A Validated Proteomic Signature of Basal-like Triple-Negative Breast Cancer Subtypes Obtained from Publicly Available Data
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Data
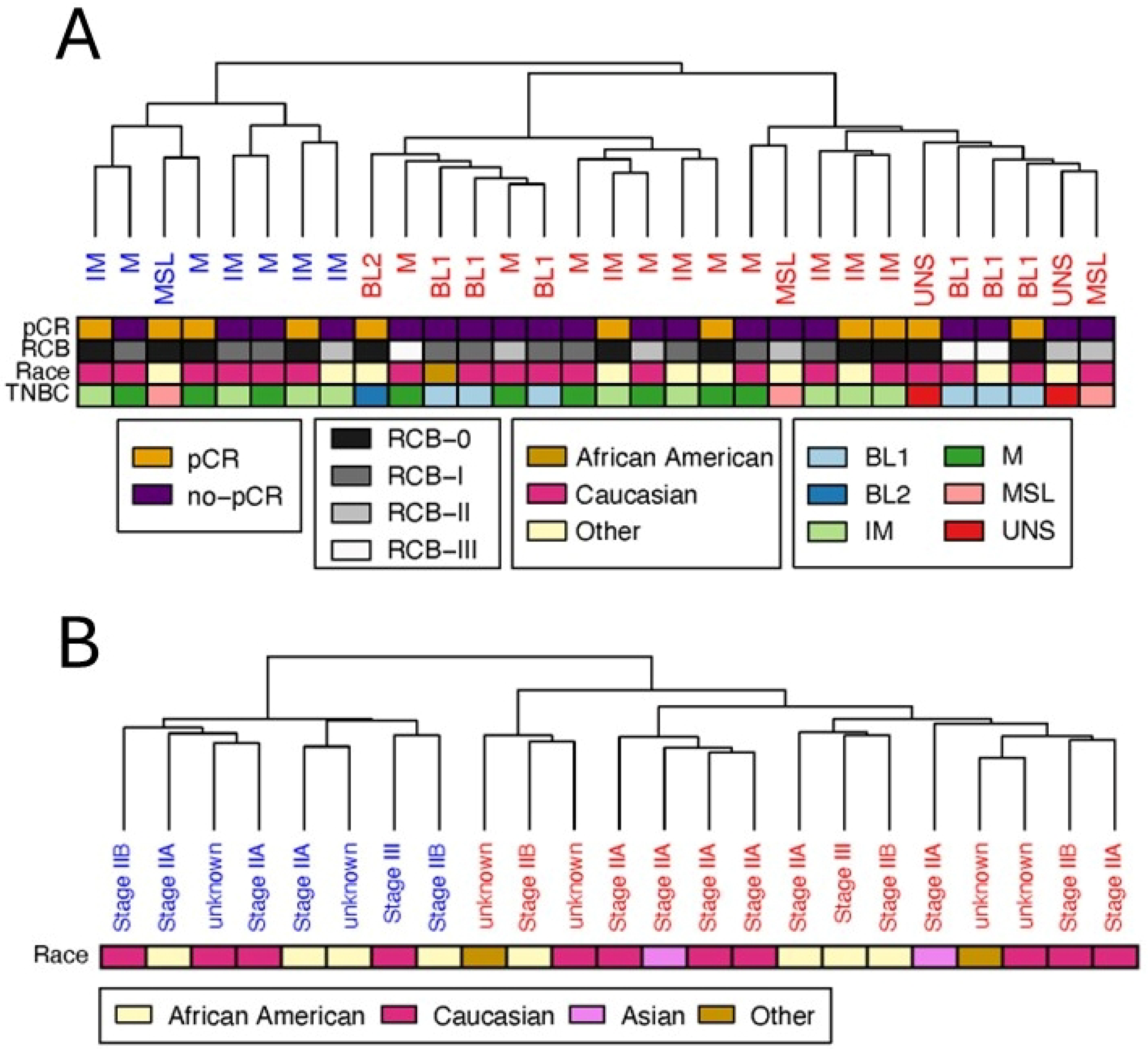
Patient and Sample Classification
2.2. Statistical Methods
2.2.1. Handling of Missing Data and Imputation
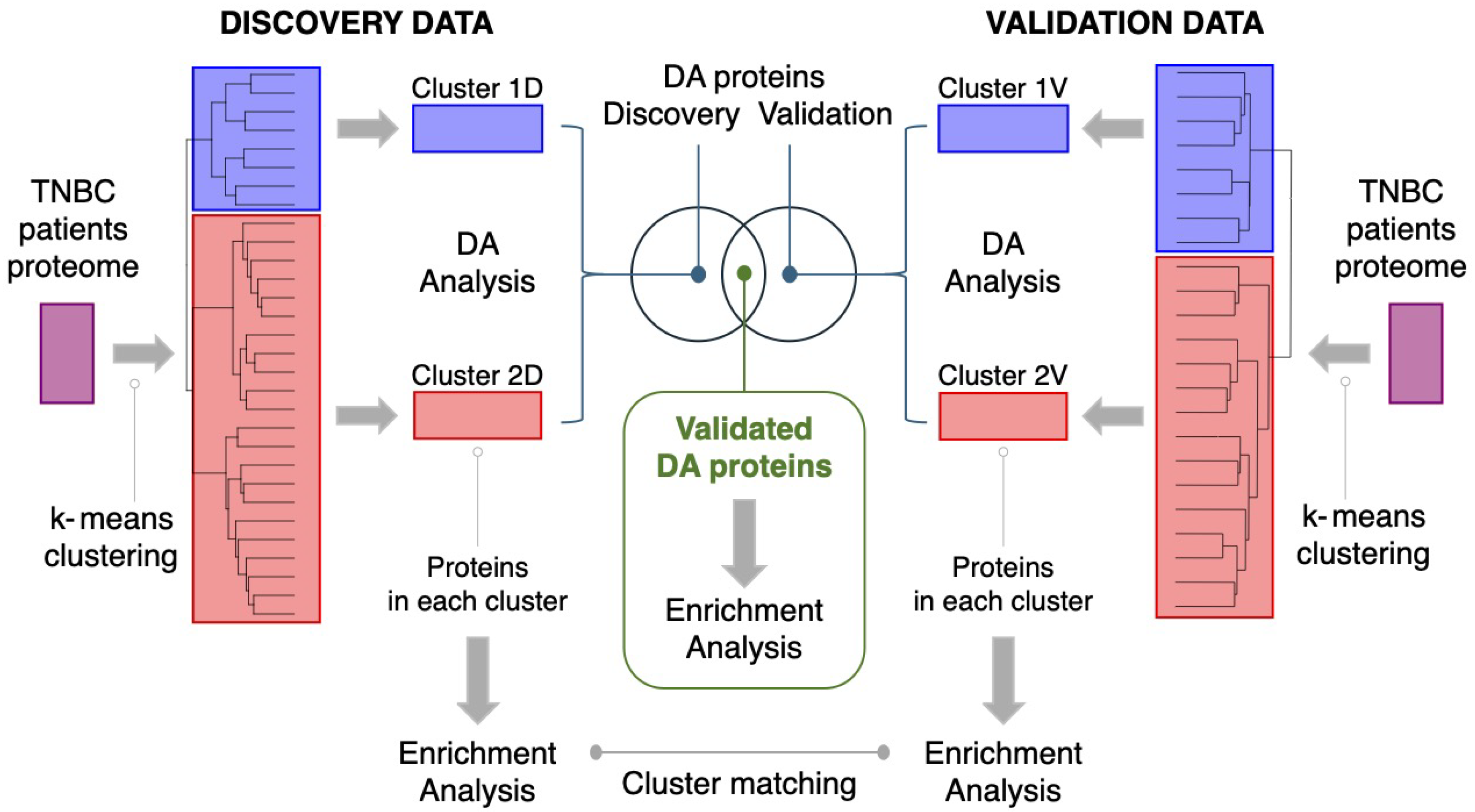
2.2.2. Clustering of Samples
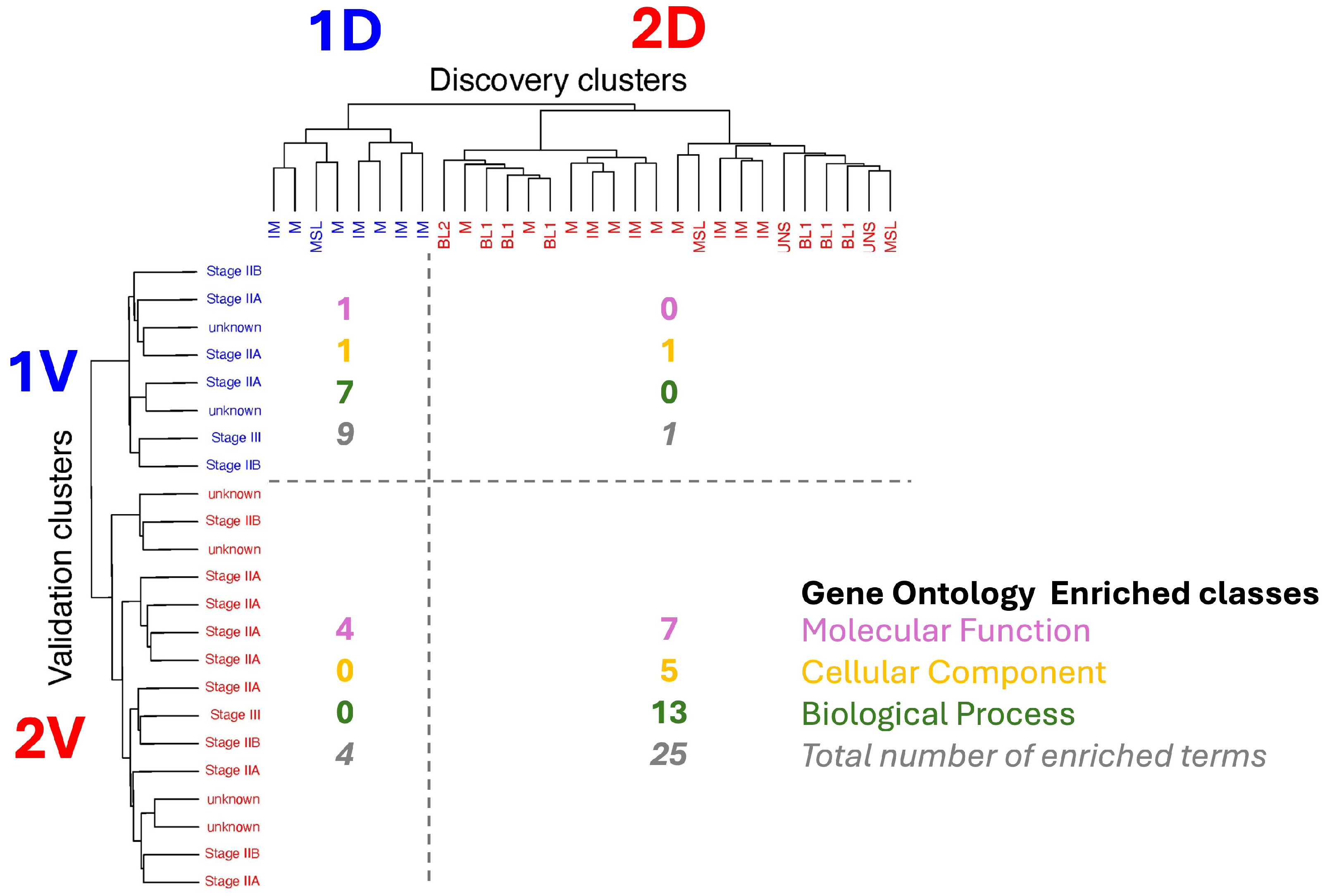
2.2.3. Matching of Clusters
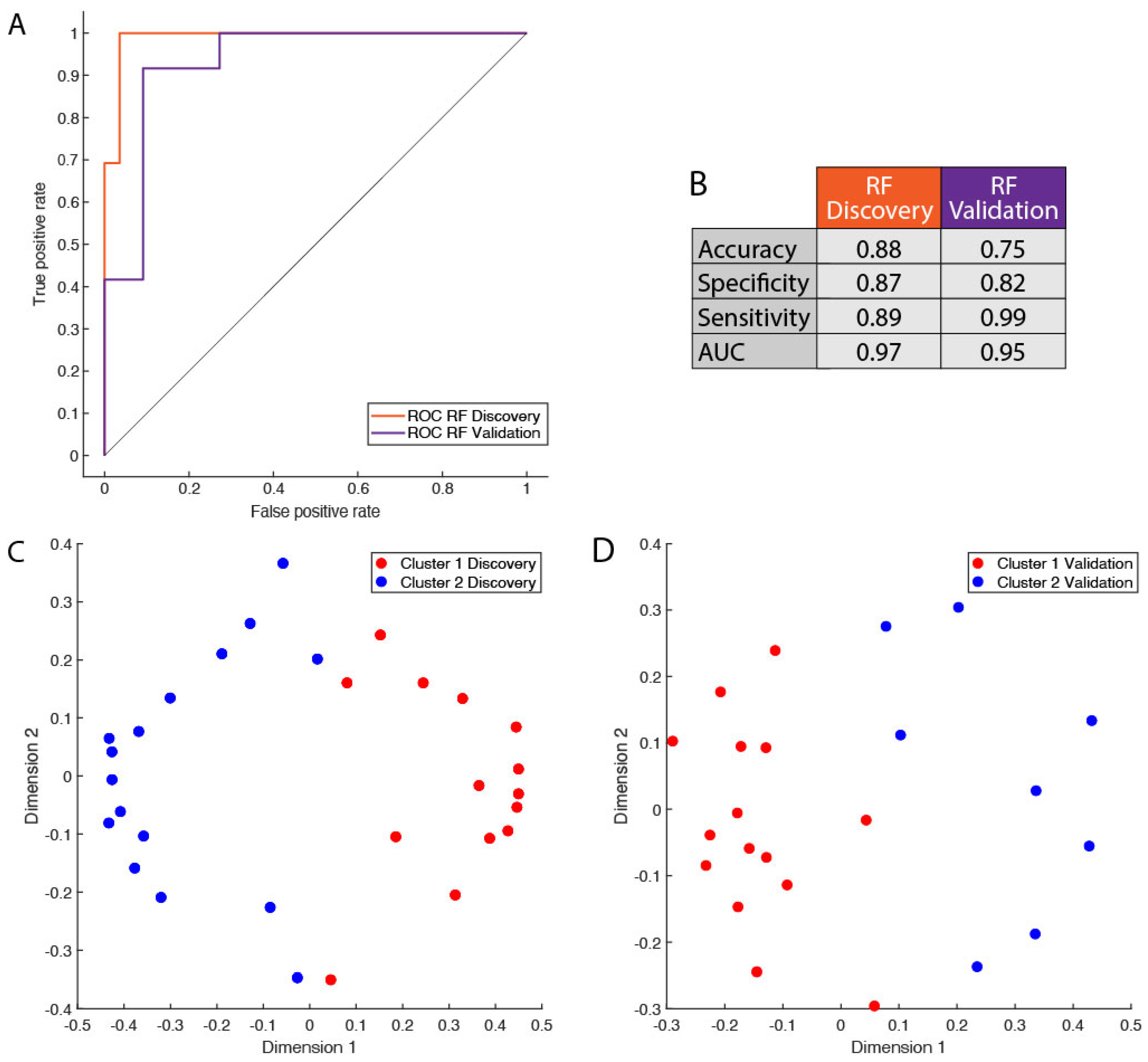
2.2.4. Random Forest Modeling
2.2.5. Differential Analysis of Protein Abundances
2.2.6. Analysis of Sample Metadata
2.2.7. Protein–Protein Interaction Analysis
2.2.8. Enrichment Analysis
2.3. Software and Data
3. Results
3.1. Basal-like Triple-Negative Breast Cancer Samples of Discovery Dataset Can Be Separated in Two Clear Clusters
3.2. Clustering of Basal-like Triple-Negative Breast Cancer Samples in the Validation Dataset Also Yields Two Clusters
3.3. Protein Signature Enables Robust Cluster Classification of Basal-like Triple-Negative Breast Cancer Patients
3.4. Identification of Reproducible Differential Proteomic Profiles Between Patient Clusters
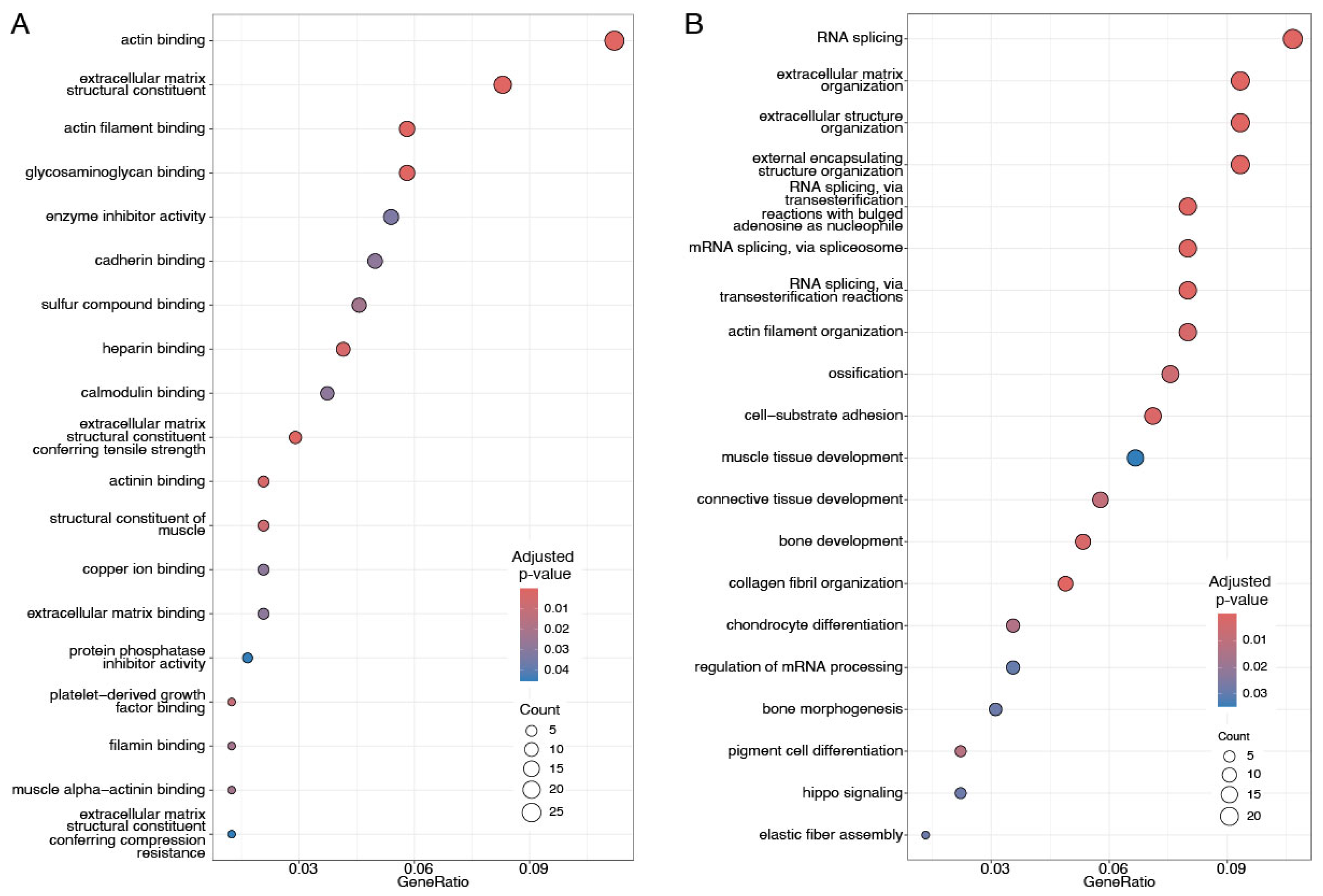
3.5. Up- and Downregulated Proteins Associated with Distinct Cellular and Molecular Pathways
3.6. Network Analysis of Differentially Abundant Proteins Reveal Functional Clusters Centered on Collagen and T-Complex Protein 1
4. Discussion
4.1. Upregulated Proteins Contributing to Cluster Separation Are Enriched for Structural and Extracellular Matrix Functions and for RNA Splicing
4.2. Functions of Downregulated Proteins
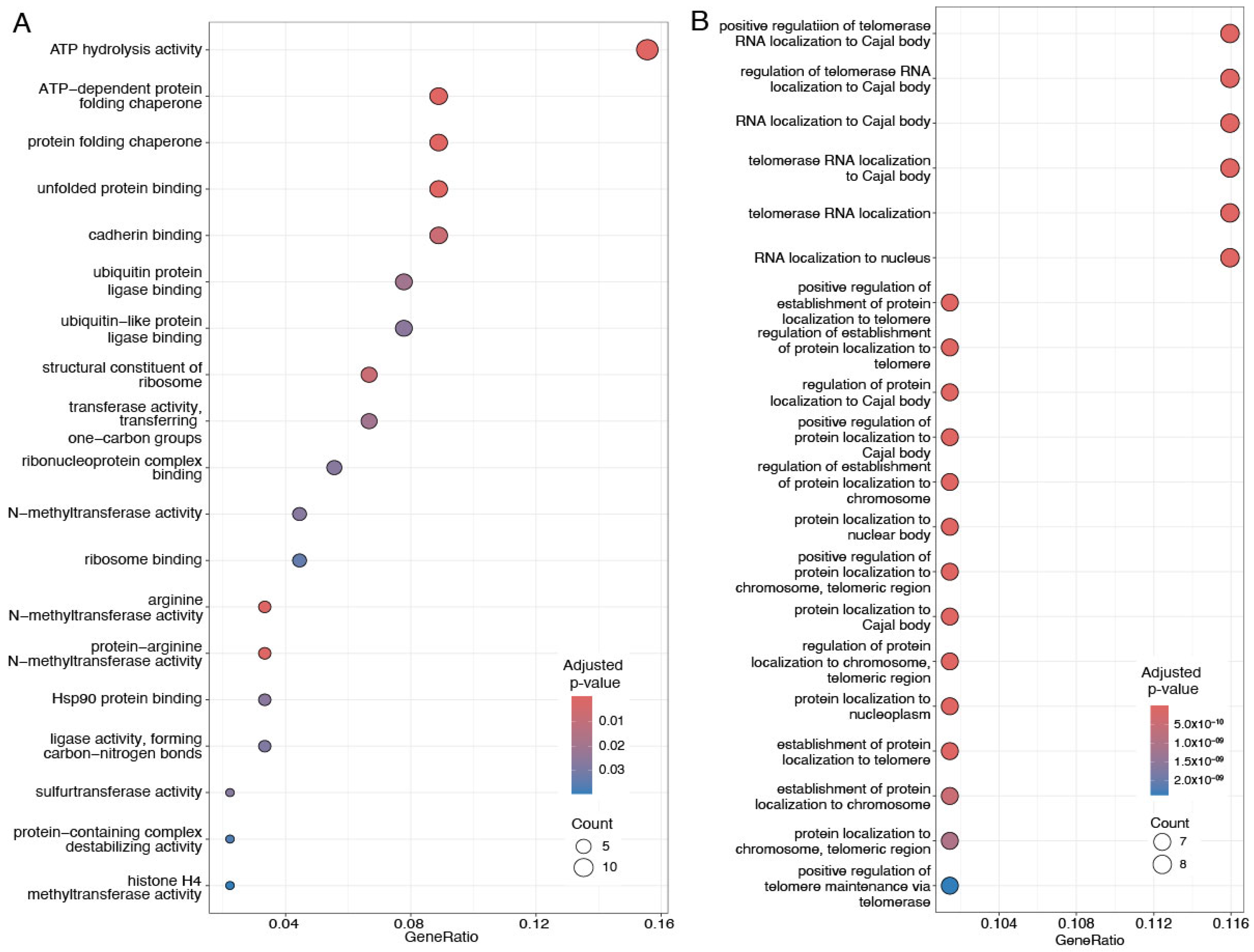
4.3. Dysregulation of Upregulated Interacting Proteins Involves SNRPG, Collagen, and PRC1 Complexes
4.4. TCP1, Microtubule, and ARS Complexes Are Affected by Downregulated Proteins
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BLBC | Basal-like breast cancer |
| CPTAC | Clinical Proteomic Tumor Analysis Consortium |
| ER | Extrogen |
| HDI | Human Development Index |
| HER2 | Human epidermal growth factor receptor 2 |
| PAM50 | Prediction Analysis of Microarray |
| PR | Progesterone |
| RF | Random Forest |
