Simple Summary
Bladder cancer can largely be divided into two types—non-muscle-invasive (NMIBC) and the more lethal form, muscle-invasive bladder cancer (MIBC). About 30% of NMIBC cases convert to MIBC. Predicting which patients with NMIBC are at risk of developing MIBC is crucial for determining the treatment course. Understanding the genetics of each of these cancer types enables the development of markers that can be used to predict conversion. In this study, we have measured genes that are upregulated by exposure to low oxygen levels (hypoxia)—a common characteristic of bladder cancers—to identify genes that may predict whether a bladder cancer is hypoxic and to identify potential markers of MIBC. We have found that different genes are increased in expression in NMIBC and MIBC, which may help to identify at-risk patients.
Abstract
Introduction: Hypoxic cancers are radioresistant, but biomarkers based on expression of multiple genes can identify patients who will benefit from hypoxia modification. Most studies identifying relevant genes exposed cells in culture to 1% oxygen, which activates hypoxia-inducible factor (HIF). However, oxygen concentrations in hypoxic tumours are heterogeneous ranging from <0.1%. As lower oxygen levels would likely affect transcriptional responses, we aimed to investigate how gene selection at different oxygen levels affects the genes identified and their prognostic capability. Methods: Four MIBC (J82, T24, UMUC3, HT1376) and two non-MIBC (RT4, RT112) bladder cancer cell lines were exposed to varying oxygen levels (20%, 1%, 0.2% and 0.1% O2) for 24 h and were then harvested and frozen. RNA was extracted and transcriptomes analysed using Clariom S microarrays. Differences in gene expression were investigated. Prognostic and predictive significance of a published 24-gene signature was compared with one generated from genes identified at lower oxygen levels. Results: The number of upregulated genes increased with decreasing O2 level. The number of biological pathways involved also increased. Differences between cell lines dominated those due to hypoxia. Some genes were commonly upregulated in MIBC and NMIBC cells and others increased exclusively in either MIBC or NMIBC cells. The median expression of a published 24-gene bladder cancer hypoxia-associated signature increased with decreasing oxygen levels. Seventy-seven genes were upregulated in at least three cell lines by exposure to 0.1% O2. The median expression of the 77 genes was of borderline prognostic significance in the bladder cancer cohort in the TCGA (The Cancer Genome Atlas). Five of the seventy-seven genes upregulated by hypoxia were present in the twenty-four-gene bladder hypoxia signature. The median expression of the 5 genes demonstrated identical prognostication to the 24-gene signature but failed to predict benefit from hypoxia modification. Conclusions: The number of genes upregulated by exposure of bladder cancer cells to hypoxia increases as O2 level is decreased from 1% to 0.2% to 0.1%. Differential upregulation of gene expression by MIBC and NMIBC cells and the associated biological pathways may be useful in understanding the genetics of bladder cancer invasiveness. Based on a search of the literature, this is the first study that assessed the expression of genes in bladder cancer using three hypoxic concentration levels to identify biomarkers for disease progression and prognosis among differentially expressed bladder cancer genes.
1. Introduction
Hypoxia is a characteristic of most solid cancers and confers an aggressive phenotype and treatment resistance, especially to radiotherapy [1]. Although radical cystectomy is considered the gold standard for treating patients with MIBC, some are treated with bladder-preserving radiotherapy in combination with a radiosensitiser [2,3]. Patients with hypoxic MIBC benefit from hypoxia modification [4]. Consequently, there is an interest in understanding the transcriptomic changes associated with hypoxia and developing biomarkers to stratify patients for treatment modification.
A bladder cancer-specific hypoxia-associated gene signature was derived by curating known hypoxia-sensitive genes, enrichment analysis in a bladder cancer cohort and final selection based on prognostication in a further clinical cohort [5]. The hypoxia score (HS) generated using the signature was validated as a prognostic marker in six independent bladder cancer cohorts [5] and was predictive of benefit from hypoxia modification in the BCON patient cohort. More recently, an in vitro study demonstrated the HS to be sensitive to hypoxia [6]. However, this study was only performed in two MIBC cell lines, and sensitivity to oxygen level was not determined.
Seed genes for hypoxia signatures can also be selected by identifying hypoxia-sensitive genes in cancer cell lines grown in vitro [7]. Previous work derived candidate gene panels by identifying genes upregulated by cells exposed to a single O2 level (1% O2) [8,9,10,11]. Tumours have a range of O2 levels from sub 1% O2. No one has investigated how the use of different oxygen levels might affect candidate gene panels and the ability to prognosticate. We hypothesised that identifying genes upregulated by exposing cells to a range of O2 levels would better represent the heterogeneity of oxygen levels in a tumour and provide a more representative gene panel for developing hypoxia-associated gene expression signatures. Therefore, we aimed to investigate how different oxygen levels affect transcriptional responses, derivation of gene signatures and signature prognostication.
In view of the propensity for NMIBC to progress to MIBC, gene candidate panels were derived based on cell lines derived from both NMIBC and MIBC [12]. As bladder cancer is a heterogeneous disease, reflected in its different molecular subtypes, multiple cell lines were used. Therefore, we generated gene expression data from four MIBC and two NMIBC cell lines exposed to different oxygen levels (20%, 1%, 0.2% and 0.1% O2) for 24 h.
2. Materials and Methods
2.1. Literature Review
A comprehensive systematic search for relevant studies and articles was conducted using keywords ((NMIBC OR MIBC OR muscle-invasive OR non-muscle-invasive) AND (bladder cancer)) AND (hypoxia AND gene expression) on PubMed. The search criteria, shown in the Supplementary Data, demonstrated that a comprehensive multiple bladder cancer cell line study of gene expression induced by cell exposure to 1%, 0.2% and 0.1% O2 levels had not previously been performed.
2.2. Cells
High-grade MIBC (J82, T24, UMUC3, HT1376) and low-grade NMIBC (RT4, RT112) bladder cancer cell lines obtained from the American Type Culture Collection were routinely screened for mycoplasma and authenticated. Characteristics of the cell lines are shown in Table 1. Cells were maintained in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% foetal bovine serum (FBS) (Gibco) and 2 mM L-glutamine in a humidified atmosphere of 95% air and 5% CO2 at 37 °C. EMEM was used for all cell lines because (1) it has a glucose content of 4.5mM, which is similar to the concentration found in human plasma, and (2) excess glucose affects the expression of glycolytic enzymes and other hypoxia-response genes [13].

Table 1.
Characteristics of the NMIBC and MIBC cells used in this study.
