MicroRNA Signatures in Lung Adenocarcinoma Metastases: Exploring the Oncogenic Targets of Tumor-Suppressive miR-195-5p and miR-195-3p
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. LUAD Patients and LUAD Cell Lines
2.2. Construction of an miRNA Expression Signature for LUAD Based on RNA Sequencing
2.3. Identification of Oncogenic Targets of miR-195-5p and miR-195-3p in LUAD Cells
2.4. Analysis of miRNA and Gene Expression and Their Clinical Significance in Patients with LUAD Using an In Silico Database
2.5. Functional Assays of miRNAs and miRNA Target Genes in LUAD Cells
2.6. Dual-Luciferase Reporter Assay
2.7. Western Blotting
2.8. Statistical Analysis
3. Results
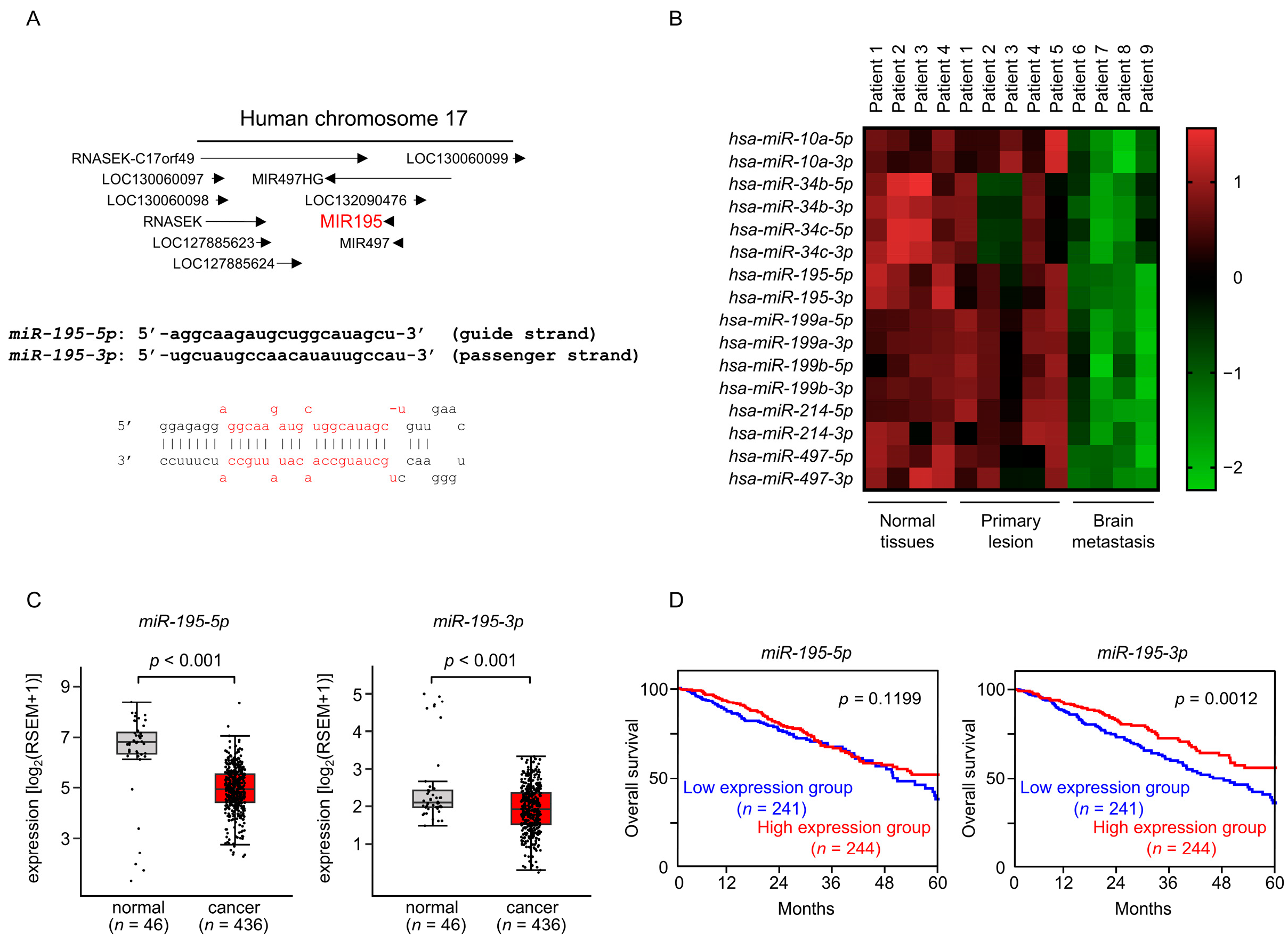
3.1. The RNA-Sequencing-Based miRNA Expression Signature Using Clinical Specimens Obtained from LUAD Brain Metastases
3.2. Expression and Clinical Significance of miR-195-5p and miR-195-3p in LUAD Clinical Specimens
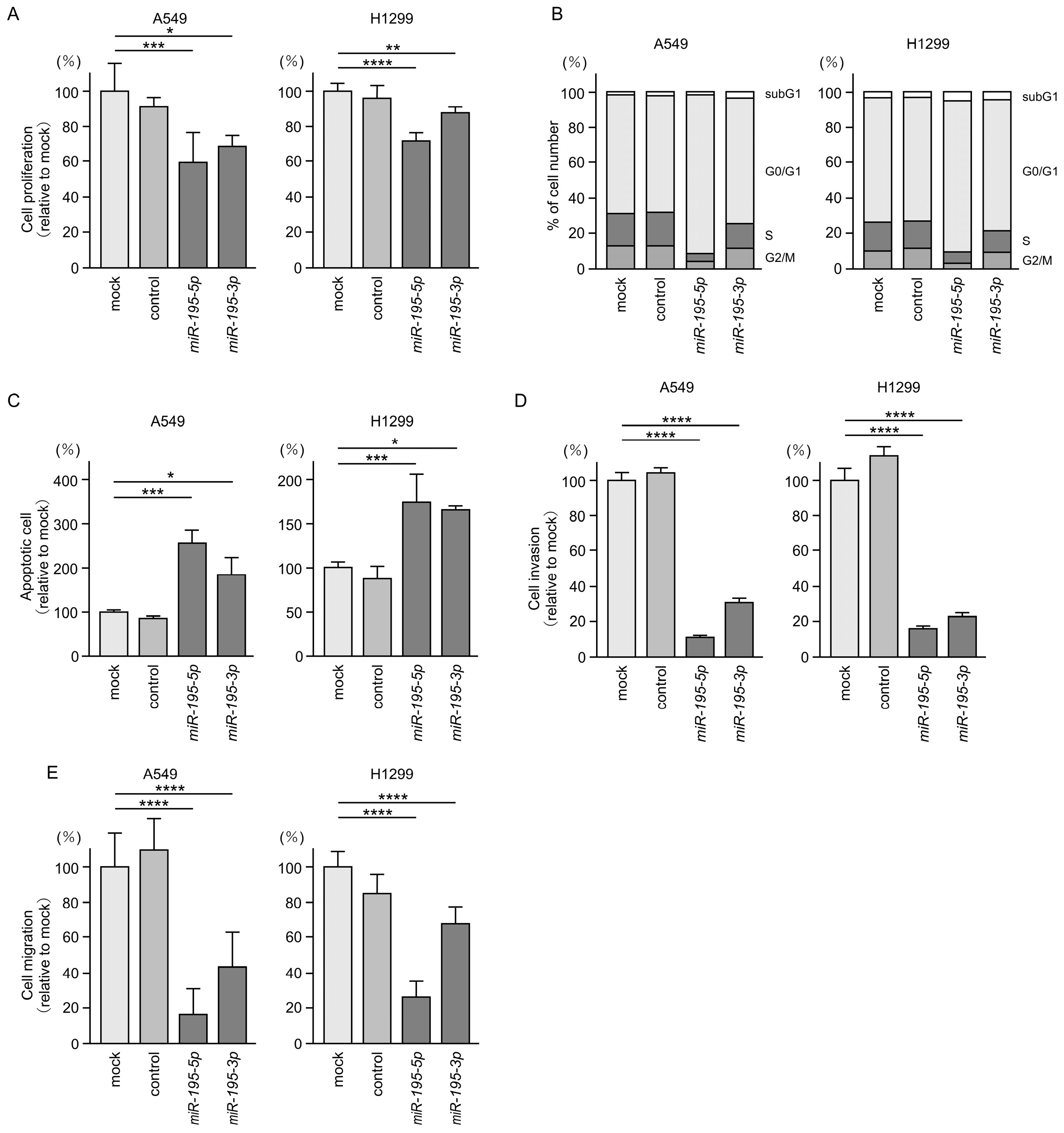
3.3. Tumor-Suppressive Functions of miR-195-5p and miR-195-3p in LUAD Cells
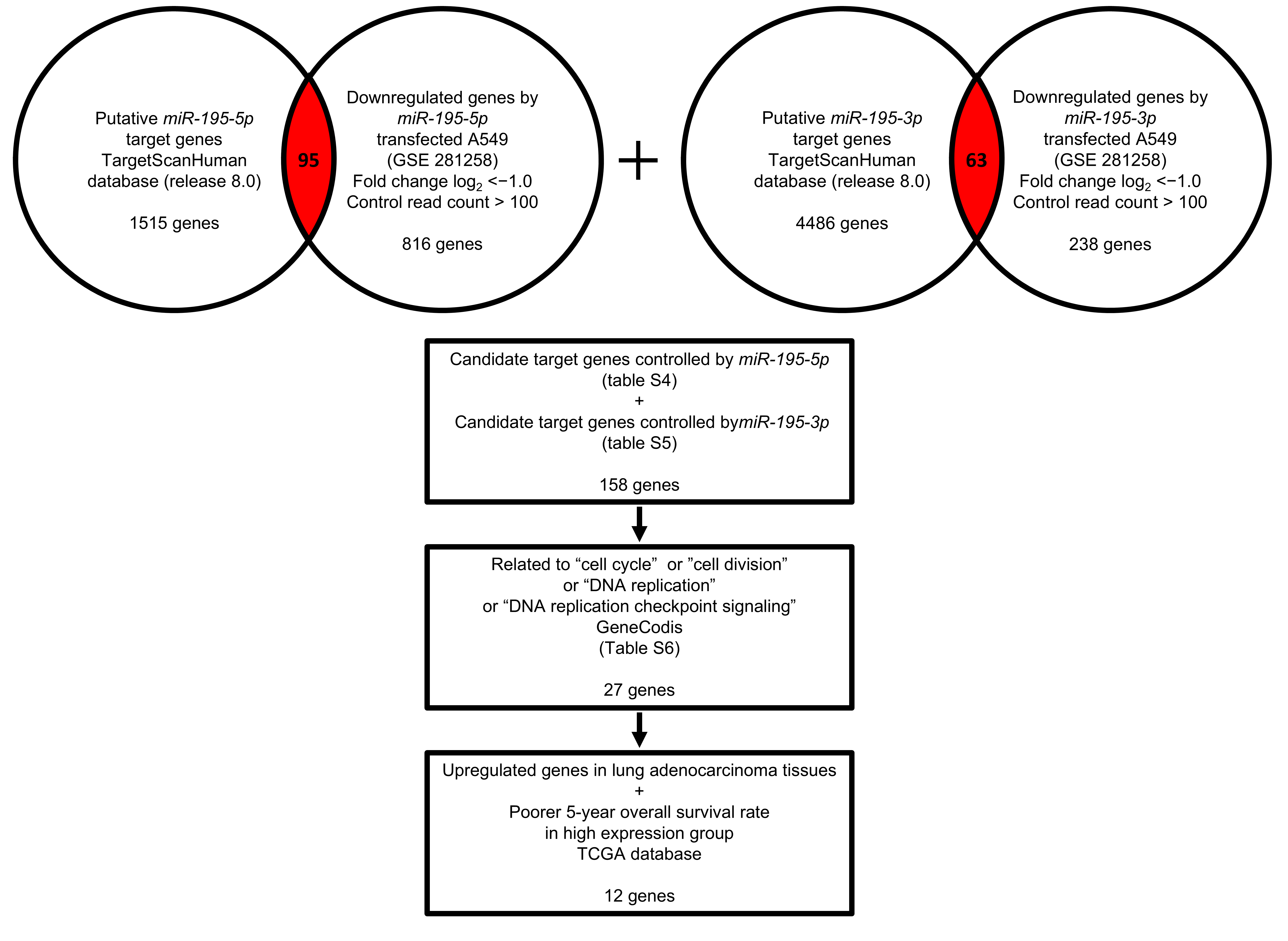
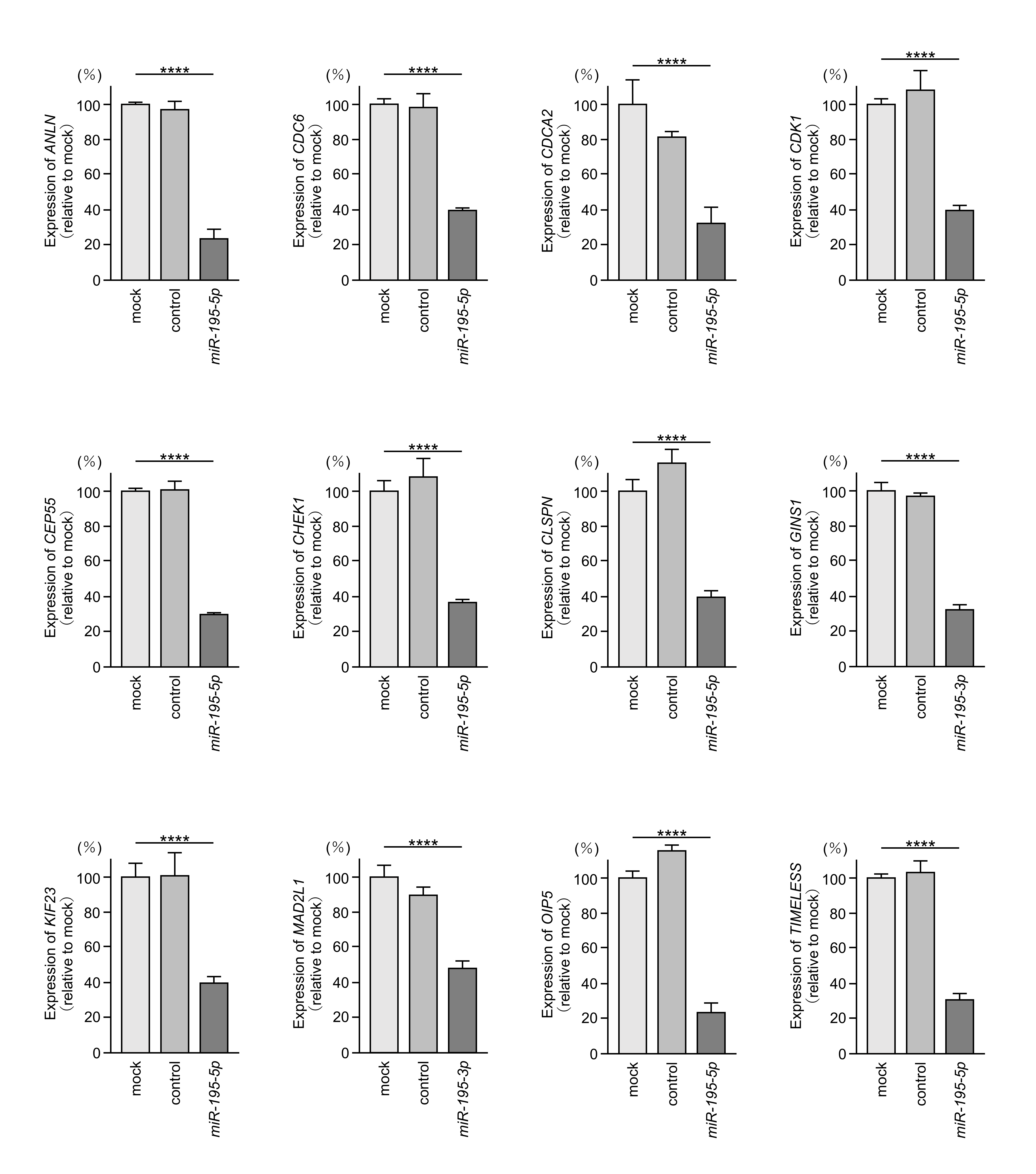
3.4. Identification of miR-195-5p- and miR-195-3p-Regulated Genes in LUAD Cells
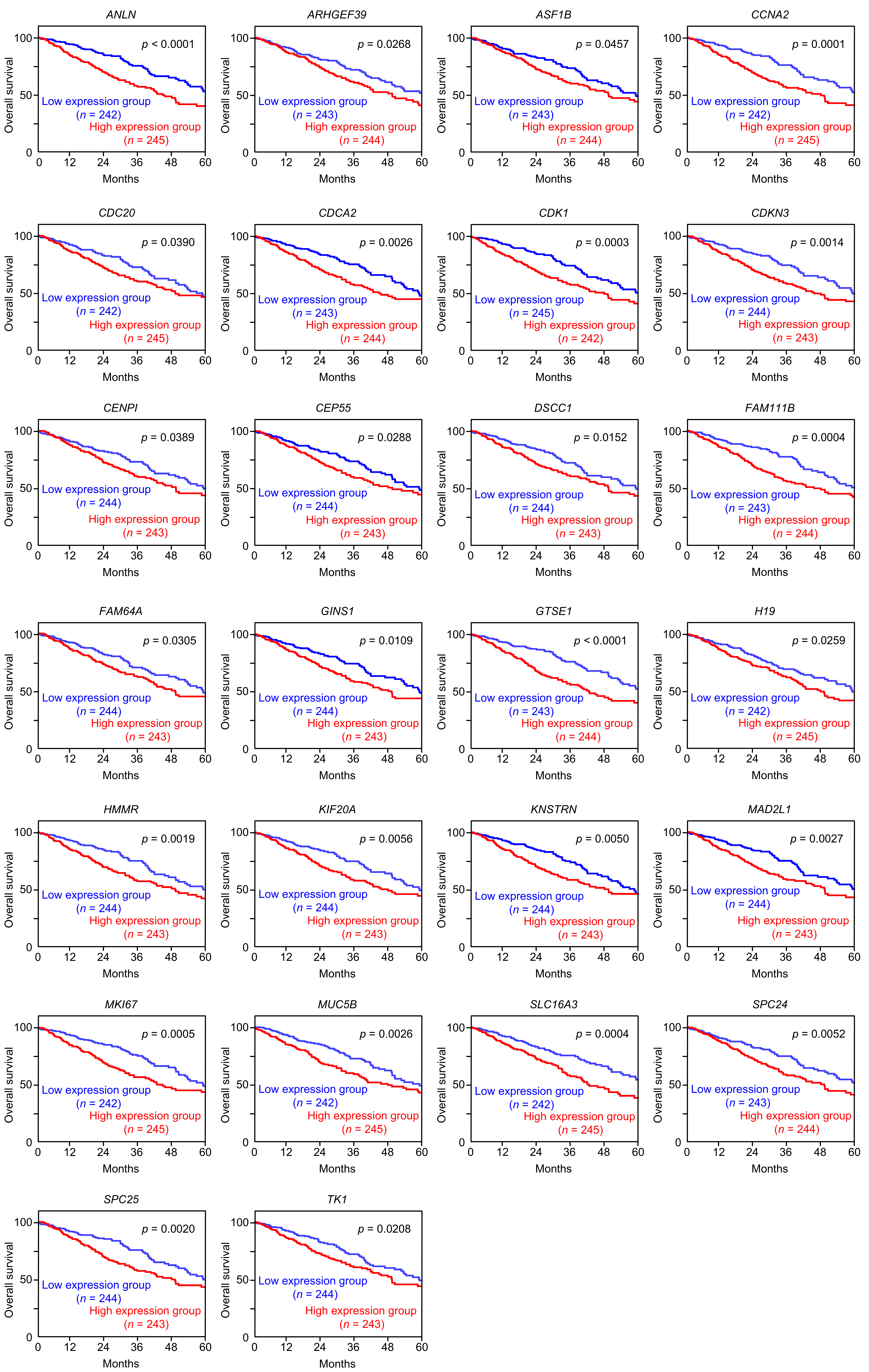
3.5. Clinical Significance of Genes Regulated by miR-195-5p or miR-195-3p in LUAD
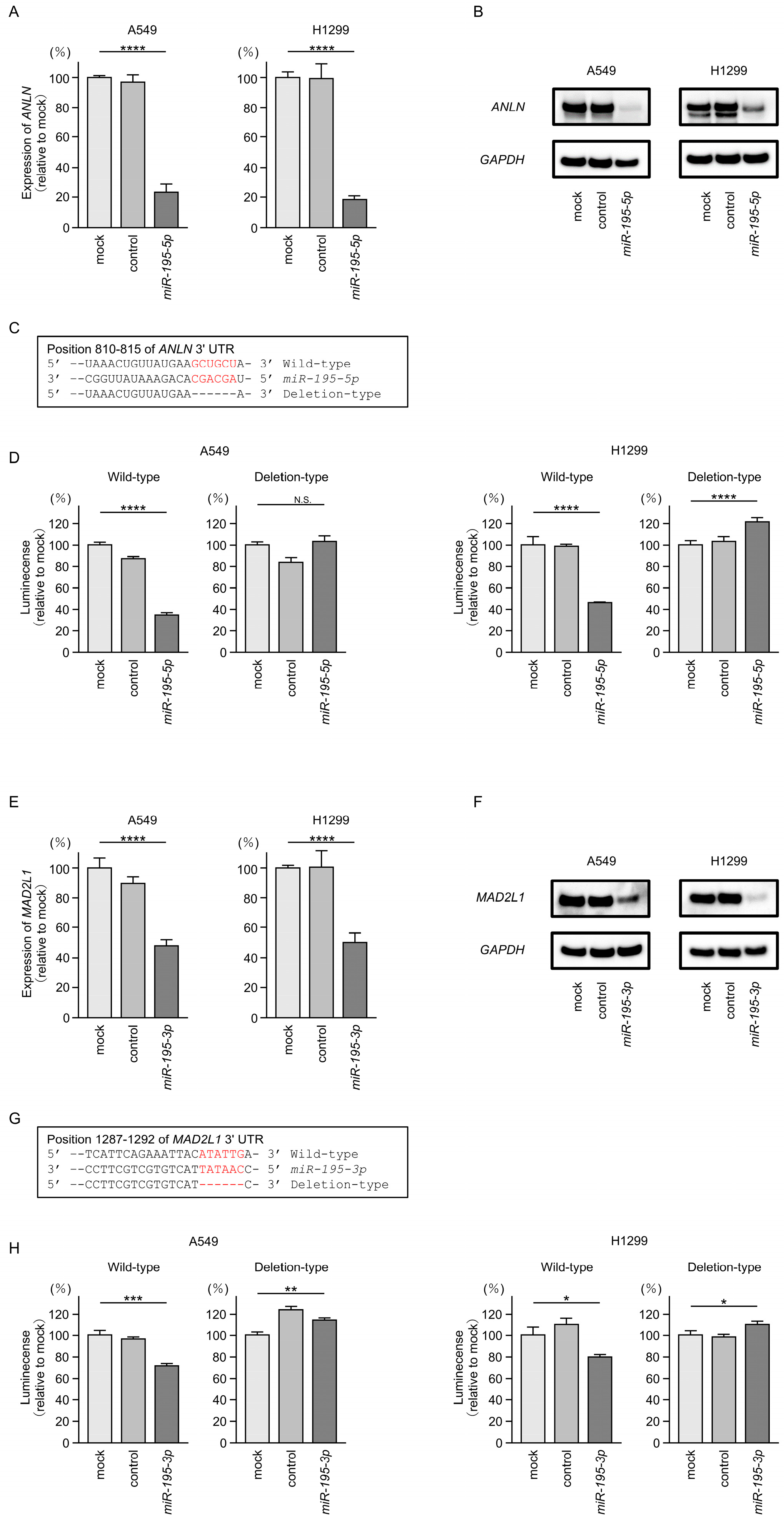
3.6. Direct Regulation of ANLN by miR-195-5p and MAD2L1 by miR-195-3p in LUAD Cells
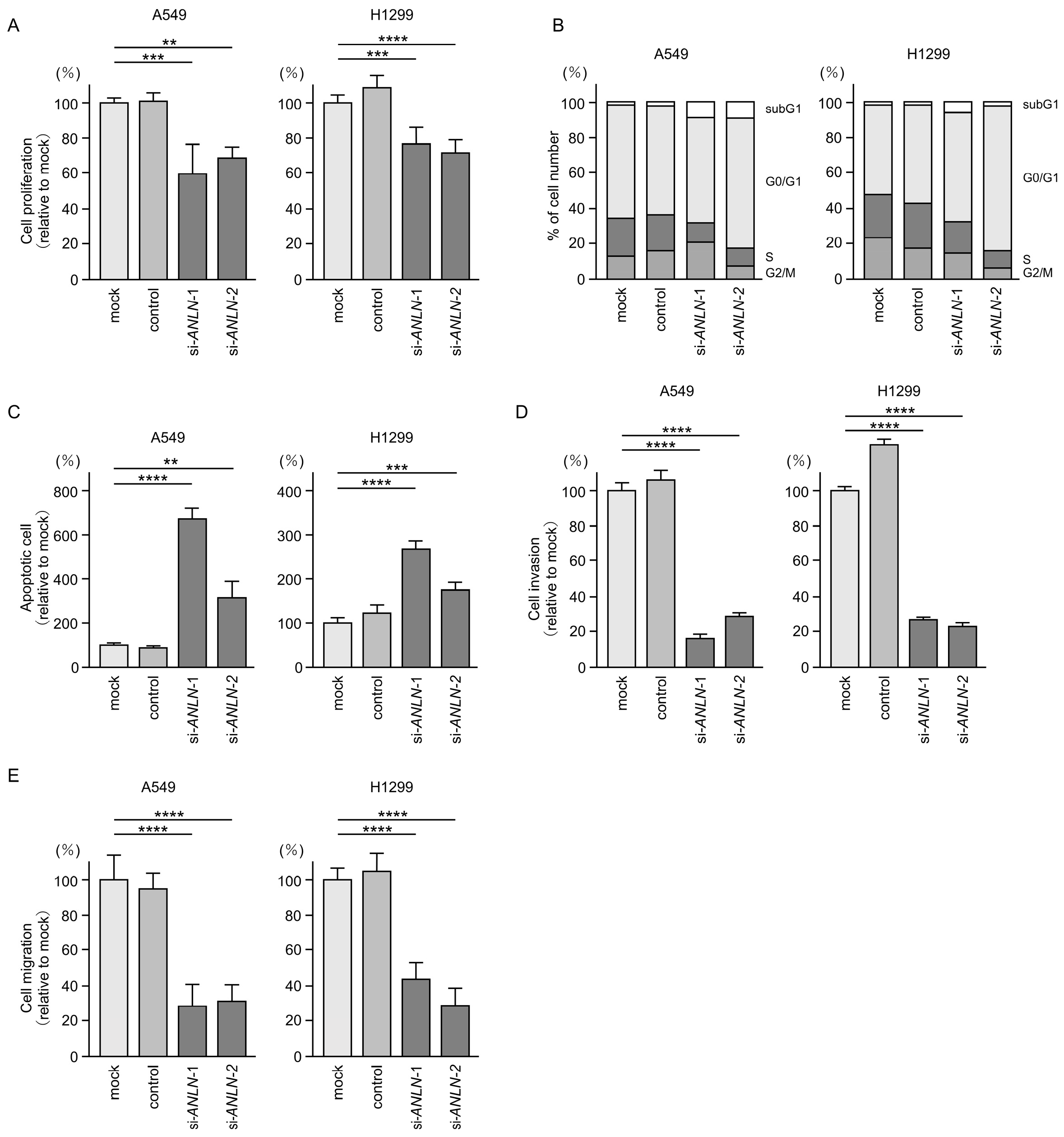
3.7. Functional Significance of ANLN in LUAD Cells
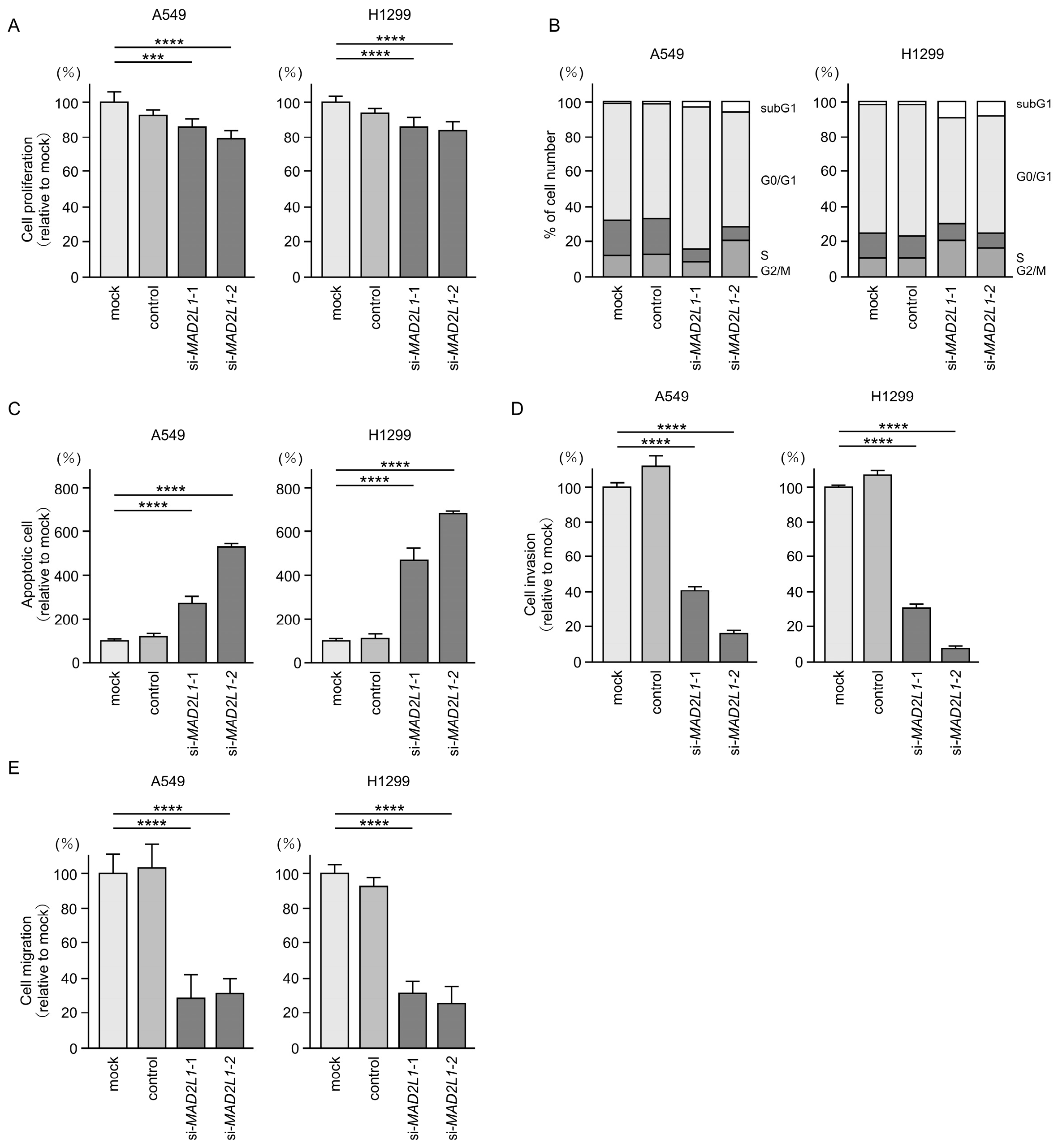
3.8. Functional Significance of MAD2L1 in LUAD Cells
3.9. ANLN- and MAD2L1-Mediated Cancer Pathways in LUAD Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Schabath, M.B.; Cote, M.L. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Tan, D.S.W. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 2022, 40, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Remon, J.; Hellmann, M.D. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 586–597. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Karimpour, M.; Ravanbakhsh, R.; Maydanchi, M.; Rajabi, A.; Azizi, F.; Saber, A. Cancer driver gene and non-coding RNA alterations as biomarkers of brain metastasis in lung cancer: A review of the literature. Biomed. Pharmacother. 2021, 143, 112190. [Google Scholar] [CrossRef]