| TNBC | Triple-negative breast cancer |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Asleh, K.; Negri, G.L.; Spencer Miko, S.E.; Colborne, S.; Hughes, C.S.; Wang, X.Q.; Gao, D.; Gilks, C.B.; Chia, S.K.; Nielsen, T.O.; et al. Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat. Commun. 2022, 13, 896. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumors. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Davey, M.G.; Kerin, M.J. Molecular profiling in contemporary breast cancer management. Br. J. Surg. 2023, 110, 743–745. [Google Scholar] [CrossRef]
- Malhotra, G.K.; Zhao, X.; Band, H.; Band, V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. Ther. 2010, 10, 955–960. [Google Scholar] [CrossRef]
- Weigelt, B.; Geyer, F.C.; Reis-Filho, J.S. Histological types of breast cancer: How special are they? Mol. Oncol. 2010, 4, 192–208. [Google Scholar] [CrossRef]
- Perez, E.A. Breast cancer management: Opportunities and barriers to an individualized approach. Oncologist 2011, 16, 20–22. [Google Scholar] [CrossRef]
- Taylor, C.; McGale, P.; Probert, J.; Broggio, J.; Charman, J.; Darby, S.C.; Kerr, A.J.; Whelan, T.; Cutter, D.J.; Mannu, G.; et al. Breast cancer mortality in 500,000 women with early invasive breast cancer in England, 1993–2015: Population based observational cohort study. BMJ 2023, 381, 1744. [Google Scholar]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Vici, P.; Pizzuti, L.; Natoli, C.; Gamucci, T.; Di Lauro, L.; Barba, M.; Sergi, D.; Botti, C.; Michelotti, A.; Moscetti, L.; et al. Triple positive breast cancer: A distinct subtype? Cancer Treat. Rev. 2015, 41, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Adamo, B.; Cheang, M.C.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Toft, D.J.; Cryns, V.L. Minireview: Basal-like breast cancer: From molecular profiles to targeted therapies. Mol. Endocrinol. 2011, 25, 199–211. [Google Scholar] [CrossRef]
- Millikan, R.C.; Newman, B.; Tse, C.K.; Moorman, P.G.; Conway, K.; Smith, L.V.; Labbok, M.H.; Geradts, J.; Bensen, J.T.; Jackson, S.; et al. Epidemiology of basal-like breast cancer. Breast Cancer Res. Treat. 2008, 109, 123–139. [Google Scholar] [CrossRef]
- Botti, G.; Cantile, M.; Collina, F.; Cerrone, M.; Sarno, S.; Anniciello, A.; Di Bonito, M. Morphological and pathological features of basal-like breast cancer. Transl. Cancer Res. 2019, 8, S503. [Google Scholar] [CrossRef]
