Cholinergic Receptor Nicotinic Beta 2 Subunit Promotes the Peritoneal Disseminating Metastasis of Colorectal Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Cell Lines Collection
2.3. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.4. shRNA Mediated CHRNB2 Knock-Down in CRC Cell Lines
2.5. Generation of Anti-CHRNB2 Antibodies
2.6. Assays of Cell Proliferation, Migration, and Invasion
2.7. Mouse Subcutaneous Xenograft Model
2.8. Mouse Peritoneal Dissemination Model
2.9. Analysis of Clinical Samples
2.10. Statistical Analysis
3. Results
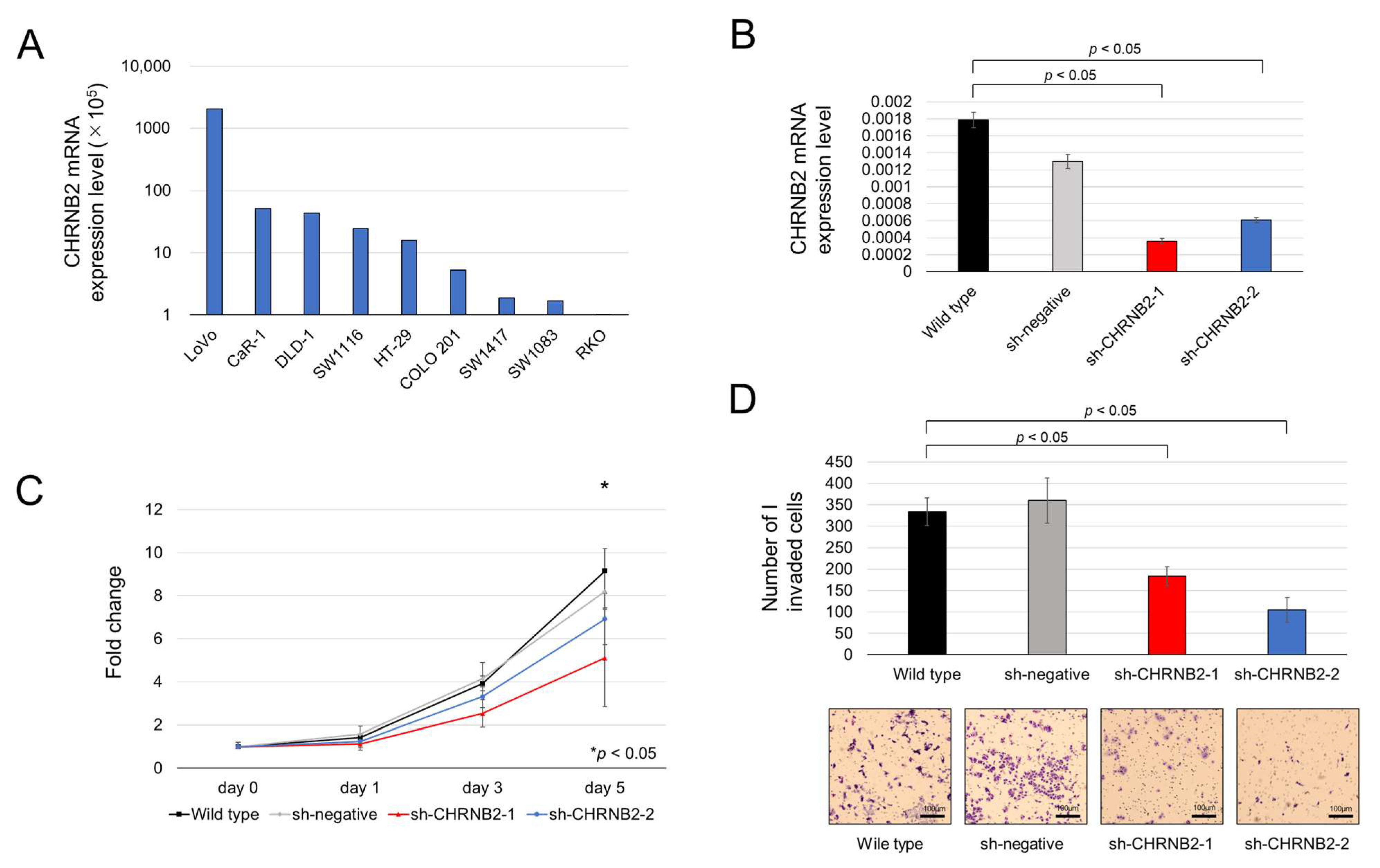
3.1. CHRNB2 mRNA Expression in CRC Cell Lines and CHRNB2 KD Efficiency
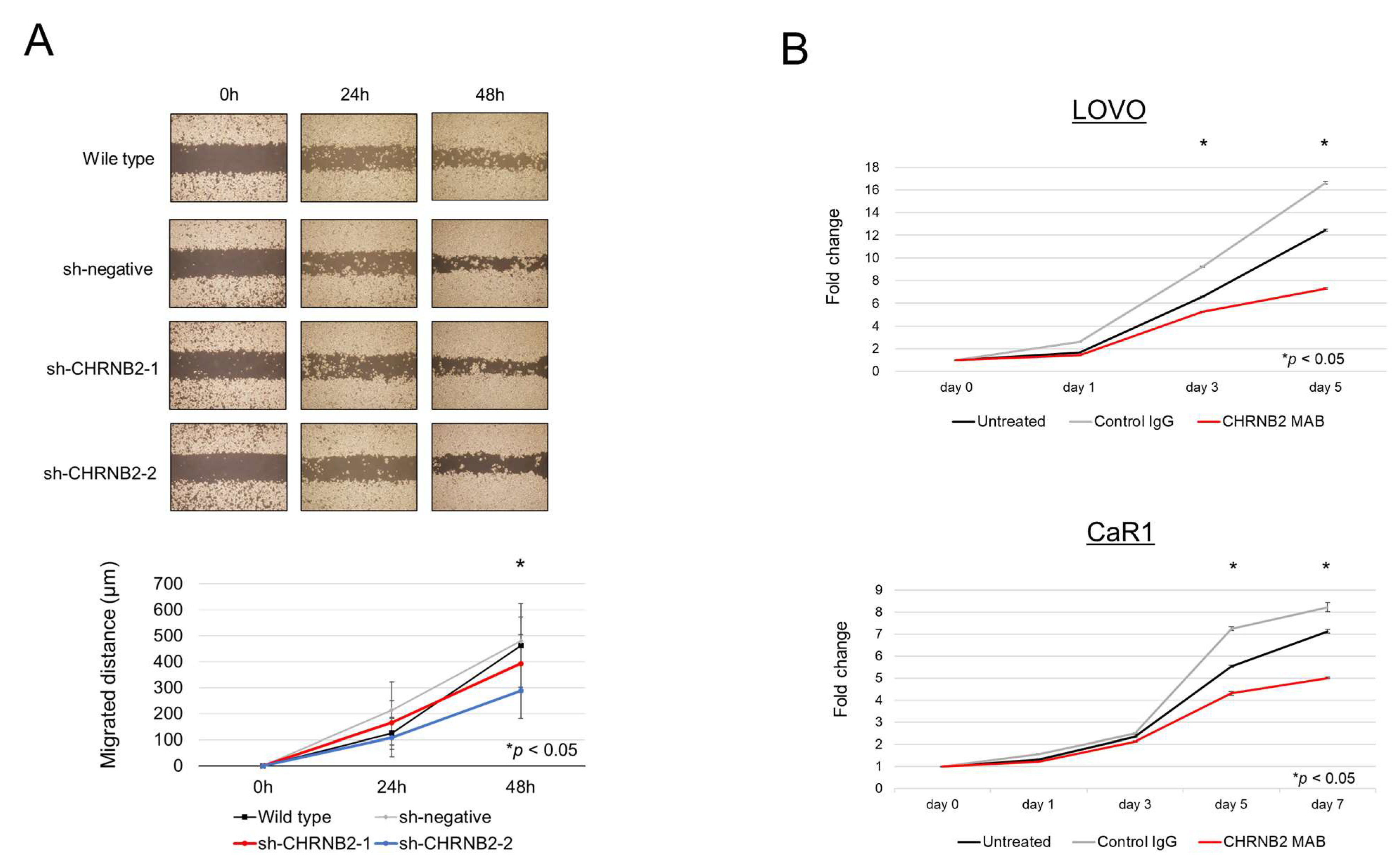
3.2. Cell Function Analysis
3.3. Effect of the CHRNB2 mAb
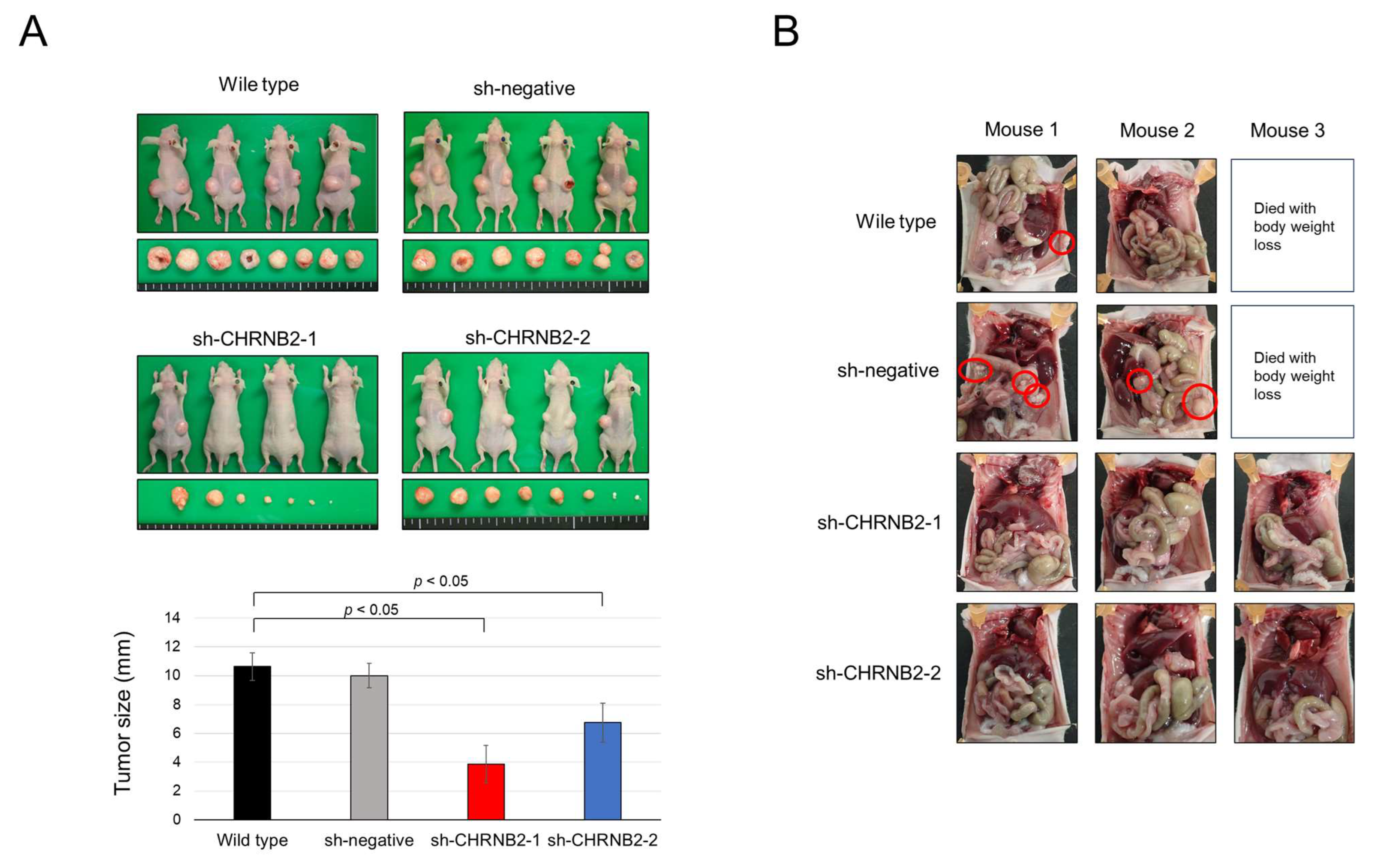
3.4. Mouse Subcutaneous Tumor Model
3.5. Mouse Peritoneal Dissemination Model
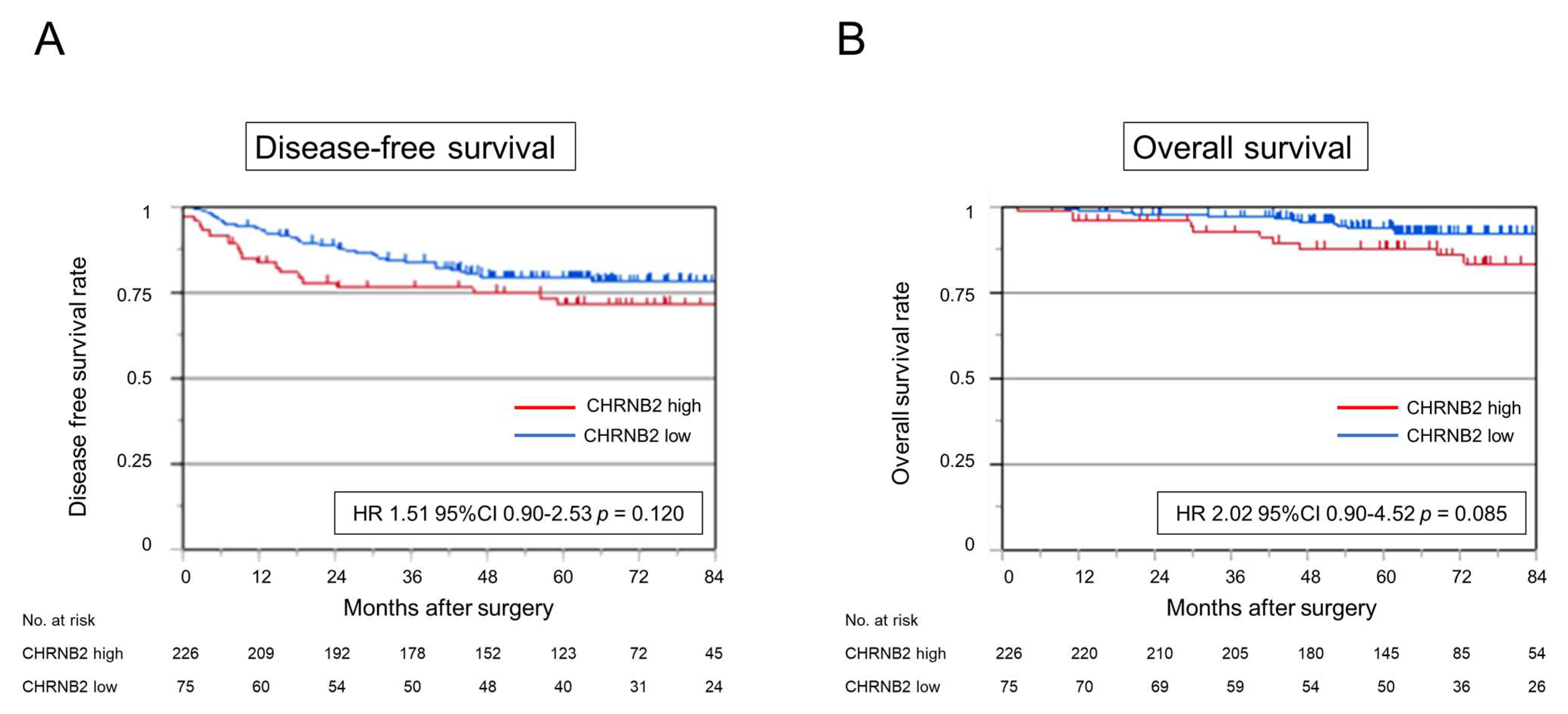
3.6. CHRNB2 mRNA Expression Analysis of Clinical Samples
3.7. Kaplan–Meier Analysis
3.8. Risk Factor for Peritoneal Recurrence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| CHRNB2 | Cholinergic receptor nicotinic beta 2 subunit |
| HIPEC | Hyperthermic intraperitoneal chemotherapy |
| CRS | Cytoreductive surgery |
| mAb | Monoclonal antibody |
| CA | Carbohydrate antigen |
| KD | Knockdown |
| qRT-PCR | Quantitative reverse-transcription polymerase chain reaction |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| DFS | Disease-free survival |
| OS | Overall survival |
References