- Martínez-Espinosa, I.; Serrato, J.A.; Ortiz-Quintero, B. MicroRNAs in Lung Cancer Brain Metastasis. Int. J. Mol. Sci. 2024, 25, 10325. [Google Scholar] [CrossRef]
- Liu, W.; Powell, C.A.; Wang, Q. Tumor microenvironment in lung cancer-derived brain metastasis. Chin. Med. J. 2022, 135, 1781–1791. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Li, M.H.; Fu, S.B.; Xiao, H.S. Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol. Sin. 2015, 36, 1200–1211. [Google Scholar] [CrossRef]
- Tomioka, Y.; Suetsugu, T.; Seki, N.; Tanigawa, K.; Hagihara, Y.; Shinmura, M.; Asai, S.; Kikkawa, N.; Inoue, H.; Mizuno, K. The Molecular Pathogenesis of Tumor-Suppressive miR-486-5p and miR-486-3p Target Genes: GINS4 Facilitates Aggressiveness in Lung Adenocarcinoma. Cells 2023, 12, 1885. [Google Scholar] [CrossRef]
- Tanigawa, K.; Misono, S.; Mizuno, K.; Asai, S.; Suetsugu, T.; Uchida, A.; Kawano, M.; Inoue, H.; Seki, N. MicroRNA signature of small-cell lung cancer after treatment failure: Impact on oncogenic targets by miR-30a-3p control. Mol. Oncol. 2023, 17, 328–343. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef]
- Tomioka, Y.; Seki, N.; Suetsugu, T.; Hagihara, Y.; Sanada, H.; Goto, Y.; Kikkawa, N.; Mizuno, K.; Tanaka, K.; Inoue, H. Identification of Tumor Suppressive miR-144-5p Targets: FAM111B Expression Accelerates the Malignant Phenotypes of Lung Adenocarcinoma. Int. J. Mol. Sci. 2024, 25, 9974. [Google Scholar] [CrossRef]
- Hagihara, Y.; Tomioka, Y.; Suetsugu, T.; Shinmura, M.; Misono, S.; Goto, Y.; Kikkawa, N.; Kato, M.; Inoue, H.; Mizuno, K.; et al. Identification of Tumor-Suppressive miR-139-3p-Regulated Genes: TRIP13 as a Therapeutic Target in Lung Adenocarcinoma. Cancers 2023, 15, 5571. [Google Scholar] [CrossRef]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Bu, L.; Tian, Y.; Wen, H.; Jia, W.; Yang, S. miR-195-5p exerts tumor-suppressive functions in human lung cancer cells through targeting TrxR2. Acta Biochim. Biophys. Sin. 2021, 53, 189–200. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Ning, Y.E.; Gu, H.; Tong, Y.; Wang, N. MiR-195-5p Inhibits Malignant Progression of Cervical Cancer by Targeting YAP1. Onco Targets Ther. 2020, 13, 931–944. [Google Scholar] [CrossRef]
