Artificial Intelligence in Advancing Inflammatory Bowel Disease Management: Setting New Standards
Simple Summary
Abstract
1. Introduction
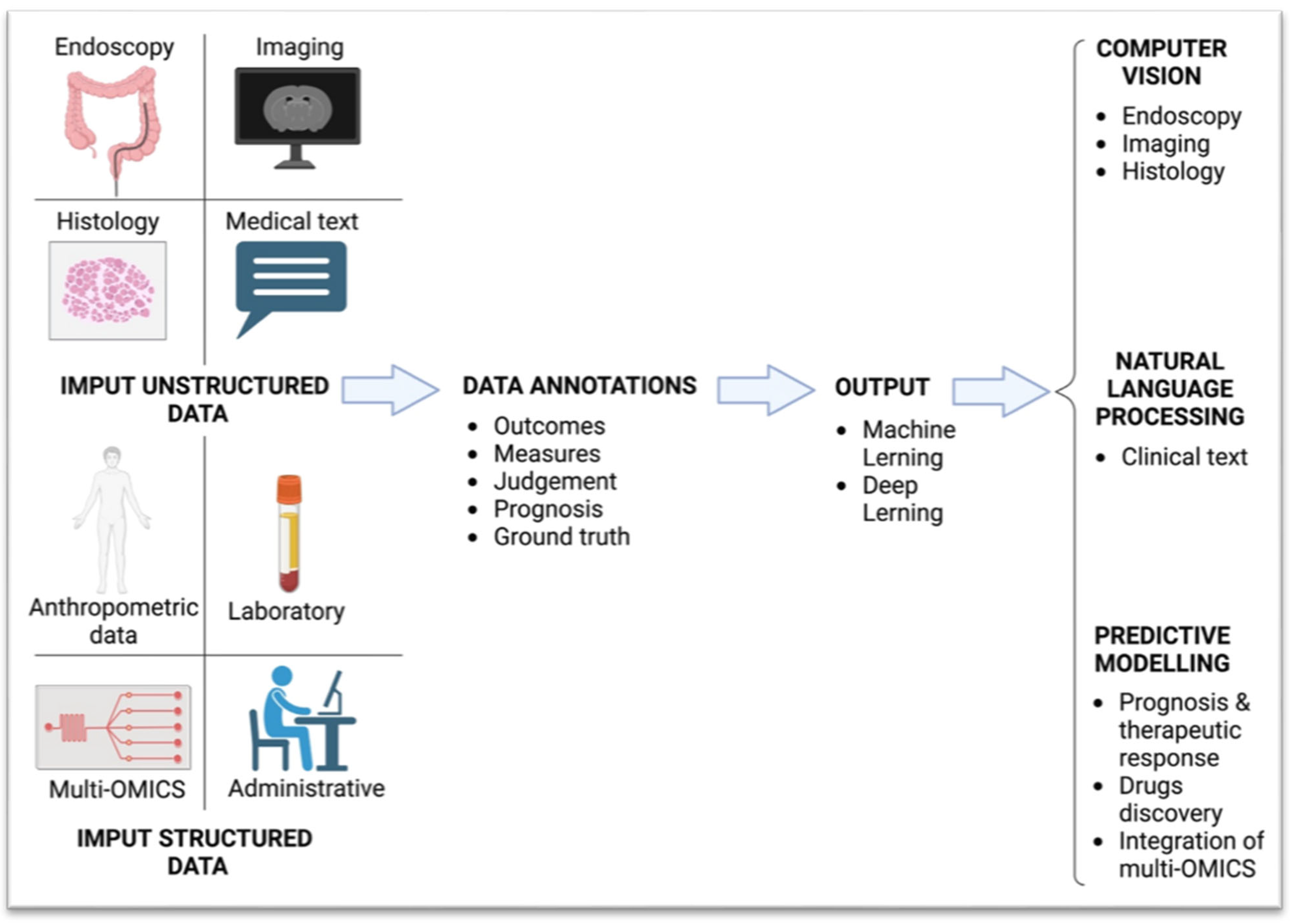
2. Methods
3. Artificial Intelligence Overview
Explanation of AI Concepts Relevant to Healthcare
4. AI Applications in Inflammatory Bowel Disease
4.1. Endoscopic Diagnosis and Assessment of UC Enabled by AI
4.2. Endoscopic Diagnosis and Assessment of Disease Activity in CD Enabled by AI
4.3. AI Drives Advanced Endoscopic Technologies
4.4. Personalising Therapy Through AI: Tailoring Treatment for Optimal Patient Outcome
4.4.1. AI in Predicting Response to Therapy
4.4.2. AI in Predicting the Course of the Disease by Determining the Histological Activity
4.4.3. AI for Continuous Monitoring of Disease Activity and Patient Self-Assessment
4.4.4. AI for Evaluation of Histological Activity and Diagnosis
4.4.5. The Role of AI in Detecting Colitis-Associated Neoplasia
4.4.6. AI in Predicting Disease Progression, Complications, and Risk Stratification
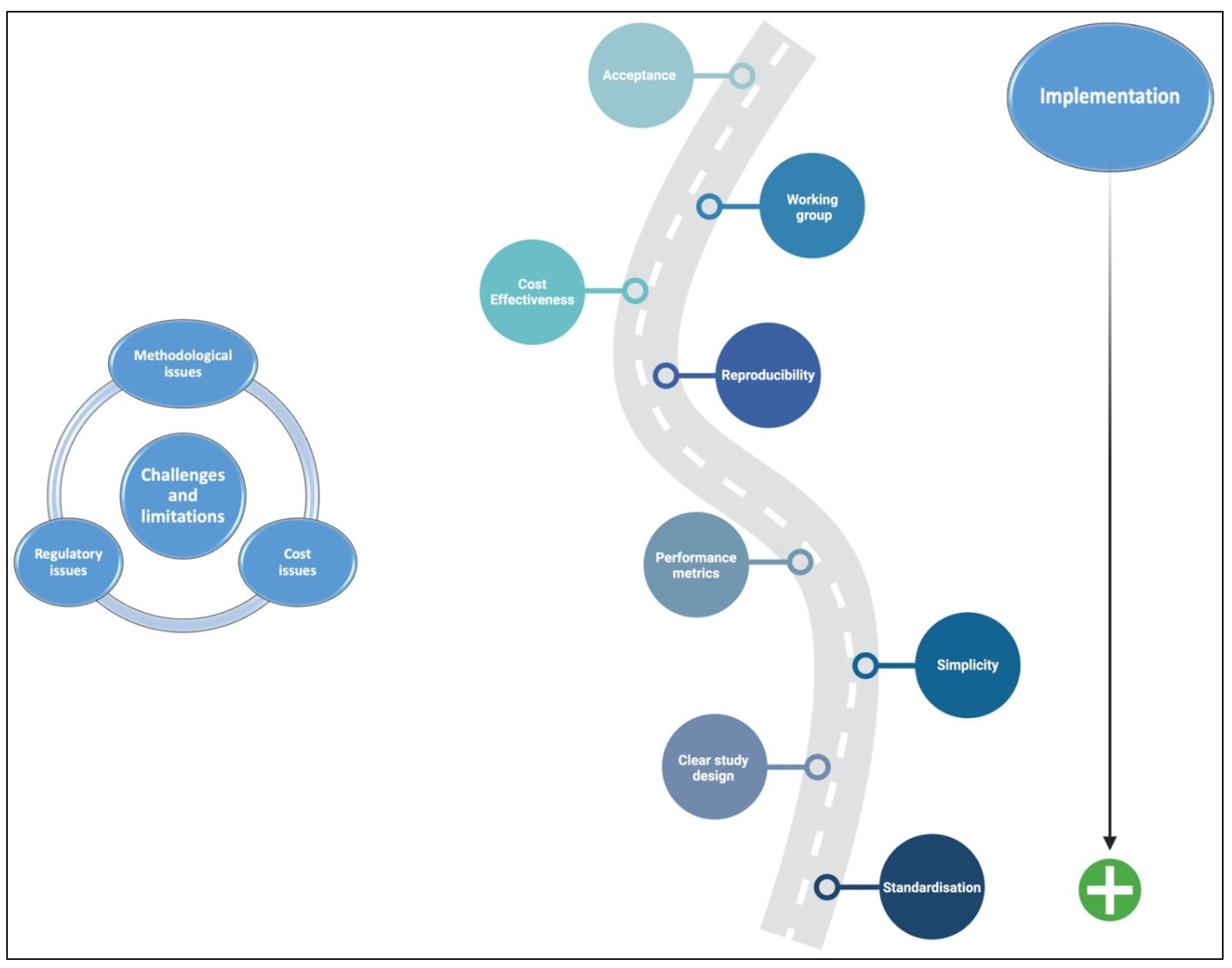
5. Challenges, Limitations, and Implementation
5.1. Methodological Issues of AI Application to IBD
5.2. Regulatory Issues of AI Application to IBD
5.3. Cost Issues of AI Application to IBD
6. Future Directions and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- Kappelman, M.D.; Rifas-Shiman, S.L.; Kleinman, K.; Ollendorf, D.; Bousvaros, A.; Grand, R.J.; Finkelstein, J.A. The prevalence and geographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2007, 5, 1424–1429. [Google Scholar] [CrossRef]