- Almansour, N.M. Triple-negative breast cancer: A brief review about epidemiology, risk factors, signaling pathways, treatment and role of artificial intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Trapani, D.; Viale, G.; Criscitiello, C.; Curigliano, G. Practical classification of triple-negative breast cancer: Intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, A.A.; Naz, S.; Hashmi, S.K.; Hussain, Z.F.; Irfan, M.; Bakar, S.M.A.; Faridi, N.; Khan, A.; Edhi, M.M. Cytokeratin 5/6 and cytokeratin 8/18 expression in triple negative breast cancers: Clinicopathologic significance in South-Asian population. BMC Res. Notes 2018, 11, 372. [Google Scholar] [CrossRef]
- Yehiely, F.; Moyano, J.V.; Evans, J.R.; Nielsen, T.O.; Cryns, V.L. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol. Med. 2006, 12, 537–544. [Google Scholar] [CrossRef]
- Schneider, B.P.; Winer, E.P.; Foulkes, W.D.; Garber, J.; Perou, C.M.; Richardson, A.; Sledge, G.W.; Carey, L.A. Triple-negative breast cancer: Risk factors to potential targets. Clin. Cancer Res. 2008, 14, 8010–8018. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, X.Z.; Xiao, Y.; Wu, S.Y.; Zuo, W.J.; Yu, Q.; Cao, A.Y.; Li, J.J.; Yu, K.D.; Liu, G.Y.; et al. Subtyping-based platform guides precision medicine for heavily pretreated metastatic triple-negative breast cancer: The FUTURE phase II umbrella clinical trial. Cell Res. 2023, 33, 389–402. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.; Turashvili, G.; Ding, J.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef]
- Doll, S.; Gnad, F.; Mann, M. The case for proteomics and phospho-proteomics in personalized cancer medicine. Proteom.–Clin. Appl. 2019, 13, 1800113. [Google Scholar] [CrossRef]
- Neagu, A.N.; Whitham, D.; Buonanno, E.; Jenkins, A.; Alexa-Stratulat, T.; Tamba, B.I.; Darie, C.C. Proteomics and its applications in breast cancer. Am. J. Cancer Res. 2021, 11, 4006–4049. [Google Scholar]
- Metwali, E.; Pennington, S. Mass Spectrometry-Based Proteomics for Classification and Treatment Optimisation of Triple Negative Breast Cancer. J. Pers. Med. 2024, 14, 944. [Google Scholar] [CrossRef]
- Bertucci, F.; Birnbaum, D.; Goncalves, A. Proteomics of breast cancer: Principles and potential clinical applications. Mol. Cell. Proteom. 2006, 5, 1772–1786. [Google Scholar] [CrossRef]