- Piccinno, E.; Scalavino, V.; Labarile, N.; Armentano, R.; Giannelli, G.; Serino, G. miR-195-5p Inhibits Colon Cancer Progression via KRT23 Regulation. Pharmaceutics 2024, 16, 1554. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, Z.; Yu, J.; Wu, J.; Zhuo, X.; Chen, Q.; Liang, Y.; Li, G.; Wan, Y. MiR-195-5p suppresses the proliferation, migration, and invasion of gallbladder cancer cells by targeting FOSL1 and regulating the Wnt/β-catenin pathway. Ann. Transl. Med. 2022, 10, 893. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, B. miR-195-5p inhibits cisplatin resistance in lung adenocarcinoma by regulating DNA damage via targeting E2F7. J. Biochem. Mol. Toxicol. 2024, 38, e70015. [Google Scholar] [CrossRef]
- Xi, J.; Xi, Y.; Zhang, Z.; Hao, Y.; Wu, F.; Bian, B.; Hao, G.; Li, W.; Zhang, S. Hsa_circ_0060937 accelerates non-small cell lung cancer progression via modulating miR-195-5p/HMGB3 pathway. Cell Cycle. 2021, 20, 2040–2052. [Google Scholar] [CrossRef] [PubMed]
- Piekny, A.J.; Maddox, A.S. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 2010, 21, 881–891. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Huang, S.; Liao, W.; Li, J.; Huang, Z.; Huang, Y.; Lian, Y. Comprehensive Analysis of ANLN in Human Tumors: A Prognostic Biomarker Associated with Cancer Immunity. Oxid. Med. Cell Longev. 2022, 2022, 5322929. [Google Scholar] [CrossRef]
- Idichi, T.; Seki, N.; Kurahara, H.; Yonemori, K.; Osako, Y.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; Mataki, Y.; et al. Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 53180–53193. [Google Scholar] [CrossRef]
- Wang, Z.; Hu, S.; Li, X.; Liu, Z.; Han, D.; Wang, Y.; Wei, L.; Zhang, G.; Wang, X. MiR-16-5p suppresses breast cancer proliferation by targeting ANLN. BMC Cancer. 2021, 21, 1188. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zheng, H.; Yuan, S.; Zhou, B.; Zhao, W.; Pan, Y.; Qi, D. Overexpression of ANLN in lung adenocarcinoma is associated with metastasis. Thorac. Cancer. 2019, 10, 1702–1709. [Google Scholar] [CrossRef] [PubMed]
- Mossaid, I.; Fahrenkrog, B. Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint. Cells 2015, 4, 706–725. [Google Scholar] [CrossRef] [PubMed]
- Michel, L.; Benezra, R.; Diaz-Rodriguez, E. MAD2 dependent mitotic checkpoint defects in tumorigenesis and tumor cell death: A double edged sword. Cell Cycle 2004, 3, 990–992. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, S.; Zhang, Y.; Li, B.; Xu, H.; Zuo, S. Identification of MAD2L1 as a Potential Biomarker in Hepatocellular Carcinoma via Comprehensive Bioinformatics Analysis. Biomed. Res. Int. 2022, 2022, 9868022. [Google Scholar] [CrossRef]