- Schrag, D.; Shi, Q.; Weiser, M.R.; Gollub, M.J.; Saltz, L.B.; Musher, B.L.; Goldberg, J.; Al Baghdadi, T.; Goodman, K.A.; McWilliams, R.R.; et al. Preoperative Treatment of Locally Advanced Rectal Cancer. N. Engl. J. Med. 2023, 389, 322–334. [Google Scholar] [CrossRef]
- Haller, D.G.; Tabernero, J.; Maroun, J.; de Braud, F.; Price, T.; Van Cutsem, E.; Hill, M.; Gilberg, F.; Rittweger, K.; Schmoll, H.J. Capecitabine plus oxaliplatin compared with fluorouracil and folinic acid as adjuvant therapy for stage III colon cancer. J. Clin. Oncol. 2011, 29, 1465–1471. [Google Scholar] [CrossRef]
- Nakayama, G.; Takano, N.; Taniguchi, H.; Ishigure, K.; Yokoyama, H.; Teramoto, H.; Hashimoto, R.; Sakai, M.; Ishiyama, A.; Kinoshita, T.; et al. Randomised phase II trial of capecitabine plus oxaliplatin with continuous versus intermittent use of oxaliplatin as adjuvant chemotherapy for stage II/III colon cancer (CCOG-1302 study). Eur. J. Cancer. 2021, 144, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Swedish Rectal Cancer Trial; Cedermark, B.; Dahlberg, M.; Glimelius, B.; Påhlman, L.; Rutqvist, L.E.; Wilking, N. Improved survival with preoperative radiotherapy in resectable rectal cancer. N. Engl. J. Med. 1997, 336, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, E.K.; Vauthey, J.N.; Ellis, L.M.; Ellis, V.; Pollock, R.; Broglio, K.R.; Hess, K.; Curley, S.A. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann. Surg. 2004, 239, 818–825. [Google Scholar] [CrossRef]
- Watanabe, K.; Nagai, K.; Kobayashi, A.; Sugito, M.; Saito, N. Factors influencing survival after complete resection of pulmonary metastases from colorectal cancer. Br. J. Surg. 2009, 96, 1058–1065. [Google Scholar] [CrossRef]