2.3. Hypoxia Treatment
Cells (0.5 × 106 cell/mL) were seeded with 10 mL of medium per Petri dish (area 56.7 cm2; Corning, Life Sciences, UK). Four Petri dishes per cell line and four oxygen conditions were used, except for the slow-growing RT112 cell line, where eight dishes per condition were used. Dishes were placed in an incubator at 95% air/5% CO2 at 37 °C for 24 h to allow for recovery from trypsin treatment. After 24 h, the medium was changed in the normoxic control dishes and returned to the incubator for 24 h. The remaining dishes were placed in the hypoxia cabinets at 1%, 0.2% and 0.1% O2 levels. Under hypoxic condition, the media in the dishes were washed twice with 5 mL of hypoxic medium (pre-incubated in the hypoxia cabinets [1%, 0.2% and 0.1% O2]). After adding 10 mL of the hypoxic medium to each dish, the dishes were incubated in hypoxia for 24 h. Each cell line/treatment experiment was caried out three times using different passages of cells (biological replicates).
2.4. Cell Harvesting
After 24 h of incubation, the cells were rinsed twice with PBS and harvested using a cell scraper (Corning®, Life Sciences, Loughborough, UK) by rotating and scraping in PBS. Ice-cold PBS hypoxia-equilibrated PBS was used. The harvested cells were transferred into 1 mL RNAse-free microfuge tubes (Eppendorf, Hamburg, Germany) and centrifuged at 4 °C for 10 min at ≥8000× g. Supernatants were removed and the cell pellets stored at −80 °C.
2.5. RNA Extraction
RNA was extracted using Qiagen QIA shredder Kit (Qiagen, Hilden, Germany) to break up the cells and the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) for RNA purification following the manufacturer’s protocol. RNA yield was determined using Nanodrop (Thermo-Scientific, Loughborough, UK) and Qubit following the manufacturers’ instructions. Mean and standard deviation of absorbance ratios 260/280 and 230/260, measures of RNA quality, for all RNA extracts were 2.07 ± 0.06 and 1.97 ± 0.12, respectively.
ClariomTM S microarray gene expression arrays were prepared using 72 ng of RNA (8 ng/μL) with the Clariom S pico HT human assay. The samples were added to Kingfisher Shallow 96-well plates (Thermo Fisher Scientific, Loughborough, UK) along with brain and human prostate reference RNA (Thermo Fisher Scientific, Loughborough, UK), and RNA extracted from a clinical bladder cancer sample (plate control). YourGene Health (Manchester, UK) generated RNA data using the Clariom S pico HT human assay (Thermo Fisher Scientific, Loughborough, UK).
2.6. Data Normalisation and Batch Correction
The data generated from Clariom S underwent quality assessment (QA) and quality control (QC). The Affymetrix CEL files were normalised by single-space transformation with probe guidance cytosine count correction using Affymetrix Array Power Tools as recommended by the manufacturer (Thermo Fisher Sciences, Waltham, MA, USA) (https://assets.thermofisher.com/TFS-Assets/LSG/brochures/sst_gccn_whitepaper.pdf, accessed on 28 July 2025). Batch correction was performed by ComBat function on the R package ‘sva’. The full expression matrix was generated and log2-transformed.
2.7. Differentially Expressed Gene (DEG) Analysis
Microarray analysis was performed in R version 4.1.1 (10 August 2021). Gene expression values and normalisation were calculated using the RMA method [14]. Gene annotation was with affycoretools_1.64.0. and clariomshumantranscriptcluster. clariomshumantranscriptcluster.db: Affymetrix clariomshuman annotation data (chip clariomshumantranscriptcluster). R package (version 8.8.0.). Differential expression was calculated with limma_3.48.3 [15]. Adjusted p-values were corrected for multiple testing (Benjamini and Hochberg method).
2.8. Gene Expression Analyses
Two significance levels were used to select upregulated genes depending on the application. Biomarker development requires stringent inclusion criteria to reduce misdiagnosis. To derive biomarker candidate gene panels, we used padj < 0.001 to select genes up- or downregulated by hypoxia to reduce the false positive rate (FDR). However, using tight criteria for identifying differentially upregulated genes between two disease subtypes may miss important genes, so a less stringent significance level (FDR), padj < 0.05, was used to identify genes differentially regulated by hypoxia in MIBC and NMIBC cell lines.
In terms of the number of cells expressing each gene, to identify potential hypoxia biomarker candidates, genes upregulated in at least 3 cell lines were determined. To identify uniquely and commonly upregulated genes between MIBC and NMIBC cells, genes upregulated in both NMIBC cell lines were compared with genes upregulated in at least 2 MIBC cell lines.
The mean for each replicate was calculated and differences between cell lines investigated using unpaired t tests and GraphPad Prism 9 software Version 9.1.2. Biological processes were investigated using the PANTHER enrichment website. Heatmaps were generated with complexHeatmap v2.12.1 (PMID: 27207943). Gene ontology enrichment was studied using clusterProfiler (PMID: 34557778) and Enrichr v3.1 (PMID: 27141961). Gene Set Enrichment Analyses (GSEA) were performed using R version 4.1.1 (10 August 2021) ‘Limma package’.
2.9. Hypoxia Sensitivity in a Published Bladder Cancer Xenograft
The sensitivity of candidate hypoxia signature panels to hypoxia in an in vivo bladder cancer model was established using published gene expression data from a bladder cancer cell line-derived xenograft [16]. The data in this model was generated by sampling tissue from regions stained (hypoxic) and unstained (non-hypoxic) with the hypoxia probe pimonidazole injected into the host prior to tumour harvesting.
2.10. Hypoxia Scores
Expression and HSs were generated for each cell line using the 24-gene bladder hypoxia signature described above to verify hypoxia sensitivity. A candidate gene panel was derived by selecting genes upregulated by hypoxia in at least three cell lines. Hypoxia scores were generated using the median expression of the 24 genes comprising the West bladder cancer hypoxia signature [5]. To derive a cut-off value for patient stratification, the median value of hypoxia scores for the analysed patient cohort was determined, and tumours with higher or lower values were classed as hypoxic and normoxic, respectively.
2.11. Analysis of the Bladder Cancer TCGA Data Base and BCON
The complete dataset of the bladder TCGA cohort (n = 406) and the accompanying clinical data were downloaded from firebrowse.org. BCON data was previously generated [5]. Kaplan–Meier (KM) plots were used to analyse overall survival (OS) from the date of surgery to the date of death.
3. Results
Figure 1 shows a principal component analysis (PCA) of the cell line data (6 cell lines, 4 oxygen levels, 3 replicates per condition). This is based on differential gene expression at 55.7% (PC1) on x-axis separation of samples into normoxic on the right, with samples exposed to 1%, 0.2% and 0.1% O2 on the left broadly clustering with decreasing O2 concentration (although with some overlap). Cell line differences are apparent with 11.3% (PC1) with the NMIBC cell line RT4 at the top; next is RT112, with some co-clustering with HT1376 and J82, and then with UMUC3 and T24 clustering at the bottom of the y-axis. There was a separation between the NMIBC (RT4, RT112) and MIBC (HT1376, J82, T24, UMUC3) cell lines.