- Zhu, X.F.; Yi, M.; He, J.; Tang, W.; Lu, M.Y.; Li, T.; Feng, Z.B. Pathological significance of MAD2L1 in breast cancer: An immunohistochemical study and meta analysis. Int. J. Clin. Exp. Pathol. 2017, 10, 9190–9201. [Google Scholar]
- Li, Q.; Tong, D.; Jing, X.; Ma, P.; Li, F.; Jiang, Q.; Zhang, J.; Wen, H.; Cui, M.; Huang, C.; et al. MAD2L1 is transcriptionally regulated by TEAD4 and promotes cell proliferation and migration in colorectal cancer. Cancer Gene Ther. 2023, 30, 727–737. [Google Scholar] [CrossRef]
- Wei, R.; Wang, Z.; Zhang, Y.; Wang, B.; Shen, N.; E, L.; Li, X.; Shang, L.; Shang, Y.; Yan, W.; et al. Bioinformatic analysis revealing mitotic spindle assembly regulated NDC80 and MAD2L1 as prognostic biomarkers in non-small cell lung cancer development. BMC Med. Genomics 2020, 13, 112. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, Y.; Zhu, L.; Cheng, J.; Li, Y. MAD2L1-mediated NANOG nuclear translocation: A critical factor in lung cancer chemoresistance. Cell Signal. 2025, 132, 111811. [Google Scholar] [CrossRef]


| MicroRNA | miRBase Accession No. | Guide or Passenger Strand | Chromosomal Location | Log2 FC | p-Value | FDR |
|---|---|---|---|---|---|---|
| hsa-miR-30d-5p | MIMAT0000245 | Guide strand | 8q24.22 | −2.51 | 0.004 | 0.018 |
| hsa-miR-34b-5p | MIMAT0000685 | Passenger strand | 11q23.1 | −2.56 | 0.047 | 0.117 |
| hsa-miR-4536-5p | MIMAT0019078 | Guide strand | Xp11.21 | −2.56 | 0.001 | 0.005 |
| hsa-miR-6785-3p | MIMAT0027471 | Passenger strand | 17q25.1 | −2.59 | 0.000 | 0.000 |
| hsa-miR-3141 | MIMAT0015010 | Guide strand | 5q33.2 | −2.61 | 0.008 | 0.031 |
| hsa-miR-30b-5p | MIMAT0000420 | Guide strand | 8q24.22 | −2.63 | 0.003 | 0.016 |
| hsa-miR-497-3p | MIMAT0004768 | Passenger strand | 17p13.1 | −2.63 | 0.000 | 0.003 |
| hsa-miR-4322 | MIMAT0016873 | Guide strand | 19p13.2 | −2.64 | 0.016 | 0.053 |
| hsa-miR-195-3p | MIMAT0004615 | Passenger strand | 17p13.1 | −2.66 | 0.000 | 0.001 |
| hsa-miR-6743-3p | MIMAT0027388 | Guide strand | 11p15.5 | −2.66 | 0.000 | 0.002 |
| hsa-miR-148a-5p | MIMAT0004549 | Passenger strand | 7p15.2 | −2.79 | 0.000 | 0.000 |
| hsa-miR-195-5p | MIMAT0000461 | Guide strand | 17p13.1 | −2.79 | 0.000 | 0.002 |
| hsa-miR-548i | MIMAT0005935 | Guide strand | 3q21.2 | −2.81 | 0.008 | 0.032 |
| hsa-miR-6788-3p | MIMAT0027477 | Guide strand | 18p11.22 | −2.82 | 0.000 | 0.000 |
| hsa-miR-363-5p | MIMAT0003385 | Passenger strand | Xq26.2 | −2.85 | 0.001 | 0.005 |
| hsa-miR-4536-3p | MIMAT0020959 | Passenger strand | Xp11.21 | −2.91 | 0.000 | 0.000 |