- Sato, H.; Maeda, K.; Kotake, K.; Sugihara, K.; Takahashi, H. Factors affecting recurrence and prognosis after R0 resection for colorectal cancer with peritoneal metastasis. J. Gastroenterol. 2016, 51, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Noda, K.; Tominaga, T.; Nonaka, T.; Ishii, M.; Fukuoka, H.; Hisanaga, M.; Takeshita, H.; To, K.; Tanaka, K.; Sawai, T.; et al. Effect of adjuvant chemotherapy after curative resection of colorectal cancer peritoneal metastasis. Int. J. Color. Dis. 2023, 38, 101. [Google Scholar] [CrossRef]
- Goéré, D.; Malka, D.; Tzanis, D.; Gava, V.; Boige, V.; Eveno, C.; Maggiori, L.; Dumont, F.; Ducreux, M.; Elias, D. Is there a possibility of a cure in patients with colorectal peritoneal carcinomatosis amenable to complete cytoreductive surgery and intraperitoneal chemotherapy? Ann. Surg. 2013, 257, 1065–1071. [Google Scholar] [CrossRef]
- Quénet, F.; Elias, D.; Roca, L.; Goéré, D.; Ghouti, L.; Pocard, M.; Facy, O.; Arvieux, C.; Lorimier, G.; Pezet, D.; et al. Cytoreductive surgery plus hyperthermic intraperitoneal chemotherapy versus cytoreductive surgery alone for colorectal peritoneal metastases (PRODIGE 7): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 256–266. [Google Scholar] [CrossRef]
- Ramsay, R.G.; Flood, M. New insights into the unique nature of colorectal cancer peritoneal metastases-rethinking HIPEC. Br. J. Cancer 2022, 127, 377–378. [Google Scholar] [CrossRef]
- Li, J.; Wang, A.R.; Chen, X.D.; Zhang, Y.X.; Pan, H.; Li, S.Q. Effect of hyperthermic intraperitoneal chemotherapy in combination with cytoreductive surgery on the prognosis of patients with colorectal cancer peritoneal metastasis: A systematic review and meta-analysis. World J. Surg. Oncol. 2022, 20, 200. [Google Scholar] [CrossRef]