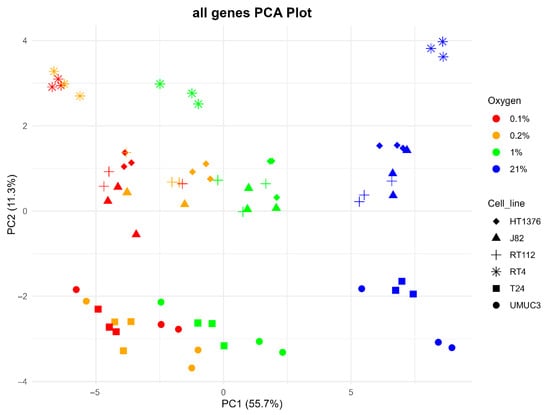
Figure 1.
Principal component analysis of all genes expressed in each cell line/condition.
Figure 2 shows the expected trend for the upregulation of widely studied hypoxia-responsive genes (CA9, SLC2A1 and VEGF) in the cell lines.
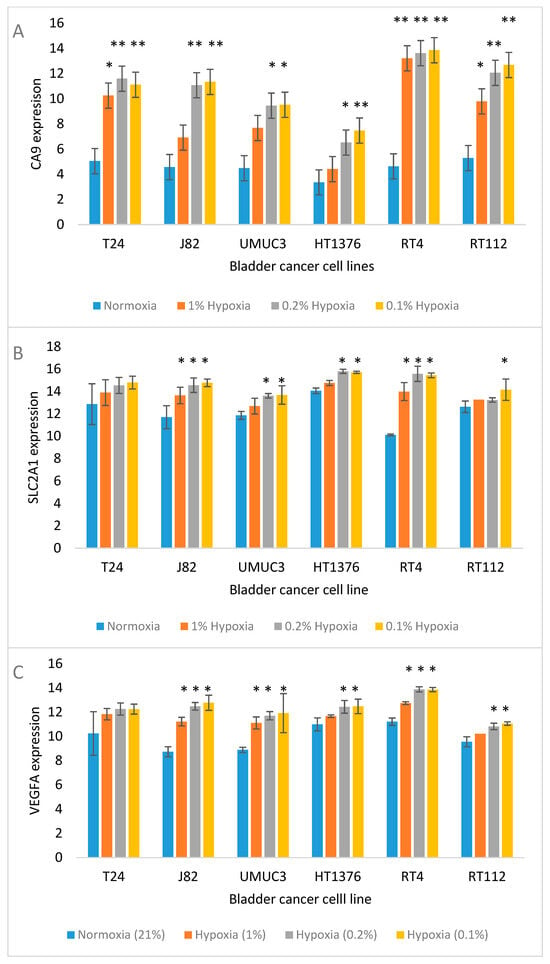
Figure 2.
Expression of known hypoxia-associated genes, CA9 (A), SLC2A1 (B) and VEGFA (C) in bladder cancer cell lines incubated for 24 h in 21% (control), 1%, 0.2% and 0.1% O2. Significant differences from control * p < 0.05, ** p < 0.01.
Table 2 summarises the number of genes affected by changes in oxygen levels, which increased with decreasing O2 level. More genes were upregulated than downregulated at all oxygen levels. Table 3 lists the genes up- or downregulated in at least three cell lines after exposure to 0.1% O2. (Supplementary Tables S1 and S2 show genes upregulated in two MIBC but no NMIBC cell lines and in both NMIBC but <2 MIBC cell lines by exposure to hypoxia, respectively). The number of processes affected by hypoxia increased with decreasing oxygen levels (Table 4). Cellular response to hypoxia was the only biological process affected by exposure to 1% O2. Additional processes affected by exposure to 0.2% O2 were carbohydrate transport and metabolism, including glycolysis, steroid hormone response and Peptidyl-proline hydroxylation to 4-hydroxy-L-proline, which is involved in collagen metabolism. More genes associated with glucose metabolism and with pregnancy were upregulated in cells exposed to 0.1% O2.

Table 2.
Number of genes affected by changes in oxygen levels in 1–6 of the cell lines: RT4, RT112, HT1367, T24, J82 and UMUC3.

Table 3.
Genes up- or downregulated in at least three cell lines exposed to 0.1% O2. Bold upregulated by 0.2% O2; bold and * upregulated by 1% O2.

Table 4.
Biological processes associated with genes upregulated by at least three bladder cancer cell lines exposed to 0.2% and 0.1% hypoxia for 24 h.
Genes also upregulated in 0.2% O2 are shown in bold and in 1% O2 in bold *.
Hypoxia scores were calculated using a 24-gene signature (HS24). HS24 tended to increase with decreasing oxygen levels (Figure 3). Figure 3B shows the median expression of the 77 genes (ME77) found to be upregulated in at least three bladder cancer cell lines after exposure to 0.1% O2 for 24 h. The median value of the 77 genes increased with decreasing oxygen levels in all cell lines. There were five genes in both signatures (SYDE1, SLC2A3, DPYSL2, FUT11, P4HA2). The median value of the five genes (ME5) also increased with decreasing oxygen level. As a further check of the hypoxia relevance of the generated signatures, we compared their expression in samples taken from an experimental tumour where hypoxia was identified using pimonidazole staining [15]. We showed previously that HS24 was higher in pimonidazole-positive areas. Supplementary Figure S1 shows ME77 and ME5 values were also higher. Supplementary Table S1 shows which of the 24 genes were increased in expression by individual cell lines and O2 levels. Two genes, CYP1B1 and GLG1, were not upregulated by hypoxia in any of the cell lines. Some cell lines differed only by one or two genes, but a core group of genes were upregulated by all cell lines.
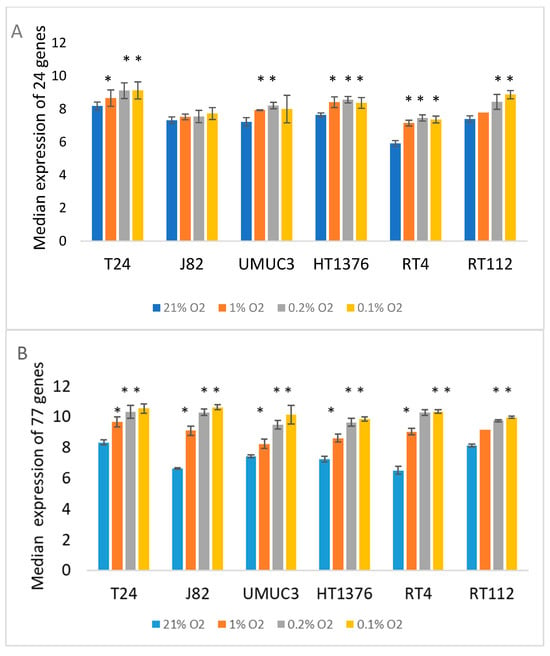
Figure 3.
Sensitivity of gene expression-based hypoxia scores HS24 (A) and the median expression of 77 genes upregulated in at least 3 cell lines (HS77) (B) in bladder cancer cell lines incubated for 24 h in 21% (control), 1%, 0.2% and 0.1% O2. Significant differences from control * p < 0.05.
