T.A. ROS induced lipid peroxidation and their role in ferroptosis. Front. Cell Dev. Biol. 2023, 11, 1226044. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, Y.; Liu, R.; Ikenoue, T.; Guan, K.L.; Liu, Y.; Zheng, P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 2008, 205, 2397–2408. [Google Scholar] [CrossRef]
- Di Nardo, A.; Kramvis, I.; Cho, N.; Sadowski, A.; Meikle, L.; Kwiatkowski, D.J.; Sahin, M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 2009, 29, 5926–5937. [Google Scholar] [CrossRef]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef]
- Emmanuel, N.; Li, H.; Chen, J.; Zhang, Y. FSP1, a novel KEAP1/NRF2 target gene regulating ferroptosis and radioresistance in lung cancers. Oncotarget 2022, 13, 1136–1139. [Google Scholar] [CrossRef]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef]
- Hayes, J.D.; McMahon, M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009, 34, 176–188. [Google Scholar] [CrossRef]
- Li, W.; Liang, L.; Liu, S.; Yi, H.; Zhou, Y. FSP1: A key regulator of ferroptosis. Trends Mol. Med. 2023, 29, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qiu, C.; Hou, M.; Wang, X.; Huang, C.; Zou, J.; Liu, T.; Qu, J. Ferroptosis in Ovarian Cancer: A Novel Therapeutic Strategy. Front. Oncol. 2021, 11, 665945. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, Y.; Jiang, Y.; Bu, J.; Gu, X. Targeting ferroptosis, the achilles’ heel of breast cancer: A review. Front. Pharmacol. 2022, 13, 1036140. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, W.K.; Bae, K.H.; Lee, S.C.; Lee, E.W. Lipid Metabolism and Ferroptosis. Biology 2021, 10, 184. [Google Scholar] [CrossRef]
- Liao, D.; Yang, G.; Yang, Y.; Tang, X.; Huang, H.; Shao, J.; Pan, Q. Identification of Pannexin 2 as a Novel Marker Correlating with Ferroptosis and Malignant Phenotypes of Prostate Cancer Cells. OncoTargets Ther. 2020, 13, 4411–4421. [Google Scholar] [CrossRef]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef]
- Anandhan, A.; Dodson, M.; Shakya, A.; Chen, J.; Liu, P.; Wei, Y.; Tan, H.; Wang, Q.; Jiang, Z.; Yang, K.; et al. NRF2 controls iron homeostasis and ferroptosis through HERC2 and VAMP8. Sci. Adv. 2023, 9, eade9585. [Google Scholar] [CrossRef]
- Malik, A.R.; Liszewska, E.; Skalecka, A.; Urbanska, M.; Iyer, A.M.; Swiech, L.J.; Perycz, M.; Parobczak, K.; Pietruszka, P.; Zarebska, M.M.; et al. Tuberous sclerosis complex neuropathology requires glutamate-cysteine ligase. Acta Neuropathol. Commun. 2015, 3, 48. [Google Scholar] [CrossRef] [PubMed]
- Bayeva, M.; Khechaduri, A.; Puig, S.; Chang, H.C.; Patial, S.; Blackshear, P.J.; Ardehali, H. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab. 2012, 16, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Champion, J.D.; Dodd, K.M.; Lam, H.C.; Alzahrani, M.A.M.; Seifan, S.; Rad, E.; Scourfield, D.O.; Fishel, M.L.; Calver, B.L.; Ager, A.; et al. Drug Inhibition of Redox Factor-1 Restores Hypoxia-Driven Changes in Tuberous Sclerosis Complex 2 Deficient Cells. Cancers 2022, 14, 6195. [Google Scholar] [CrossRef]