- Tonello, M.; Cenzi, C.; Pizzolato, E.; Fiscon, R.; Del Bianco, P.; Pilati, P.; Sommariva, A. Systemic Chemotherapy in Colorectal Peritoneal Metastases Treated with Cytoreductive Surgery: Systematic Review and Meta-Analysis. Cancers 2024, 16, 1182. [Google Scholar] [CrossRef]
- Patel, C.M.; Sahdev, A.; Reznek, R.H. CT, MRI and PET imaging in peritoneal malignancy. Cancer Imaging 2011, 11, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Shimizu, D.; Nakamura, S.; Sawaki, K.; Umeda, S.; Miwa, T.; Tanaka, H.; Inokawa, Y.; Hattori, N.; Hayashi, M.; et al. Blockade of CHRNB2 signaling with a therapeutic monoclonal antibody attenuates the aggressiveness of gastric cancer cells. Oncogene 2021, 40, 5495–5504. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, J.M. Nicotinic acetylcholine receptors of muscles and nerves: Comparison of their structures, functional roles, and vulnerability to pathology. Ann. N. Y. Acad. Sci. 2003, 998, 41–52. [Google Scholar] [CrossRef]
- Xu, T.; Luong, D.; Zhong, N. Familial Epilepsy Associated With Concurrent CHRNB2 Mutation and RBFOX1 Exon Deletion: A Case Report. Cureus 2023, 15, e35845. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L. Alzheimer’s disease genetics current status and future perspectives. Int. Rev. Neurobiol. 2009, 84, 167–184. [Google Scholar] [CrossRef]
- Ehringer, M.A.; Clegg, H.V.; Collins, A.C.; Corley, R.P.; Crowley, T.; Hewitt, J.K.; Hopfer, C.J.; Krauter, K.; Lessem, J.; Rhee, S.H.; et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144B, 596–604. [Google Scholar] [CrossRef]