| hsa-miR-497-5p | MIMAT0002820 | Guide strand | 17p13.1 | −2.91 | 0.000 | 0.002 |
| hsa-miR-214-5p | MIMAT0004564 | Passenger strand | 1q24.3 | −2.92 | 0.000 | 0.001 |
| hsa-miR-95-3p | MIMAT0000094 | Guide strand | 4p16.1 | −2.92 | 0.008 | 0.033 |
| hsa-miR-4662a-5p | MIMAT0019731 | Guide strand | 8q24.13 | −2.94 | 0.008 | 0.031 |
| hsa-miR-145-5p | MIMAT0000437 | Guide strand | 5q32 | −2.95 | 0.000 | 0.002 |
| hsa-miR-130a-3p | MIMAT0000425 | Guide strand | 11q12.1 | −3.02 | 0.000 | 0.002 |
| hsa-miR-504-5p | MIMAT0002875 | Guide strand | Xq26.3 | −3.02 | 0.012 | 0.042 |
| hsa-miR-199a-3p | MIMAT0000232 | Guide strand | 19p13.2 | −3.03 | 0.000 | 0.002 |
| hsa-miR-34c-3p | MIMAT0004677 | Passenger strand | 11q23.1 | −3.06 | 0.009 | 0.036 |
| hsa-miR-6763-5p | MIMAT0027426 | Guide strand | 12q24.33 | −3.07 | 0.000 | 0.000 |
| hsa-miR-214-3p | MIMAT0000271 | Guide strand | 1q24.3 | −3.14 | 0.000 | 0.002 |
| hsa-miR-199b-3p | MIMAT0004563 | Guide strand | 9q34.11 | −3.15 | 0.000 | 0.002 |
| hsa-miR-218-5p | MIMAT0000275 | Guide strand | 4p15.31 | −3.17 | 0.001 | 0.006 |
| hsa-miR-34c-5p | MIMAT0000686 | Guide strand | 11q23.1 | −3.22 | 0.042 | 0.107 |
| hsa-miR-4470 | MIMAT0018997 | Guide strand | 8q12.3 | −3.30 | 0.000 | 0.001 |
| hsa-miR-1281 | MIMAT0005939 | Guide strand | 22q13.2 | −3.32 | 0.000 | 0.000 |
| hsa-miR-4259 | MIMAT0016880 | Guide strand | 1q23.2 | −3.33 | 0.000 | 0.000 |
| hsa-miR-3064-3p | MIMAT0019865 | Passenger strand | 17q23.3 | −3.41 | 0.000 | 0.000 |
| hsa-miR-3926 | MIMAT0018201 | Guide strand | 8p23.1 | −3.44 | 0.000 | 0.000 |
| hsa-miR-5091 | MIMAT0021083 | Guide strand | 4p15.33 | −3.46 | 0.000 | 0.001 |
| hsa-miR-199b-5p | MIMAT0000263 | Passenger strand | 9q34.11 | −3.56 | 0.001 | 0.007 |
| hsa-miR-602 | MIMAT0003270 | Guide strand | 9q34.3 | −3.61 | 0.000 | 0.003 |
| hsa-miR-548ar-3p | MIMAT0022266 | Passenger strand | 13q34 | −3.62 | 0.000 | 0.000 |
| hsa-miR-10a-3p | MIMAT0004555 | Passenger strand | 17q21.32 | −3.82 | 0.001 | 0.004 |
| hsa-miR-10a-5p | MIMAT0000253 | Guide strand | 17q21.32 | −3.82 | 0.000 | 0.002 |
| hsa-miR-548w | MIMAT0015060 | Guide strand | 16p12.1 | −3.85 | 0.000 | 0.000 |
| hsa-miR-199a-5p | MIMAT0000231 | Passenger strand | 19p13.2 | −3.91 | 0.000 | 0.000 |
| hsa-miR-150-5p | MIMAT0000451 | Guide strand | 19q13.33 | −4.00 | 0.000 | 0.001 |
| hsa-miR-548a | MIMAT0003251 | Guide strand | 6p22.3 | −4.19 | 0.000 | 0.000 |
| hsa-miR-34b-3p | MIMAT0004676 | Guide strand | 11q23.1 | −4.32 | 0.005 | 0.023 |