- Shukla, H.D. Comprehensive analysis of cancer-proteogenome to identify biomarkers for the early diagnosis and prognosis of cancer. Proteomes 2017, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, B.; Li, J. Integrating proteomic and phosphoproteomic data for pathway analysis in breast cancer. BMC Syst. Biol. 2018, 12, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Han, B.; Zhang, J.T. Identification of 14-3-3σ as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res. 2006, 66, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Haibe-Kains, B.; Majjaj, S.; Lallemand, F.; Durbecq, V.; Larsimont, D.; Gonzalez-Angulo, A.M.; Pusztai, L.; Symmans, W.F.; Bardelli, A.; et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor–positive breast cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 10208–10213. [Google Scholar] [CrossRef]
- Azevedo, A.L.K.; Gomig, T.H.B.; Batista, M.; Marchini, F.K.; Spautz, C.C.; Rabinovich, I.; Sebastião, A.P.M.; Oliveira, J.C.; Gradia, D.F.; Cavalli, I.J.; et al. High-throughput proteomics of breast cancer subtypes: Biological characterization and multiple candidate biomarker panels to patients’ stratification. J. Proteom. 2023, 285, 104955. [Google Scholar] [CrossRef]
- Shenoy, A.; Belugali Nataraj, N.; Perry, G.; Loayza Puch, F.; Nagel, R.; Marin, I.; Balint, N.; Bossel, N.; Pavlovsky, A.; Barshack, I.; et al. Proteomic patterns associated with response to breast cancer neoadjuvant treatment. Mol. Syst. Biol. 2020, 16, e9443. [Google Scholar] [CrossRef]
- Thangudu, R.R.; Rudnick, P.A.; Holck, M.; Singhal, D.; MacCoss, M.J.; Edwards, N.J.; Ketchum, K.A.; Kinsinger, C.R.; Kim, E.; Basu, A. Abstract LB-242: Proteomic Data Commons: A resource for proteogenomic analysis. Cancer Res. 2020, 80, LB-242. [Google Scholar] [CrossRef]
- Anurag, M.; Jaehnig, E.J.; Krug, K.; Lei, J.T.; Bergstrom, E.J.; Kim, B.J.; Vashist, T.D.; Huynh, A.M.T.; Dou, Y.; Gou, X.; et al. Proteogenomic markers of chemotherapy resistance and response in triple-negative breast cancer. Cancer Discov. 2022, 12, 2586–2605. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 2020, 183, 1436–1456. [Google Scholar] [CrossRef] [PubMed]
- Thangudu, R.R.; Holck, M.; Singhal, D.; Pilozzi, A.; Edwards, N.; Rudnick, P.A.; Domagalski, M.J.; Chilappagari, P.; Ma, L.; Xin, Y.; et al. NCI’s Proteomic Data Commons: A Cloud-Based Proteomics Repository Empowering Comprehensive Cancer Analysis through Cross-Referencing with Genomic and Imaging Data. Cancer Res. Commun. 2024, 4, 2480–2488. [Google Scholar] [CrossRef] [PubMed]