- Kelley, M.R.; Georgiadis, M.M.; Fishel, M.L. APE1/Ref-1 role in redox signaling: Translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr. Mol. Pharmacol. 2012, 5, 36–53. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Ramakrishnan, S.K.; Weisz, K.; Triner, D.; Xie, L.; Attili, D.; Pant, A.; Győrffy, B.; Zhan, M.; Carter-Su, C.; et al. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Guo, W.; Ling, J.; Xu, D.; Liao, Y.; Zhao, H.; Du, X.; Wang, H.; Xu, M.; Song, H.; et al. Iron-dependent CDK1 activity promotes lung carcinogenesis via activation of the GP130/STAT3 signaling pathway. Cell Death Dis. 2019, 10, 297. [Google Scholar] [CrossRef]
- Zhang, W.; Gong, M.; Zhang, W.; Mo, J.; Zhang, S.; Zhu, Z.; Wang, X.; Zhang, B.; Qian, W.; Wu, Z.; et al. Thiostrepton induces ferroptosis in pancreatic cancer cells through STAT3/GPX4 signalling. Cell Death Dis. 2022, 13, 630. [Google Scholar] [CrossRef]
- Peyssonnaux, C.; Nizet, V.; Johnson, R.S. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle 2008, 7, 28–32. [Google Scholar] [CrossRef]
- Xue, D.; Zhou, X.; Qiu, J. Emerging role of NRF2 in ROS-mediated tumor chemoresistance. Biomed. Pharmacother. 2020, 131, 110676. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, S.; Abe, Y.; Nishi, M.; Hamamoto, A.; Inoue, Y.; Ohnishi, Y.; Nishime, C.; Matsumoto, H.; Yamazaki, H.; Kijima, H.; et al. The subunits of glutamate cysteine ligase enhance cisplatin resistance in human non-small cell lung cancer xenografts in vivo. Int. J. Oncol. 2004, 25, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kim, M.J.; Han, T.H.; Lee, J.Y.; Kim, S.; Kim, H.; Oh, K.J.; Kim, W.K.; Han, B.S.; Bae, K.H.; et al. FSP1 confers ferroptosis resistance in KEAP1 mutant non-small cell lung carcinoma in NRF2-dependent and -independent manner. Cell Death Dis. 2023, 14, 567. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahsin, T.; McPhail, D.K.; Champion, J.D.; Alzahrani, M.A.M.; Hilditch, M.L.; Faris-Orr, A.; Calver, B.L.; Cronin, J.G.; Mareque-Rivas, J.C.; Sexton, D.W.; et al. Targeting NRF2 and FSP1 to Overcome Ferroptosis Resistance in TSC2-Deficient and Cancer Cells. Cancers 2025, 17, 2714. https://doi.org/10.3390/cancers17162714
Tahsin T, McPhail DK, Champion JD, Alzahrani MAM, Hilditch ML, Faris-Orr A, Calver BL, Cronin JG, Mareque-Rivas JC, Sexton DW, et al. Targeting NRF2 and FSP1 to Overcome Ferroptosis Resistance in TSC2-Deficient and Cancer Cells. Cancers. 2025; 17(16):2714. https://doi.org/10.3390/cancers17162714
Chicago/Turabian StyleTahsin, Tasmia, Darius K. McPhail, Jesse D. Champion, Mohammad A. M. Alzahrani, Madeleine L. Hilditch, Alexandre Faris-Orr, Brian L. Calver, James G. Cronin, Juan C. Mareque-Rivas, Darren W. Sexton, and et al. 2025. "Targeting NRF2 and FSP1 to Overcome Ferroptosis Resistance in TSC2-Deficient and Cancer Cells" Cancers 17, no. 16: 2714. https://doi.org/10.3390/cancers17162714
APA StyleTahsin, T., McPhail, D. K., Champion, J. D., Alzahrani, M. A. M., Hilditch, M. L., Faris-Orr, A., Calver, B. L., Cronin, J. G., Mareque-Rivas, J. C., Sexton, D. W., Hughes, S. F., Conlan, R. S., Davies, D. M., & Tee, A. R. (2025). Targeting NRF2 and FSP1 to Overcome Ferroptosis Resistance in TSC2-Deficient and Cancer Cells. Cancers, 17(16), 2714. https://doi.org/10.3390/cancers17162714