| Gene ID | Gene Symbol | Gene Name | Downregulated by miR-195-5p or miR-195-3p | miR-195-5p Transfectant Log2 Fold Change | miR-195-3p Transfectant Log2 Fold Change |
|---|---|---|---|---|---|
| 402 | ARL2 | ADP ribosylation factor-like GTPase 2 | miR-195-5p | −3.31 | 0.11 |
| 896 | CCND3 | cyclin D3 | miR-195-5p | −3.00 | 0.74 |
| 11339 | OIP5 | Opa-interacting protein 5 | miR-195-5p | −2.49 | −0.62 |
| 54443 | ANLN | anillin, actin-binding protein | miR-195-5p | −2.35 | −0.36 |
| 55165 | CEP55 | centrosomal protein 55 | miR-195-5p | −2.27 | −0.67 |
| 983 | CDK1 | cyclin-dependent kinase 1 | miR-195-5p | −1.86 | −0.63 |
| 63967 | CLSPN | claspin | miR-195-5p | −1.85 | −0.70 |
| 993 | CDC25A | cell division cycle 25A | miR-195-5p | −1.80 | 0.18 |
| 990 | CDC6 | cell division cycle 6 | miR-195-5p | −1.78 | −0.54 |
| 157313 | CDCA2 | cell division cycle- associated 2 | miR-195-5p | −1.78 | −0.53 |
| 55038 | CDCA4 | cell division cycle- associated 4 | miR-195-5p | −1.75 | −0.28 |
| 1111 | CHEK1 | checkpoint kinase 1 | miR-195-5p | −1.56 | −0.31 |
| 6867 | TACC1 | transforming acidic coiled-coil-containing protein 1 | miR-195-5p | −1.52 | 0.27 |
| 9493 | KIF23 | kinesin family member 23 | miR-195-5p | −1.52 | −0.11 |
| 80010 | RMI1 | RecQ-mediated genome instability 1 | miR-195-3p | −1.52 | −1.07 |
| 144455 | E2F7 | E2F transcription factor 7 | miR-195-5p | −1.46 | −0.01 |
| 4085 | MAD2L1 | mitotic arrest deficient 2-like 1 | miR-195-3p | −1.44 | −2.13 |
| 8914 | TIMELESS | timeless circadian clock | miR-195-5p | −1.39 | −0.23 |
| 79187 | FSD1 | fibronectin type III and SPRY domain-containing 1 | miR-195-5p | −1.26 | −0.72 |
| 6197 | RPS6KA3 | ribosomal protein S6 kinase A3 | miR-195-5p | −1.24 | 0.62 |
| 27183 | VPS4A | vacuolar protein sorting 4 homolog A | miR-195-5p | −1.16 | −0.07 |
| 9874 | TLK1 | tousled-like kinase 1 | miR-195-5p | −1.12 | 0.13 |
| 9837 | GINS1 | GINS complex subunit 1 | miR-195-3p | −1.08 | −1.85 |
| 996 | CDC27 | cell division cycle 27 | miR-195-5p | −1.06 | −0.11 |
| 8243 | SMC1A | structural maintenance of chromosomes 1A | miR-195-3p | −0.47 | −1.32 |
| 7329 | UBE2I | ubiquitin-conjugating enzyme E2 I | miR-195-3p | −0.03 | −1.00 |
| 57804 | POLD4 | DNA polymerase delta 4 | miR-195-3p | 0.19 | −1.35 |
| Entrez Gene ID | Gene Symbol | Gene Name | siANLN Transfectant Log2 Fold Change | siMAD2L1 Transfectant Log2 Fold Change |
|---|---|---|---|---|