- Edwards, N.J.; Oberti, M.; Thangudu, R.R.; Cai, S.; McGarvey, P.B.; Jacob, S.; Madhavan, S.; Ketchum, K.A. The CPTAC data portal: A resource for cancer proteomics research. J. Proteome Res. 2015, 14, 2707–2713. [Google Scholar] [CrossRef]
- Fix, E. Discriminatory Analysis: Nonparametric Discrimination, Consistency Properties; USAF School of Aviation Medicine: Wright-Patterson AFB, OH, USA, 1985; Volume 1. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Narasimhan, B.; Chu, G. Impute: Imputation for Microarray Data, R package version 1.80.0; Bioconductor: Addis Ababa, Ethiopia, 2024. [Google Scholar]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June–18 July 1967; University of California: Berkeley, CA, USA, 1967; pp. 281–297. [Google Scholar]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C (Applied Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Thorndike, R.L. Who belongs in the family? Psychometrika 1953, 18, 267–276. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G. Cluster validation by prediction strength. J. Comput. Graph. Stat. 2005, 14, 511–528. [Google Scholar] [CrossRef]
- Hennig, C. Cluster-wise assessment of cluster stability. Comput. Stat. Data Anal. 2007, 52, 258–271. [Google Scholar] [CrossRef]
- Hennig, C. Dissolution point and isolation robustness: Robustness criteria for general cluster analysis methods. J. Multivar. Anal. 2008, 99, 1154–1176. [Google Scholar] [CrossRef]
- Hennig, C. fpc: Flexible Procedures for Clustering, R package version 2.2-13; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The gene ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Suarez-Diez, M.; Saccenti, E. Effects of sample size and dimensionality on the performance of four algorithms for inference of association networks in metabonomics. J. Proteome Res. 2015, 14, 5119–5130. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Wilson, D.J. The harmonic mean p-value for combining dependent tests. Proc. Natl. Acad. Sci. USA 2019, 116, 1195–1200. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Los Angeles, CA, USA, 27 December 1965–7 January 1996; pp. 226–231. [Google Scholar]
- R Core Team R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2010.