Figure 4 shows a heatmap of the expression of the 78 (77 upregulated and 1 downregulated) genes found to be differentially expressed in at least three cell lines exposed to 0.1% O2 for 24 h among individual cell lines exposed to 1%, 0.2% and 0.1% O2 for 24 h. Most genes showed a gradual increase in expression with decreasing oxygen concentration (except TAF8A, which decreased).
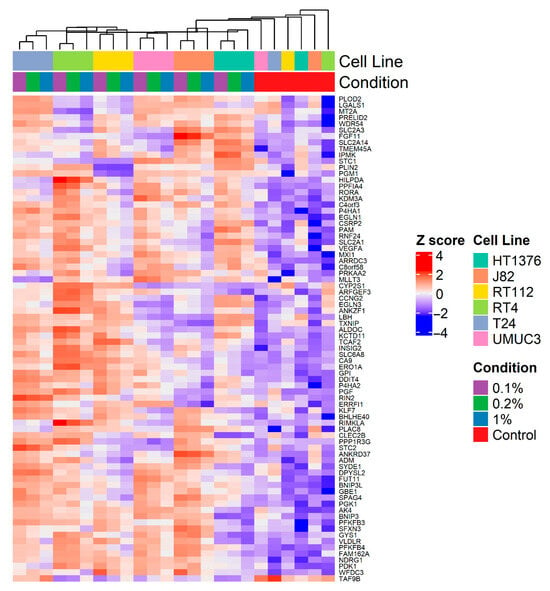
Figure 4.
Heat map of expression of 78 genes (77 upregulated, 1 downregulated) in ≥3 bladder cancer cell lines exposed to 0.1% O2 for 24 h demonstrating gradation in expression in each cell line exposed to 1%, 0.2% and 0.1% O2 for 24 h.
Figure 5 shows Kaplan–Meier curves stratifying patients in the TCGA-BLCA cohort (n = 413) according to median HS24, ME77 and ME5. HS and ME scores were derived as the median gene expression of 24 genes (published signature), the 77 genes upregulated in ≥3 cell lines exposed to 0.1% O2 and the 5 genes common to both signatures, respectively. Both HS24 and ME5 performed similarly as prognostic biomarkers (p < 0.001) and were superior to ME77 (p = 0.056). Further Kaplan–Meier curves using several blader cancer cohorts are shown in Supplementary Figure S2, demonstrating prognostication in some. However, ME5 did not predict a benefit of hypoxia modification (p = 0.77). ME scores derived from the 11 genes and 47 genes upregulated by exposure to 1% and 0.2% O2, respectively, were not prognostic for patients in the TCGA BLCA cohort (p = 0.5 and 0.32, respectively).
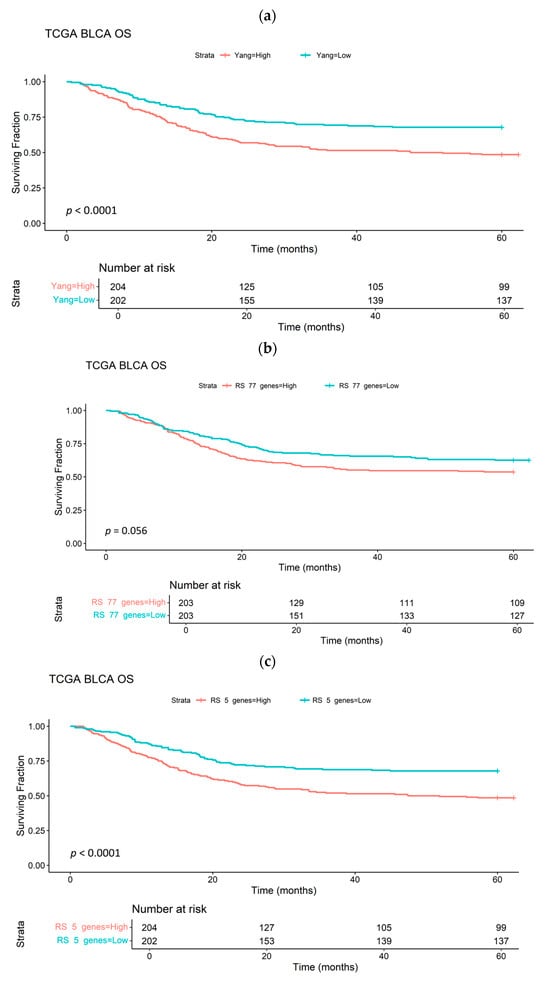
Figure 5.
Kaplan–Meier survival curves for patients with bladder cancer in the TGCA-BLCA cohort (n = 408) stratified by median expression of a 24-gene signature (a), 77 genes upregulated in at least 3 bladder cancer cell lines exposed to 0.1% O2 (b) or 5 genes common to a and b (c).
Biological processes associated with genes upregulated by at least two MIBC and both NMIBC cell lines or genes uniquely expressed by MIBC (at least two) or both NMIBC cell lines following exposure to hypoxia (0.1, 0.2 and 1% O2 for 24 h) were identified. Eight common genes were upregulated in ≥ 2 MIBC and in both NMIBC cell lines after exposure to 1% O2 (Figure 5). Additional genes were upregulated by exposure to 0.2% and 0.1% O2 (Table 5).

Table 5.
Genes that are upregulated in at least two MIBC and both NMIBC cell lines exposed to 1%, 0.2% and 0.1% O2 for 24 h compared with cells maintained in 21% O2.
Table 6 shows the biological processes upregulated by exposure to each hypoxic O2 level A common to both NMIBC cell lines and at least two MIBC cell lines and (B) no NMIBC and at least two MIBC cell lines (exclusive to MIBC). Supplementary Tables S2 and S3 shows genes upregulated in at least 2 MIBC but not NMIBC cells and genes upregulated in both NMIBC cells but not in at least 2 MIBC cells exposed to (A) 0.1%, (B) 0.2% or (C) 1% O2 for 24. No biological processes were upregulated exclusively by NMIBC cells. Processes that were upregulated in ≥2 MIBC and both NMIBC cell lines by exposure to 1%, 0.2% and 0.1% O2 included response to hypoxia, protein hydroxylation, angiogenesis, VEGF signalling, processes related to glucose metabolism and amino acid metabolism, leucocyte chemotaxis and epithelial cell proliferation. Biological processes that were uniquely upregulated by MIBC cells were primarily pathways related to the negative control of kinases associated with intracellular signalling pathways.

Table 6.
Biological pathways associated with genes upregulated by exposure to 1%, 0.2% and 0.1% O2 for 24 h in (A) at least 2 MIBC and both NMIBC cell lines and (B) at least 2 MIBC but no NMIBC cell lines.