| 54443 | ANLN | anillin actin-binding protein | −6.14 | −2.32 |
| 84904 | ARHGEF39 | Rho guanine nucleotide exchange factor 39 | −3.61 | −2.28 |
| 55723 | ASF1B | anti-silencing function 1B histone chaperone | −4.52 | −2.71 |
| 890 | CCNA2 | cyclin A2 | −3.64 | −2.09 |
| 991 | CDC20 | cell division cycle 20 | −4.26 | −2.67 |
| 157313 | CDCA2 | cell division cycle-associated 2 | −3.01 | −2.03 |
| 55536 | CDCA7L | cell division cycle-associated 7-like | −1.55 | −2.61 |
| 983 | CDK1 | cyclin-dependent kinase 1 | −4.60 | −1.96 |
| 1033 | CDKN3 | cyclin-dependent kinase inhibitor 3 | −2.92 | −2.43 |
| 2491 | CENPI | centromere protein I | −3.73 | −2.60 |
| 55165 | CEP55 | centrosomal protein 55 | −3.59 | −2.03 |
| 1164 | CKS2 | CDC28 protein kinase regulatory subunit 2 | −2.68 | −1.55 |
| 1719 | DHFR | dihydrofolate reductase | −4.01 | −2.18 |
| 79075 | DSCC1 | DNA replication and sister chromatid cohesion 1 | −2.76 | −2.39 |
| 374393 | FAM111B | family with sequence similarity 111 member B | −4.26 | −3.13 |
| 54478 | FAM64A | family with sequence similarity 64 member A | −3.89 | −1.92 |
| 9837 | GINS1 | GINS complex subunit 1 | −2.36 | −3.05 |
| 51512 | GTSE1 | G2 and S-phase-expressed 1 | −4.21 | −2.36 |
| 283120 | H19 | H19, imprinted maternally expressed transcript | −2.58 | −2.98 |
| 8479 | HIRIP3 | HIRA-interacting protein 3 | −2.21 | −1.93 |
| 3161 | HMMR | hyaluronan-mediated motility receptor | −3.78 | −2.60 |
| 55806 | HR | hair growth-associated | −1.94 | −1.91 |
| 56992 | KIF15 | kinesin family member 15 | −4.13 | −2.51 |
| 10112 | KIF20A | kinesin family member 20A | −3.86 | −2.05 |
| 90417 | KNSTRN | kinetochore localized astrin/SPAG5-binding protein | −1.65 | −1.25 |
| 4085 | MAD2L1 | MAD2 mitotic arrest deficient-like 1 | −2.91 | −5.58 |
| 4288 | MKI67 | marker of proliferation Ki-67 | −4.47 | −3.08 |
| 727897 | MUC5B | mucin 5B, oligomeric mucus/gel-forming | −1.82 | −1.00 |
| 23397 | NCAPH | non-SMC condensin I complex subunit H | −3.76 | −2.26 |
| 9768 | PCLAF | PCNA clamp-associated factor | −4.43 | −2.75 |
| 5933 | RBL1 | RB transcriptional corepressor like 1 | −1.87 | −2.54 |
| 10535 | RNASEH2A | ribonuclease H2 subunit A | −2.93 | −2.08 |
| 200916 | RPL22L1 | ribosomal protein L22 like 1 | −1.07 | −2.64 |
| 9123 | SLC16A3 | solute carrier family 16 member 3 | −1.13 | −1.57 |
| 147841 | SPC24 | SPC24, NDC80 kinetochore complex component | −3.75 | −2.40 |
| 57405 | SPC25 | SPC25, NDC80 kinetochore complex component | −4.89 | −2.89 |