- Perkins, K.A.; Lerman, C.; Mercincavage, M.; Fonte, C.A.; Briski, J.L. Nicotinic acetylcholine receptor beta2 subunit (CHRNB2) gene and short-term ability to quit smoking in response to nicotine patch. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2608–2612. [Google Scholar] [CrossRef]
- Qin, C.; Li, T.; Wang, Y.; Zhao, B.; Li, Z.; Li, T.; Yang, X.; Zhao, Y.; Wang, W. CHRNB2 represses pancreatic cancer migration and invasion via inhibiting beta-catenin pathway. Cancer Cell Int. 2022, 22, 340. [Google Scholar] [CrossRef]
- Umeda, S.; Kanda, M.; Miwa, T.; Tanaka, H.; Tanaka, C.; Kobayashi, D.; Hayashi, M.; Yamada, S.; Nakayama, G.; Koike, M.; et al. Fraser extracellular matrix complex subunit 1 promotes liver metastasis of gastric cancer. Int. J. Cancer 2020, 146, 2865–2876. [Google Scholar] [CrossRef] [PubMed]
- Henstra, C.; van Praagh, J.; Olinga, P.; Nagelkerke, A. The gastrointestinal microbiota in colorectal cancer cell migration and invasion. Clin. Exp. Metastasis 2021, 38, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, S.A.; Eide, P.W.; Nesbakken, A.; Guren, T.; Leithe, E.; Lothe, R.A. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim. Biophys. Acta 2015, 1855, 104–121. [Google Scholar] [CrossRef]
- Matly, A.; Quinn, J.A.; McMillan, D.C.; Park, J.H.; Edwards, J. The relationship between β-catenin and patient survival in colorectal cancer systematic review and meta-analysis. Crit. Rev. Oncol./Hematol. 2021, 163, 103337. [Google Scholar] [CrossRef] [PubMed]