4. Discussion
Cancer cells adapt to environmental stress such as hypoxia by upregulating the expression of genes encoding survival pathways. Identifying genes differentially regulated by hypoxia enables the derivation of hypoxia biomarkers that allows for the selection of patients for hypoxia modification. A hypoxia-sensitive panel of 77 genes commonly upregulated by at least three bladder cancer cell lines identified by exposing MIBC and NMIBC cell lines to 0.1% O2 compared unfavourably with the validated HS24 hypoxia biomarker. Five genes were identified, common to both the HS77 and HS24 gene panels, which proved to be equally as prognostic as the HS24 biomarker in the TCGA BLCA cohort but was not predictive for the selection of patients for hypoxia modification.
This study also determined biological pathways that were upregulated by exposure of NMIBC and MIBC cells to hypoxia. The biological process most significantly upregulated by both MIBC and NMIBC cells exposed to hypoxia was protein hydroxylation, driven by the expression of EGLN3, PLOD2, P4HA2, P4HA1 and EGLN1. Previous studies have demonstrated that the propyl-4-hydoxylases P4HA1 and 2 are upregulated by hypoxia in epithelial ovarian cancer cells exposed to hypoxia, and high expression is associated with decreased survival in patients with ovarian cancer. P4HA1 and 2 play a central role in collagen synthesis, EMT, collective cancer cell migration and angiogenesis [17,18,19]. PLOD2 encodes lysyl hydroxylase, which modifies collagen IV (COLVI), which modifies the extracellular matrix to weaken the lung endothelial barrier, is highly expressed in lung sarcoma metastasis [20]. PLOD2 is included in gene hypoxia signatures for RCC [21] and is a prognostic marker for patients with oesophageal cancer [22].
An important clinical requirement is the development of biomarkers that can identify NMIBC patients who are likely to progress to MIBC for early radical cystectomy or neoadjuvant/adjuvant chemotherapy to improve outcomes. Biological processes associated with genes upregulated uniquely by MIBC cells were predominantly negative regulation of kinase components of signalling pathways and driven by DUSP1, SPRY1, SPRY3, CAMK2N1 and PRKAR2B. The expression of the hypoxia-sensitive gene DUSP, which codes for dual specificity phosphatase, prevents HIF-1 overactivation by MAPK inhibition [23]. DUSP1 is prognostic for patients with gastric cancer [24] and important in immune cell regulation [23,24,25]. DUSP1 expression is decreased in glioblastoma-derived tumour stem cells (TSCs), but increased expression induces TSC differentiation [26]. PRKAR2B, which encodes Protein Kinase CAMP-Dependent Type II Regulatory Subunit Beta, is crucial for maintaining the growth of prostate cancer cells by activation of HIF-1α [27]. In lung cancer, calcium/calmodulin-dependent protein kinase II (CAMK2) inhibitor 1 (CAMK2N1) is a tumour suppressor gene which acts through inhibition of the Akt/mTOR pathway [28]. Overexpression of CAMK2N1 in lung cancer cells inhibits cancer cell proliferation and metastasis and increases cell death mechanisms [28]. However, in gastric cancer, CAMK2N1 expression has been shown to have a carcinogenic effect and to be negatively correlated with immune infiltrate [29], whilst in advanced prostate cancer (PCa) CAMK2N1 overexpression was associated with aggressiveness [30]. SPRY1 and 3 genes encode Sprouty 1 and 3, inhibitors of fibroblast growth factor (FGF). Expression of SPRY1 is associated with decreased growth and invasiveness of prostate and breast cancer [31,32]. However, SPRY3 is a designated tumour promoter for glioblastoma [33].
The expression of STC1 and LOX, which are only increased by hypoxia in MIBC cells, has previously been shown to have significant roles in human carcinogenesis [34,35,36]. STC1 expression is associated with poor postoperative survival [34], tumour growth and metastasis [35] and has been proposed as a potential drug target [34]. LOX expression has been shown to be associated with metastatic potential [36].
Expression of CA9 is commonly shown to be upregulated by hypoxia [37]. Whilst some studies have demonstrated association with prognosis [38,39], others have shown that the HIF-regulated genes CA9 and HIF-specific prolyl hydroxylases (hypoxia-inducible PHDs) genes have limitations as hypoxia biomarkers [40,41,42,43]. The HIF-inducible gene VEGF was generally found to be increased in expression by hypoxia treatment, in agreement with previous work [44]. Several studies have demonstrated improvement in treatment response by inclusion of a VEGF inhibitor in several cancer types, including breast cancer [45], NSCLC [46], metastatic colorectal cancer [47] and renal cell carcinoma [48].
A metabolic switch in response to hypoxia indicates cancer progression [49]. The expression of glucose transporter members, including SLC2A1 (GLUT1) and SLC2A3 (GLUT3), was upregulated by hypoxia in most MIBC and NMIBC cells. Hypoxia-induced increase in GLUT1 protein expression has previously been shown to correspond with increased uptake of the PET glucose analogue FDG in hypoxic cancer cells [50]. The creatine transporter SLC6A8 was consistently upregulated in all cell lines. SLC6A8 promotes intracellular phosphocreatine levels in hypoxia, increasing ATP concentration, suppressing apoptosis and promoting survival [51]. An in vivo study has shown that inhibition of SLC6A8 inhibits colorectal cancer cell growth [52].
Based on a search in PubMed, no previous study has investigated the effects of exposure to 1%, 0.2% and 0.1% O2 concentration levels in multiple bladder cancer cell lines on gene expression. The three levels of O2 are associated with HIF1 induction (1% O2), UTR gene expression (0.2% O2) and of radiobiological relevance (0.1% O2). A possible criticism is that the study only examined cells exposed to hypoxia for a single time point, 24 h. However, by this time point, transcriptional adaptation to hypoxia has taken place, and 24 h represents biologically relevant hypoxia [53,54,55]. A fixed time point of 24 h is classed as chronic hypoxia, but some cells in solid cancers will be exposed to cycling hypoxia [56]. However, generally, cycling hypoxia induces similar but weaker changes in gene expression profiles than chronic hypoxia does [56]. The exception in some cells is the greater induction of immune-associated genes associated with cycling hypoxia [56].
This study examined changes in the expression of genes in established rather than primary bladder cancer cell lines in response to hypoxia. One study that examined gene expression in both established and primary bladder cancer cells lines demonstrated eight upregulated genes in primary bladder tissue of particular interest due to their sensitivity to hypoxia in vitro [57]. We found that seven of these genes, IGFBP3, VEGF, CCNG2, NDRG1, PFKFB3, ADM and SLC2A1, were also upregulated in MIBC and/or NMIBC in our study, suggesting parallel hypoxia-induced expression changes with primary bladder tumour tissue.
Two NMIBC cell lines were used in this study. There is one more NMIBC cell line available, SW780, which could be analysed in future work to verify the PCA clustering. Genomic analysis using in vivo models of NMIBC and MIBC cell lines would help verify differential gene expression.