| 7083 | TK1 | thymidine kinase 1 | −4.42 | −2.45 |
| 8458 | TTF2 | transcription termination factor 2 | −1.57 | −1.46 |
| 89891 | WDR34 | WD repeat domain 34 | −1.21 | −1.80 |
| Description | p-Value | Genes |
|---|---|---|
| cell division | <0.001 | MAD2L1, CDC20, NCAPH, CCNA2, ANLN, SPC25, FAM64A, KNSTRN, CDCA2, CEP55, CDK1, SPC24, CDCA7L, CKS2 |
| DNA replication | <0.001 | FAM111B, RNASEH2A, KIAA0101, GINS1, DSCC1, CDK1 |
| mitotic spindle assembly checkpoint signaling | <0.001 | MAD2L1, CDC20, SPC25, SPC24 |
| chromosome segregation | <0.001 | SPC25, KNSTRN, CDCA2, SPC24, CENPI |
| mitotic cell cycle phase transition | <0.001 | CCNA2, CDK1, CKS2 |
| mitotic sister chromatid segregation | <0.001 | MAD2L1, KNSTRN, CENPI |
| regulation of chromosome segregation | <0.001 | MKI67, CDCA2 |
| mitotic cytokinesis | <0.001 | KIF20A, ANLN, CEP55 |
| G1/S transition in the mitotic cell cycle | <0.001 | CCNA2, CDK1, CDKN3 |
| mitotic sister chromatid cohesion | <0.001 | CDC20, DSCC1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomioka, Y.; Seki, N.; Mizuno, K.; Suetsugu, T.; Tsuruzono, K.; Hagihara, Y.; Kato, M.; Minemura, C.; Yonezawa, H.; Tanaka, K.; et al. MicroRNA Signatures in Lung Adenocarcinoma Metastases: Exploring the Oncogenic Targets of Tumor-Suppressive miR-195-5p and miR-195-3p. Cancers 2025, 17, 2348. https://doi.org/10.3390/cancers17142348
Tomioka Y, Seki N, Mizuno K, Suetsugu T, Tsuruzono K, Hagihara Y, Kato M, Minemura C, Yonezawa H, Tanaka K, et al. MicroRNA Signatures in Lung Adenocarcinoma Metastases: Exploring the Oncogenic Targets of Tumor-Suppressive miR-195-5p and miR-195-3p. Cancers. 2025; 17(14):2348. https://doi.org/10.3390/cancers17142348
Chicago/Turabian StyleTomioka, Yuya, Naohiko Seki, Keiko Mizuno, Takayuki Suetsugu, Kentaro Tsuruzono, Yoko Hagihara, Mayuko Kato, Chikashi Minemura, Hajime Yonezawa, Kentaro Tanaka, and et al. 2025. "MicroRNA Signatures in Lung Adenocarcinoma Metastases: Exploring the Oncogenic Targets of Tumor-Suppressive miR-195-5p and miR-195-3p" Cancers 17, no. 14: 2348. https://doi.org/10.3390/cancers17142348
APA StyleTomioka, Y., Seki, N., Mizuno, K., Suetsugu, T., Tsuruzono, K., Hagihara, Y., Kato, M., Minemura, C., Yonezawa, H., Tanaka, K., & Inoue, H. (2025). MicroRNA Signatures in Lung Adenocarcinoma Metastases: Exploring the Oncogenic Targets of Tumor-Suppressive miR-195-5p and miR-195-3p. Cancers, 17(14), 2348. https://doi.org/10.3390/cancers17142348