- Sugarbaker, P.H.; Deng, T.; Chang, D. Peritoneal cytology as an indicator of peritoneal metastases in colorectal cancer. J. Surg. Oncol. 2021, 124, 361–366. [Google Scholar] [CrossRef]
- Baaten, I.C.P.A.; West, N.P.; Quyn, A.J.; Seymour, M.T.; Seligmann, J.F. Colorectal cancer peritoneal metastases: Biology, treatment and next steps. Eur. J. Surg. Oncol. 2020, 46, 675–683. [Google Scholar] [CrossRef]
- Van’t Erve, I.; Rovers, K.P.; Constantinides, A.; Bolhuis, K.; Wassenaar, E.C.; Lurvink, R.J.; Huysentruyt, C.J.; Snaebjornsson, P.; Boerma, D.; van den Broek, D.; et al. Detection of tumor-derived cell-free DNA from colorectal cancer peritoneal metastases in plasma and peritoneal fluid. J. Pathol. Clin. Res. 2021, 7, 203–208. [Google Scholar] [CrossRef]

| Univariable | Multivariable | |||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p | Hazard Ratio | 95% CI | p | |
| Age (≥65) | 0.92 | 0.33–2.55 | 0.883 | |||
| Gender (male) | 2.29 | 0.83–6.89 | 0.105 | |||
| Tumor location (right) | 2.19 | 0.82–5.84 | 0.116 | |||
| Tumor size (≥40 mm) | 4.03 | 1.15–14.2 | 0.029 * | 2.25 | 0.60–8.48 | 0.226 |
| Carcinoembryonic antigen (>5 ng/mL) | 3.44 | 1.25–9.49 | 0.016 * | 2.30 | 0.81–6.50 | 0.115 |
| Carbohydrate antigen 19-9 (>37 IU/mL) | 2.13 | 0.77–5.87 | 0.142 | |||
| Approach (open) | 5.01 | 1.87–13.3 | 0.001 * | 1.90 | 0.66–5.54 | 0.234 |
| Lymph node dissection (<D3) | 2.65 | 0.96–7.30 | 0.059 | |||
| Tumor depth (pT4) | 9.27 | 3.36–25.6 | <0.001 * | 5.69 | 1.95–16.6 | 0.001 * |
| Lymph node metastasis | 1.14 | 0.43–3.05 | 0.783 | |||
| Tumor differentiation (undifferentiated) | 0.96 | 0.13–7.29 | 0.970 | |||
| Lymphatic involvement (ly+) | 1.63 | 0.46–5.73 | 0.444 | |||