5. Conclusions
In summary, this work has identified genes that are upregulated by exposure of multiple bladder cancer cells to hypoxia and demonstrated that some genes are upregulated in both MIBC and NMIBC cells but others uniquely in either MIBC or NMIBC cells. The study has also corroborated the hypoxia sensitivity of the HS24 bladder cancer gene signature in multiple cancer cell lines. The importance of gene expression platforms in biomarker design is also demonstrated.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cancers17162624/s1: Figure S1: Median gene expression values for 77 (76) and 5 gene panels identified as prognostic in the TCGA cohort in pimonidazole-low and -high stained regions in a bladder cancer cell-derived xenograft. * p < 0.05; ** p < 0.01. Figure S2: Overall survival curves for patients with bladder cancer stratified by median expression of the 77 genes (ME77) upregulated at least 3 bladder cancer cell lines or 5 genes common to the ME77 and Yang 24-gene bladder cancer hypoxia signature, respectively, in BCON (A,B), GSE38164 (C,D), R19915 GLP8333 (E,F), R19915 GLP5186 (G,H) and IMvigo210 (I,J); Table S1: Genes upregulated in at least 2 MIBC but not NMIBC cells exposed to (A) 0.1%, (B) 0.2% or (C) 1% O2 for 24 h. Table S2 Genes upregulated in both NMIBC cells but not in at least 2 MIBC cells exposed to (A) 0.1%, (B) 0.2% or (C) 1% O2 for 24 h. Table S3 Genes up-regulated in both NMIBC cancer cells but not in at least 2 MIBC cancer cells exposed to (A) 0.1% (B) 0.2% or (C) 1% O2 for 24 h.
Author Contributions
Conceptualisation, T.A.D.S. and C.M.L.W.; Methodology, T.A.D.S., C.M.L.W. and R.S.; Validation, L.Z. and B.L.; Formal Analysis, B.L. and L.Z.; Investigation, R.S.; Data Curation, R.S.; Writing—Original Draft Preparation, R.S. and T.A.D.S.; Writing—Review and Editing, All authors; Visualisation, R.S., C.G.Q. and T.A.D.S.; Supervision, T.A.D.S., P.J.H. and A.C.; Project Administration, C.M.L.W., T.A.D.S. and A.C.; Funding Acquisition, C.M.L.W. and T.A.D.S. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for this work was provided by the Ministry of Higher Education and the Ministry of Health, Saudi Arabian Government (Riyadh, Saudi Arabia) and the Saudi Arabian Cultural Bureau (London, United Kingdom), [KSP12018294/1440-7582-2]. The research was carried out at the National Institute for Health and Care Research (NIHR) Manchester Biomedical Research Centre (BRC) (NIHR203308)].
Data Availability Statement
Cell line transcriptomic data generated in this study will be made publicly available on the GEO repository (accession number GSE302555).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- West, C.M.L.; Slevin, F. Tumour hypoxia. Clin. Oncol. R. Coll. Radiol. 2019, 31, 595–599. [Google Scholar] [CrossRef]
- Flaig, T.W. NCCN Guidelines Updates: Management of Muscle-Invasive Bladder Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 591–593. [Google Scholar]
- Flegar, L.; Kraywinkel, K.; Zacharis, A.; Aksoy, C.; Koch, R.; Eisenmenger, N.; Groeben, C.; Huber, J. Treatment trends for muscle-invasive bladder cancer in Germany from 2006 to 2019. World J. Urol. 2022, 40, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.P.; Mistry, H.; Irlam, J.; Valentine, H.; Yang, L.; Lane, B.; West, C.; Choudhury, A.; Hoskin, P.J. Long-Term Outcomes of Radical Radiation Therapy with Hypoxia Modification with Biomarker Discovery for Stratification: 10-Year Update of the BCON (Bladder Carbogen Nicotinamide) Phase 3 Randomized Trial (ISRCTN45938399). Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Taylor, J.; Eustace, A.; Irlam, J.J.; Denley, H.; Hoskin, P.J.; Alsner, J.; Buffa, F.M.; Harris, A.L.; Choudhury, A.; et al. A Gene Signature for Selecting Benefit from Hypoxia Modification of Radiotherapy for High-Risk Bladder Cancer Patients. Clin. Cancer Res. 2017, 23, 4761–4768. [Google Scholar] [CrossRef]
- Smith, T.A.D.; Lane, B.; More, E.; Valentine, H.; Lunj, S.; Abdelkarem, O.A.; Irlam, J.J.; Shabbir, R.; Vora, S.; Denley, H.; et al. Comparison of multiple gene expression platforms for measuring a bladder cancer hypoxia signature. Mol. Med. Rep. 2022, 26, 261. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; West, C.M.L. Hypoxia gene expression signatures as predictive biomarkers for personalising radiotherapy. Br. J. Radiol. 2018, 92, 20180036. [Google Scholar] [CrossRef]
- Lane, B.; Khan, M.T.; Choudhury, A.; Salem, A.; West, C.M.L. Development and validation of a hypoxia-associated signature for lung adenocarcinoma. Sci. Rep. 2022, 12, 1290. [Google Scholar] [CrossRef]
- Calvo Tardón, M.; Marinari, E.; Migliorini, D.; Bes, V.; Tankov, S.; Charrier, E.; McKee, T.A.; Dutoit, V.; Dietrich, P.Y.; Cosset, E.; et al. An Experimentally Defined Hypoxia Gene Signature in Glioblastoma and Its Modulation by Metformin. Biology 2020, 9, 264. [Google Scholar] [CrossRef]
- Trong, P.D.; Rösch, S.; Mairbäurl, H.; Pusch, S.; Unterberg, A.; Herold-Mende, C.; Warta, R. Identification of a Prognostic Hypoxia-Associated Gene Set in IDH-Mutant Glioma. Int. J. Mol. Sci. 2018, 19, 2903. [Google Scholar] [CrossRef]
- Yang, L.; Forker, L.; Irlam, J.J.; Pillay, N.; Choudhury, A.; West, C.M.L. Validation of a hypoxia related gene signature in multiple soft tissue sarcoma cohorts. Oncotarget 2017, 9, 3946–3955. [Google Scholar] [CrossRef] [PubMed]
- Boegemann, M.; Krabbe, L.M. Prognostic Implications of Immunohistochemical Biomarkers in Non-muscle-invasive Blad Cancer and Muscle-invasive Bladder Cancer. Mini. Rev. Med. Chem. 2020, 20, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jia, X.; Duan, Y.; Xiao, H.; Sundqvist, K.G.; Permert, J.; Wang, F. Excess glucose induces hypoxia-inducible factor-1α in pancreatic cancer cells and stimulates glucose metabolism and cell migration. Cancer Biol. Ther. 2013, 14, 428–435. [Google Scholar] [CrossRef][Green Version]