- Studio, R. Integrated Development Environment for R; R Studio Inc: Boston, MA, USA, 2018. [Google Scholar]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumor purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling tumor infiltrating immune cells with CIBERSORT. In Cancer Systems Biology: Methods and Protocols; Springer: New York, NY, USA, 2018; pp. 243–259. [Google Scholar]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.H.O.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The immune landscape of cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Q.; Stingl, C.; Look, M.P.; Smid, M.; Braakman, R.B.; De Marchi, T.; Sieuwerts, A.M.; Span, P.N.; Sweep, F.C.; Linderholm, B.K.; et al. Comparative proteome analysis revealing an 11-protein signature for aggressive triple-negative breast cancer. J. Natl. Cancer Inst. 2014, 106, djt376. [Google Scholar] [CrossRef]
- Lawrence, R.T.; Perez, E.M.; Hernández, D.; Miller, C.P.; Haas, K.M.; Irie, H.Y.; Lee, S.I.; Blau, C.A.; Villén, J. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015, 11, 630–644. [Google Scholar] [CrossRef]
- Gromova, I.; Espinoza, J.A.; Grauslund, M.; Santoni-Rugiu, E.; Møller Talman, M.L.; van Oostrum, J.; Moreira, J.M. Functional proteomic profiling of triple-negative breast cancer. Cells 2021, 10, 2768. [Google Scholar] [CrossRef]
- Gong, T.Q.; Jiang, Y.Z.; Shao, C.; Peng, W.T.; Liu, M.W.; Li, D.Q.; Zhang, B.Y.; Du, P.; Huang, Y.; Li, F.F.; et al. Proteome-centric cross-omics characterization and integrated network analyses of triple-negative breast cancer. Cell Rep. 2022, 38, 110460. [Google Scholar] [CrossRef]
- Izdebska, M.; Zielińska, W.; Hałas-Wiśniewska, M.; Grzanka, A. Involvement of actin and actin-binding proteins in carcinogenesis. Cells 2020, 9, 2245. [Google Scholar] [CrossRef]
- Hao, R.; Liu, Y.; Du, Q.; Liu, L.; Chen, S.; You, H.; Dong, Y. Transgelin-2 expression in breast cancer and its relationships with clinicopathological features and patient outcome. Breast Cancer 2019, 26, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Jimenez Jimenez, A.M.; Haddad, Y.; Jemelikova, V.; Adam, V.; Merlos Rodrigo, M.A. Multifaceted role of transgelin isoforms in cancer hallmarks. Carcinogenesis 2025, 46, bgaf014. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodeling in tumor progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zheng, X.; Zheng, Y.; Chen, Y.; Fei, W.; Wang, F.; Zheng, C. Extracellular matrix: Emerging roles and potential therapeutic targets for breast cancer. Front. Oncol. 2021, 11, 650453. [Google Scholar] [CrossRef]
- Insua-Rodríguez, J.; Oskarsson, T. The extracellular matrix in breast cancer. Adv. Drug Deliv. Rev. 2016, 97, 41–55. [Google Scholar] [CrossRef]
- Jena, M.K.; Janjanam, J. Role of extracellular matrix in breast cancer development: A brief update. F1000Research 2018, 7, 274. [Google Scholar] [CrossRef]
- Manou, D.; Caon, I.; Bouris, P.; Triantaphyllidou, I.E.; Giaroni, C.; Passi, A.; Karamanos, N.K.; Vigetti, D.; Theocharis, A.D. The complex interplay between extracellular matrix and cells in tissues. Methods Mol. Biol. 2019, 1952, 1–20. [Google Scholar] [CrossRef]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef]
- Ren, J.; Smid, M.; Iaria, J.; Salvatori, D.C.; van Dam, H.; Zhu, H.J.; Martens, J.W.; Ten Dijke, P. Cancer-associated fibroblast-derived Gremlin 1 promotes breast cancer progression. Breast Cancer Res. 2019, 21, 109. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Patel, A.A.; Steitz, J.A. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003, 4, 960–970. [Google Scholar] [CrossRef]
- Hegele, A.; Kamburov, A.; Grossmann, A.; Sourlis, C.; Wowro, S.; Weimann, M.; Will, C.L.; Pena, V.; Lührmann, R.; Stelzl, U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 2012, 45, 567–580. [Google Scholar] [CrossRef]
- Anczuków, O.; Krainer, A.R. The spliceosome, a potential Achilles heel of MYC-driven tumors. Genome Med. 2015, 7, 107. [Google Scholar] [CrossRef]
- Elghobashi-Meinhardt, N. ATP hydrolysis captured in atomic detail. Nat. Chem. 2024, 16, 306–307. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation. Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Arismendi-Morillo, G.; Mukherjee, P.; Chinopoulos, C. On the origin of ATP synthesis in cancer. Iscience 2020, 23, 101761. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Srinivasan, S.; Raman, P.; Jiang, Y.; Kaufman, B.A.; Taylor, D.; Dong, D.; Chakrabarti, R.; Picard, M.; Carstens, R.P.; et al. Aggressive triple negative breast cancers have unique molecular signature on the basis of mitochondrial genetic and functional defects. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1864, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Pelicano, H.; Zhang, W.; Liu, J.; Hammoudi, N.; Dai, J.; Xu, R.H.; Pusztai, L.; Huang, P. Mitochondrial dysfunction in some triple-negative breast cancer cell lines: Role of mTOR pathway and therapeutic potential. Breast Cancer Res. 2014, 16, 434. [Google Scholar] [CrossRef]
- Van Drie, J.H. Protein folding, protein homeostasis, and cancer. Chin. J. Cancer 2011, 30, 124–137. [Google Scholar] [CrossRef]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef]
- Langer, T.; Neupert, W. Heat shock proteins hsp60 and hsp70: Their roles in folding, assembly and membrane translocation of proteins. Curr. Top. Microbiol. Immunol. 1990, 167, 3–30. [Google Scholar]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Brackley, K.I.; Grantham, J. Activities of the chaperonin containing TCP-1 (CCT): Implications for cell cycle progression and cytoskeletal organisation. Cell Stress Chaperones 2009, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ong, M.S.; Deng, S.; Halim, C.E.; Cai, W.; Tan, T.Z.; Huang, R.Y.J.; Sethi, G.; Hooi, S.C.; Kumar, A.P.; Yap, C.T. Cytoskeletal proteins in cancer and intracellular stress: A therapeutic perspective. Cancers 2020, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, A.; Subramani, R.; Lakshmanaswamy, R. Involvement of actin cytoskeletal modifications in the inhibition of triple-negative breast cancer growth and metastasis by nimbolide. Mol.-Ther.-Oncolytics 2021, 20, 596–606. [Google Scholar] [CrossRef]
- Morris, G.E. The cajal body. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2008, 1783, 2108–2115. [Google Scholar] [CrossRef]