| Vascular invasion (v+) | 2.86 | 0.81–10.0 | 0.100 | |||
| Postoperative adjuvant chemotherapy | 1.21 | 0.43–3.32 | 0.713 | |||
| High CHRNB2 expression | 3.29 | 1.23–8.79 | 0.017 * | 2.83 | 1.03–16.6 | 0.041 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umeda, S.; Tanaka, K.; Kishida, T.; Hattori, N.; Tanaka, H.; Shimizu, D.; Takami, H.; Hayashi, M.; Tanaka, C.; Nakayama, G.; et al. Cholinergic Receptor Nicotinic Beta 2 Subunit Promotes the Peritoneal Disseminating Metastasis of Colorectal Cancer. Cancers 2025, 17, 2485. https://doi.org/10.3390/cancers17152485
Umeda S, Tanaka K, Kishida T, Hattori N, Tanaka H, Shimizu D, Takami H, Hayashi M, Tanaka C, Nakayama G, et al. Cholinergic Receptor Nicotinic Beta 2 Subunit Promotes the Peritoneal Disseminating Metastasis of Colorectal Cancer. Cancers. 2025; 17(15):2485. https://doi.org/10.3390/cancers17152485
Chicago/Turabian StyleUmeda, Shinichi, Kenshiro Tanaka, Takayoshi Kishida, Norifumi Hattori, Haruyoshi Tanaka, Dai Shimizu, Hideki Takami, Masamichi Hayashi, Chie Tanaka, Goro Nakayama, and et al. 2025. "Cholinergic Receptor Nicotinic Beta 2 Subunit Promotes the Peritoneal Disseminating Metastasis of Colorectal Cancer" Cancers 17, no. 15: 2485. https://doi.org/10.3390/cancers17152485
APA StyleUmeda, S., Tanaka, K., Kishida, T., Hattori, N., Tanaka, H., Shimizu, D., Takami, H., Hayashi, M., Tanaka, C., Nakayama, G., & Kanda, M. (2025). Cholinergic Receptor Nicotinic Beta 2 Subunit Promotes the Peritoneal Disseminating Metastasis of Colorectal Cancer. Cancers, 17(15), 2485. https://doi.org/10.3390/cancers17152485