- Carvalho, B.S.; Irizarry, R.A. A Framework for Oligonucleotide Microarray Preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Iimma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Shabbir, R.; Telfer, B.A.; Dickie, B.; Reardon, M.; Babur, M.; Williams, K.; West, C.M.L.; Choudhury, A.; Smith, T.A.D. Implementation of Oxygen Enhanced Magnetic Resonance Imaging (OE-MRI) and a Pilot Genomic Study of Hypoxia in Bladder Cancer Xenografts. Cancer Genom. Proteom. 2024, 21, 380–387. [Google Scholar] [CrossRef]
- Song, M.; Schnettler, E.; Venkatachalam, A.; Wang, Y.; Feldman, L.; Argenta, P.; Rodriguez-Rodriguez, L.; Ramakrishnan, S. Increased expression of collagen prolyl hydroxylases in ovarian cancer is associated with cancer growth and metastasis. Am. J. Cancer Res. 2023, 13, 6051–6062. [Google Scholar]
- Aggarwal, V.; Sahoo, S.; Donnenberg, V.S.; Chakraborty, P.; Jolly, M.K.; Sant, S. P4HA2: A link between tumor-intrinsic hypoxia, partial EMT and collective migration. Adv. Cancer Biol. Metastasis 2022, 5, 100057. [Google Scholar] [CrossRef]
- Xu, Y.; Xia, D.; Huang, K.; Liang, M. Hypoxia-induced P4HA1 overexpression promotes post-ischemic angiogenesis by enhancing endothelial glycolysis through downregulating FBP1. J. Transl. Med. 2024, 22, 74. [Google Scholar] [CrossRef]
- Liu, Y.; Murazzi, I.; Fuller, A.M.; Pan, H.; Irizarry-Negron, V.M.; Devine, A.; Katti, R.; Skuli, N.; Ciotti, G.E.; Pak, K.; et al. Sarcoma Cells Secrete Hypoxia-Modified Collagen VI to Weaken the Lung Endothelial Barrier and Promote Metastasis. Cancer Res. 2024, 84, 977–993. [Google Scholar] [CrossRef]
- Wu, X.; Xie, W.; Gong, B.; Fu, B.; Chen, W.; Zhou, L.; Luo, L. Development and validation of a combined hypoxia- and metabolism-related prognostic signature to predict clinical prognosis and immunotherapy responses in clear cell renal cell carcinoma. Front. Oncol. 2023, 13, 1162846. [Google Scholar] [CrossRef]
- Gong, X.; Wang, A.; Song, W. Clinicopathological significances of PLOD2, epithelial-mesenchymal transition markers, and cancer stem cells in patients with esophageal squamous cell carcinoma. Medicine 2022, 101, e30112. [Google Scholar] [CrossRef]
- Liu, C.; Shi, Y.; Du, Y.; Ning, X.; Liu, N.; Huang, D.; Liang, J.; Xue, Y.; Fan, D. Dual-specificity phosphatase DUSP1 protects overactivation of hypoxia-inducible factor 1 through inactivating ERK MAPK. Exp. Cell Res. 2005, 309, 410–418. [Google Scholar] [CrossRef]
- Yu, J.; Li, H.; Huang, C.; Chen, H. Identification and characterization of ferroptosis-related genes in therapy-resistant gastric cancer. Medicine 2024, 103, e38193. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Chen, Y. Development of a Comprehensive Gene Signature Linking Hypoxia, Glycolysis, Lactylation, and Metabolomic Insights in Gastric Cancer through the Integration of Bulk and Single-Cell RNA-Seq Data. Biomedicines 2023, 11, 2948. [Google Scholar] [CrossRef] [PubMed]
- Mills, B.N.; Albert, G.P.; Halterman, M.W. Expression Profiling of the MAP Kinase Phosphatase Family Reveals a Role for DUSP1 in the Glioblastoma Stem Cell Niche. Cancer Microenviron. 2017, 10, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Sun, J.; Xie, S.; Chi, C.; Zhu, Y.; Pan, J.; Dong, B.; Huang, Y.; Xia, W.; Sha, J.; et al. PRKAR2B-HIF-1α loop promotes aerobic glycolysis and tumour growth in prostate cancer. Cell Prolif. 2020, 53, e12918. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, C.; Jiang, H.; He, W. Targeting CAMK2N1/CAMK2 inhibits invasion, migration and angiogenesis of non-small cell lung cancer by promoting autophagy and apoptosis via AKT/mTOR signaling pathway. Gene 2024, 913, 148375. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Ren, X.; Ren, Q. NcRNA-mediated upregulation of CAMK2N1 is associated with poor prognosis and tumor immune infiltration of gastric cancer. Front. Genet. 2022, 13, 888672. [Google Scholar] [CrossRef]
- Carneiro, I.; Quintela-Vieira, F.; Lobo, J.; Moreira-Barbosa, C.; Menezes, F.D.; Martins, A.T.; Oliveira, J.; Silva, R.; Jerónimo, C.; Henrique, R. Expression of EMT-related genes CAMK2N1 and WNT5A is increased in locally invasive and metastatic prostate cancer. J. Cancer 2019, 10, 5915. [Google Scholar] [CrossRef]
- Mekkawy, A.H.; Pourgholami, M.H.; Morris, D.L. Human Sprouty1 suppresses growth, migration, and invasion in human breast cancer cells. Tumour. Biol. 2014, 35, 5037–5048. [Google Scholar] [CrossRef]
- Kwabi-Addo, B.; Wang, J.; Erdem, H.; Vaid, A.; Castro, P.; Ayala, G.; Ittmann, M. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004, 64, 4728–4735. [Google Scholar] [CrossRef]
- Celik-Selvi, B.E.; Stütz, A.; Mayer, C.E.; Salhi, J.; Siegwart, G.; Sutterlüty, H. Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells. Cells 2019, 8, 808. [Google Scholar] [CrossRef]
- Tamura, S.; Kaike, M.; Kunisaki, C.; Tanaka, K.; Masuda, M.; Imada, T. Clinical significance of STC1 gene expression in patients with colorectal cancer. Anticancer Res. 2011, 31, 325–329. [Google Scholar]
- Chang, A.C.; Doherty, J.; Huschtscha, L.I.; Redvers, R.; Restall, C.; Reddel, R.R.; Anderson, R.L. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin. Exp. Metastasis 2015, 32, 15–27. [Google Scholar] [CrossRef]
- Xiao, Q.; Ge, G. Lysyl, Oxidase, Extracellular Matrix Remodeling and Cancer Metastasis Cancer. Microenviron 2012, 5, 261–273. [Google Scholar] [CrossRef]