- Hsu, T.Y.T.; Simon, L.M.; Neill, N.J.; Marcotte, R.; Sayad, A.; Bland, C.S.; Echeverria, G.V.; Sun, T.; Kurley, S.J.; Tyagi, S.; et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015, 525, 384–388. [Google Scholar] [CrossRef]
- Yang, H.; Beutler, B.; Zhang, D. Emerging roles of spliceosome in cancer and immunity. Protein Cell 2022, 13, 559–579. [Google Scholar] [CrossRef]
- Niño, C.A.; Scotto di Perrotolo, R.; Polo, S. Recurrent spliceosome mutations in cancer: Mechanisms and consequences of aberrant splice site selection. Cancers 2022, 14, 281. [Google Scholar] [CrossRef]
- Ivanova, O.M.; Anufrieva, K.S.; Kazakova, A.N.; Malyants, I.K.; Shnaider, P.V.; Lukina, M.M.; Shender, V.O. Non-canonical functions of spliceosome components in cancer progression. Cell Death Dis. 2023, 14, 77. [Google Scholar] [CrossRef]
- El Marabti, E.; Younis, I. The cancer spliceome: Reprograming of alternative splicing in cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Gahete, M.D.; Herman-Sanchez, N.; Fuentes-Fayos, A.C.; Lopez-Canovas, J.L.; Luque, R.M. Dysregulation of splicing variants and spliceosome components in breast cancer. Endocr.-Relat. Cancer 2022, 29, R123–R142. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Kong, X.; Yang, X.; Cheng, R.; Zhang, W.; Gao, P.; Chen, L.; Wang, Z.; Fang, Y.; et al. Prognostic alternative mRNA splicing signature and a novel biomarker in triple-negative breast cancer. DNA Cell Biol. 2020, 39, 1051–1063. [Google Scholar] [CrossRef]
- Ezkurdia, I.; Juan, D.; Rodriguez, J.M.; Frankish, A.; Diekhans, M.; Harrow, J.; Vazquez, J.; Valencia, A.; Tress, M.L. Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum. Mol. Genet. 2014, 23, 5866–5878. [Google Scholar] [CrossRef] [PubMed]
- Blijlevens, M.; van der Meulen-Muileman, I.H.; de Menezes, R.X.; Smit, E.F.; van Beusechem, V.W. High-throughput RNAi screening reveals cancer-selective lethal targets in the RNA spliceosome. Oncogene 2019, 38, 4142–4153. [Google Scholar] [CrossRef]
- Mabonga, L.; Kappo, A.P. The oncogenic potential of small nuclear ribonucleoprotein polypeptide G: A comprehensive and perspective view. Am. J. Transl. Res. 2019, 11, 6702. [Google Scholar]
- Prusty, A.B.; Meduri, R.; Prusty, B.K.; Vanselow, J.; Schlosser, A.; Fischer, U. Impaired spliceosomal UsnRNP assembly leads to Sm mRNA down-regulation and Sm protein degradation. J. Cell Biol. 2017, 216, 2391–2407. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.X.; Yang, W.X.; Pei, Y.C.; Luo, H.; Li, X.G.; Wang, Y.J.; Zhang, G.L.; Ling, H.; Shao, Z.M.; Hu, X. An in vivo CRISPR screen identifies that SNRPC promotes triple-negative breast cancer progression. Cancer Res. 2023, 83, 2000–2015. [Google Scholar] [CrossRef]
- Dai, X.; Cai, L.; Zhang, Z.; Li, J. SNRPD1 conveys prognostic value on breast cancer survival and is required for anthracycline sensitivity. BMC Cancer 2023, 23, 376. [Google Scholar] [CrossRef]
- Yu, S.; Si, Y.; Yu, J.; Jiang, C.; Cheng, F.; Xu, M.; Fan, Z.; Liu, F.; Liu, C.; Wang, Y.; et al. SNRPB2 promotes triple-negative breast cancer progression by controlling alternative splicing of MDM4 pre-mRNA. Cancer Sci. 2024, 115, 3915–3927. [Google Scholar] [CrossRef]
- Yang, W.; Hong, L.; Guo, L.; Wang, Y.; Han, X.; Han, B.; Xing, Z.; Zhang, G.; Zhou, H.; Chen, C.; et al. Targeting SNRNP200-induced splicing dysregulation offers an immunotherapy opportunity for glycolytic triple-negative breast cancer. Cell Discov. 2024, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, J.X.; Wu, H.T.; Li, X.L.; Wen, X.F.; Du, C.W.; Zhang, G.J. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov. Med. 2018, 25, 211–223. [Google Scholar] [PubMed]
- Li, X.; Jin, Y.; Xue, J. Unveiling Collagen’s Role in Breast Cancer: Insights into Expression Patterns, Functions and Clinical Implications. Int. J. Gen. Med. 2024, 17, 1773–1787. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, H.; Luo, Y.; Jiao, Y.; Wu, J.; Dong, S.; Du, D. An integrated analysis of bulk and single-cell sequencing data reveals that EMP1+/COL3A1+ fibroblasts contribute to the bone metastasis process in breast, prostate, and renal cancers. Front. Immunol. 2023, 14, 1313536. [Google Scholar] [CrossRef]
- Luo, Q.; Li, J.; Su, X.; Tan, Q.; Zhou, F.; Xie, S. COL11A1 serves as a biomarker for poor prognosis and correlates with immune infiltration in breast cancer. Front. Genet. 2022, 13, 935860. [Google Scholar] [CrossRef]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.J. Collagen type XI alpha 1 (COL11A1): A novel biomarker and a key player in cancer. Cancers 2021, 13, 935. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Li, Z.; Zhang, T.; Liu, W.; Liu, Z.; Yuan, Y.C.; Su, F.; Xu, L.; Wang, Y.; et al. YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC cancer 2015, 15, 50. [Google Scholar] [CrossRef]
- Agarwal, N.; Theodorescu, D. The role of transcription factor YY1 in the biology of cancer. Crit. Rev. Oncog. 2017, 22, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Huang, W.; Kute, T.E.; Miller, L.D.; Zhang, Q.; Hatcher, H.; Wang, J.; Stovall, D.B.; Russell, G.B.; Cao, P.D.; et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012, 180, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lahusen, T.; Wang, R.; Xiao, C.; Xu, X.; Hwang, Y.; He, W.; Shi, Y.; Deng, C. Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene 2012, 31, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Ishii, H.; Hulett, M.D.; Li, J.M.; Santiago, F.S.; Parish, C.R.; Khachigian, L.M. Yin Yang-1 inhibits tumor cell growth and inhibits p21WAF1/Cip1 complex formation with cdk4 and cyclin D1. Int. J. Oncol. 2012, 40, 1575–1580. [Google Scholar]
- Vidal, M.; Starowicz, K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp. Hematol. 2017, 48, 12–31. [Google Scholar] [CrossRef]
- Xu, J.; Wang, R.; Han, Y. RING1 and YY1 binding protein inhibits breast cancer progression by regulating cell cycle and apoptosis-related genes. Oncotarget 2018, 9, 10134–10147. [Google Scholar]
- Miretti, S.; Allavena, G.; Mazzucchelli, G. RING1 and YY1 binding protein interacts with p53 and modulates its activity in breast cancer cells. Mol. Cancer Res. 2009, 7, 552–564. [Google Scholar]
- García, E.; Marcos-Gutiérrez, C.; del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef]
- Liu, J.; Fan, H.; Liang, X.; Chen, Y. Polycomb repressor complex: Its function in human cancer and therapeutic target strategy. Biomed. Pharmacother. 2023, 169, 115897. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Björk, L.; Mulder, J.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Spiess, C.; Meyer, A.; Reissmann, S.; Frydman, J. Diversity of the chaperonin-assisted protein folding system. Nat. Rev. Mol. Cell Biol. 2004, 5, 199–210. [Google Scholar]
- Guest, S.; Kratche, Z.; Bollig-Fischer, A.; Haddad, R.; Ethier, S. The chaperonin CCT2 promotes breast cancer growth by regulating the cytoskeleton and cell proliferation. Oncogene 2015, 34, 2303–2311. [Google Scholar]
- Amit, M.; Alcalay, Y.; Meir, K.; Pasmanik-Chor, M.; Liran, O.; Horowitz, S.; Schneider, V.; Berman, B.; Rivlin, N.; Rotter, V. Gene expression of CCT subunits and its association with tumor progression in breast cancer. PLoS ONE 2013, 8, e64210. [Google Scholar]
- Gundersen, G.G.; Cook, T.A. Microtubules and signal transduction. Curr. Opin. Cell Biol. 1999, 11, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cui, H.; Yang, X.; Peng, L. TUBA1C: A new potential target of LncRNA EGFR-AS1 promotes gastric cancer progression. BMC Cancer 2023, 23, 258. [Google Scholar] [CrossRef]
- Swanton, C.; Nicke, B.; Schuett, M.; Eklund, A.C.; Ng, C.; Li, Q.; Hardcastle, T.; Lee, A.; Roy, R.; East, P.; et al. Chromosomal instability determines taxane response. Proc. Natl. Acad. Sci. USA 2009, 106, 8671–8676. [Google Scholar] [CrossRef]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef]
- Kim, S.; You, S.; Hwang, D. Aminoacyl-tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. USA 2014, 111, E1909–E1917. [Google Scholar]
- Liu, X.; Zhang, G.; Yu, T.; He, J.; Liu, J.; Chai, X.; Zhao, G.; Yin, D.; Zhang, C. Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis. Cancer Lett. 2022, 543, 215781. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Nie, A.; Yu, D.; Bian, M. Roles of aminoacyl-tRNA synthetases in cancer. Front. Cell Dev. Biol. 2020, 8, 599765. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Pan, C.; Li, Y.; Lin, L.; Jin, Y.; Zheng, J.; Yu, Z. A comprehensive study based on exosome-related immunosuppression genes and tumor microenvironment in hepatocellular carcinoma. BMC Cancer 2022, 22, 1344. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Suh, J.H.; Park, M.C.; Goughnour, P.C.; Min, B.S.; Kim, S.B.; Lee, W.Y.; Cho, Y.B.; Cheon, J.H.; Lee, K.Y.; Nam, D.H.; et al. Plasma lysyl-tRNA synthetase 1 (KARS1) as a novel diagnostic and monitoring biomarker for colorectal cancer. J. Clin. Med. 2020, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.G.; Choi, J.W.; Lee, J.Y.; Kim, H.; Oh, Y.S.; Lee, J.W.; Tak, Y.K.; Song, J.M.; Razin, E.; Yun, S.H.; et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 2012, 26, 4142–4159. [Google Scholar] [CrossRef]
- Lee, S.; Kwon, N.H.; Seo, B.; Lee, J.Y.; Cho, H.Y.; Kim, K.; Kim, H.S.; Jung, K.; Jeon, Y.H.; Kim, S.; et al. Discovery of novel potent migrastatic Thiazolo [5, 4-b] pyridines targeting Lysyl-tRNA synthetase (KRS) for treatment of Cancer metastasis. Eur. J. Med. Chem. 2021, 218, 113405. [Google Scholar] [CrossRef]

| Metadata | Cluster 1D | Cluster 2D | Pval | FDR |
|---|---|---|---|---|
| Average Tumor Content (%) for biopsy | 68.125 | 63.438 | 0.252 | 0.378 |
| Chromosomal instability | 6.637 | 4.058 | 0.003 | 0.080 |
| Mutation load HG38 v2 | 668.071 | 150.438 | 0.151 | 0.280 |
| Microsatellite instability score | 195.357 | 24.188 | 0.013 | 0.083 |
| Signature 3 | 0.171 | 0.152 | 0.712 | 0.777 |
| Signature 6 | 0.016 | 0.045 | 0.236 | 0.378 |
| Signature 15 | 0.035 | 0.086 | 0.324 | 0.435 |
| Signature 10 | 0.000 | 0.006 | 0.385 | 0.486 |
| Signature 12 | 0.044 | 0.009 | 0.132 | 0.280 |
| Signature 4 | 0.063 | 0.000 | 0.011 | 0.083 |
| Signature 7 | 0.005 | 0.004 | 0.923 | 0.923 |
| Signature 9 | 0.009 | 0.018 | 0.549 | 0.628 |
| Signature 13 | 0.015 | 0.029 | 0.326 | 0.435 |
| Signature 21 | 0.048 | 0.005 | 0.923 | 0.923 |
| Stimulatory immune modulator proteins | −0.448 | −0.092 | 0.077 | 0.185 |
| Inhibitory immune modulator proteins | −0.310 | 0.002 | 0.146 | 0.280 |
| HLA immune modulator proteins | −0.821 | −0.669 | 0.506 | 0.607 |
| ESTIMATE ImmuneScore | 1388.4 | 2200.3 | 0.038 | 0.102 |
| ESTIMATE StromalScore | 238.5 | 740.7 | 0.025 | 0.083 |
| ESTIMATE TumorPurity | 0.654 | 0.497 | 0.022 | 0.083 |
| Cibersort absolute immune score | 1.769 | 2.724 | 0.017 | 0.083 |
| xCell ImmuneScore | 0.099 | 0.277 | 0.019 | 0.083 |
| xCell StromaScore | 0.034 | 0.046 | 0.228 | 0.378 |
| xCell MicroenvironmentScore | 0.133 | 0.323 | 0.028 | 0.083 |
| Metadata | Cluster 1V | Cluster 2V | p-Value | FDR |
|---|---|---|---|---|
| Chromosome Instability Index | 2.765 | 2.370 | 0.664 | 0.800 |
| CIBERSORT AbsoluteScore | 1.040 | 0.889 | 0.738 | 0.800 |
| ESTIMATE ImmuneScore | 1574.727 | 1506.019 | 0.881 | 0.881 |
| ESTIMATE StromalScore | 184.157 | −433.674 | 0.045 | 0.584 |
| ESTIMATE TumorPurity | 0.637 | 0.709 | 0.220 | 0.714 |
| Number of non-synonymous mutations | 106.250 | 115.500 | 0.399 | 0.800 |
| Stemness Score | 0.704 | 0.796 | 0.192 | 0.714 |
| xCell ImmuneScore | 0.094 | 0.080 | 0.734 | 0.800 |
| xCell StromalScore | 0.004 | 0.001 | 0.300 | 0.780 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furlan, C.; Suarez-Diez, M.; Saccenti, E. A Validated Proteomic Signature of Basal-like Triple-Negative Breast Cancer Subtypes Obtained from Publicly Available Data. Cancers 2025, 17, 2601. https://doi.org/10.3390/cancers17162601
Furlan C, Suarez-Diez M, Saccenti E. A Validated Proteomic Signature of Basal-like Triple-Negative Breast Cancer Subtypes Obtained from Publicly Available Data. Cancers. 2025; 17(16):2601. https://doi.org/10.3390/cancers17162601
Chicago/Turabian StyleFurlan, Cristina, Maria Suarez-Diez, and Edoardo Saccenti. 2025. "A Validated Proteomic Signature of Basal-like Triple-Negative Breast Cancer Subtypes Obtained from Publicly Available Data" Cancers 17, no. 16: 2601. https://doi.org/10.3390/cancers17162601
APA StyleFurlan, C., Suarez-Diez, M., & Saccenti, E. (2025). A Validated Proteomic Signature of Basal-like Triple-Negative Breast Cancer Subtypes Obtained from Publicly Available Data. Cancers, 17(16), 2601. https://doi.org/10.3390/cancers17162601