- Said, H.M.; Staab, A.; Hagemann, C.; Vince, G.H.; Katzer, A.; Flentje, M.; Vordermark, D. Distinct patterns of hypoxic expression of carbonic anhydrase IX (CA IX) in human malignant glioma cell lines. J. Neuro-Oncol. 2007, 81, 27–38. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Simopoulos, K.; Pastorek, J.; Wykoff, C.C.; Gatter, K.C.; Harris, A.L. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin. Cancer Res. 2001, 7, 3399–3403. [Google Scholar] [PubMed]
- Tanaka, N.; Kato, H.; Inose, T.; Kimura, H.; Faried, A.; Sohda, M.; Nakajima, M.; Fukai, Y.; Miyazaki, T.; Masuda, N.; et al. Expression of carbonic anhydrase 9, a potential intrinsic marker of hypoxia, is associated with poor prognosis in oesophageal squamous cell carcinoma. Br. J. Cancer 2008, 99, 1468–1475. [Google Scholar] [CrossRef] [PubMed]
- Willam, C.; Warnecke, C.; Schefold, J.C.; Kügler, J.; Koehne, P.; Frei, U.; Wiesener, M.; Eckardt, K.U. Inconsistent effects of acidosis on HIF-α protein and its target genes. Pflügers Arch. 2006, 451, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, J.G.; Overgaard, J. Lack of prognostic and predictive value of CA IX in radiotherapy of squamous cell carcinoma of the head and neck with known modifiable hypoxia: An evaluation of the DAHANCA 5 study. Radiother. Oncol. 2007, 83, 383–388. [Google Scholar] [CrossRef]
- Douglas, C.M.; Bernstein, J.M.; Ormston, V.E.; Hall, R.C.; Merve, A.; Swindell, R.; Valentine, H.R.; Slevin, N.J.; West, C.M.L.; Homer, J.J. Lack of prognostic effect of carbonic anhydrase-9, hypoxia inducible factor-1α and bcl-2 in 286 patients with early squamous cell carcinoma of the glottic larynx treated with radiotherapy. Clin. Oncol. R. Coll. Radiol. 2013, 25, 59–65. [Google Scholar] [CrossRef]
- Jonathan, R.A.; Wijffels, K.I.; Peeters, W.; de Wilde, P.C.; Marres, H.A.; Merkx, M.A.; Oosterwijk, E.; van der Kogel, A.J.; Kaanders, J.H. The prognostic value of endogenous hypoxia-related markers for head and neck squamous cell carcinomas treated with ARCON. Radiother. Oncol. 2006, 79, 288–297. [Google Scholar] [CrossRef]
- Jubb, A.M.; Pham, T.Q.; Hanby, A.M.; Frantz, G.D.; Peale, F.V.; Wu, T.D.; Koeppen, H.W.; Hillan, K.J. Expression of vascular endothelial growth factor, hypoxia inducible factor 1α, and carbonic anhydrase IX in human tumours. J. Clin. Pathol. 2004, 57, 504–512. [Google Scholar] [CrossRef]
- Miller, K.W. A randomized phase III trial of paclitaxel versus paclitaxel plus bevacizumab as first-line therapy for locally recurrent or metastatic breast cancer. Proc. Am. Soc. Clin. Oncol. 2005, 23, 4. [Google Scholar] [CrossRef]
- Sandler, A.; Gray, R.; Perry, M.C.; Brahmer, J.; Schiller, J.H.; Dowlati, A.; Lilenbaum, R.; Johnson, D.H. Paclitaxel–carboplatin alone or with bevacizumab for non–small-cell lung cancer. N. Engl. J. Med. 2006, 355, 2542–2550. [Google Scholar] [CrossRef]
- Hurwitz, H.; Fehrenbacher, L.; Novotny, W.; Cartwright, T.; Hainsworth, J.; Heim, W.; Berlin, J.; Baron, A.; Griffing, S.; Holmgren, E.; et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004, 350, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Haworth, L.; Sherry, R.M.; Hwu, P.; Schwartzentruber, D.J.; Topalian, S.L.; Steinberg, S.M.; Chen, H.X.; Rosenberg, S.A. A randomized trial of bevacizumab, an anti–vascular endothelial growth factor antibody, for metastatic renal cancer. N. Engl. J. Med. 2003, 349, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Chabaud, S.; Pellerin, È.; Caneparo, C.; Ringuette-Goulet, C.; Pouliot, F.; Bolduc, S. Bladder cancer cell lines adapt their aggressiveness profile to oxygen tension. Oncol. Lett. 2022, 24, 220. [Google Scholar] [CrossRef]
- Smith, T.A.D.; AbdelKarem, O.A.; Irlam, J.J.; Lane, B.; Valentine, H.; Bibby, B.A.S.; Denley, H.; Choudhury, A.; West, C.M.L. Selection of endogenous control genes for normalising gene expression data derived from formalin-fixed paraffin-embedded tumour tissue. Sci. Rep. 2020, 10, 17258. [Google Scholar] [CrossRef]
- Li, Q.; Liu, M.; Sun, Y.; Jin, T.; Zhu, P.; Wan, X.; Hou, Y.; Tu, G. SLC6A8-mediated intracellular creatine accumulation enhances hypoxic breast cancer cell survival via ameliorating oxidative stress. J. Exp. Clin. Cancer Res. 2021, 40, 168. [Google Scholar] [CrossRef]
- Kurth, I.; Yamaguchi, N.; Andreu-Agullo, C.; Tian, H.S.; Sridhar, S.; Takeda, S.; Gonsalves, F.C.; Loo, J.M.; Barlas, A.; Manova-Todorova, K.; et al. Therapeutic targeting of SLC6A8 creatine transporter suppresses colon cancer progression and modulates human creatine levels. Sci. Adv. 2021, 7, eabi7511. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Liu, X.; Yan, N.; Li, S.; Cao, G.; Cheng, Q.; Xia, Q.; Wang, H. Hypoxia-inducible transcription factor-1alpha promotes hypoxia-induced A549 apoptosis via a mechanism that involves the glycolysis pathway. BMC Cancer 2006, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Schwab, L.P.; Peacock, D.L.; Majumdar, D.; Ingels, J.F.; Jensen, L.C.; Smith, K.D.; Cushing, R.C.; Seagroves, T.N. Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012, 14, R6. [Google Scholar] [CrossRef] [PubMed]
- Tátrai, E.; Bartal, A.; Gacs, A.; Paku, S.; Kenessey, I.; Garay, T.; Hegedűs, B.; Molnár, E.; Cserepes, M.T.; Hegedűs, Z. Cell type-dependent HIF1 α-mediated effects of hypoxia on proliferation, migration and metastatic potential of human tumor cells. Oncotarget 2017, 8, 44498–44510. [Google Scholar] [CrossRef]
- Olbryt, M.; Habryka, A.; Student, S.; Jarząb, M.; Tyszkiewicz, T.; Lisowska, K.M. Global gene expression profiling in three tumor cell lines subjected to experimental cycling and chronic hypoxia. PLoS ONE 2014, 9, e105104. [Google Scholar] [CrossRef]
- Ord, J.J.; Streeter, E.H.; Roberts, I.S.; Cranston, D.; Harris, A.L. Comparison of hypoxia transcriptome in vitro with in vivo gene expression in human bladder cancer. Br. J. Cancer 2005, 93, 346–354. [Google Scholar] [CrossRef][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).