- Loftus, E.V. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 2004, 126, 1504–1517. [Google Scholar] [CrossRef]
- Herauf, M.; Coward, S.; Peña-Sánchez, J.N.; Bernstein, C.N.; Benchimol, E.I.; Kaplan, G.G.; Canadian Gastro-Intestinal Epidemiology Consortium. Commentary on the Epidemiology of Inflammatory Bowel Disease in Compounding Prevalence Nations: Toward Sustaining Healthcare Delivery. Gastroenterology 2024, 166, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Orlando, A.; Guglielmi, F.W.; Cottone, M.; Orlando, E.; Romano, C.; Sinagra, E. Clinical implications of mucosal healing in the management of patients with inflammatory bowel disease. Dig. Liver Dis. 2013, 45, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Le Berre, C.; Danese, S.; Peyrin-Biroulet, L. Can we change the natural course of inflammatory bowel disease? Ther. Adv. Gastroenterol. 2023, 16, 17562848231163118. [Google Scholar] [CrossRef] [PubMed]
- Pakdin, M.; Zarei, L.; Bagheri Lankarani, K.; Ghahramani, S. The cost of illness analysis of inflammatory bowel disease. BMC Gastroenterol. 2023, 23, 21. [Google Scholar] [CrossRef]
- Maaser, C.; Sturm, A.; Vavricka, S.R.; Kucharzik, T.; Fiorino, G.; Annese, V.; Calabrese, E.; Baumgart, D.C.; Bettenworth, D.; Borralho Nunes, P.; et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J. Crohns Colitis. 2019, 13, 144–164. [Google Scholar] [CrossRef]
- Sturm, A.; Maaser, C.; Calabrese, E.; Annese, V.; Fiorino, G.; Kucharzik, T.; Vavricka, S.R.; Verstockt, B.; van Rheenen, P.; Tolan, D.; et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 2: IBD scores and general principles and technical aspects. J. Crohns Colitis. 2019, 13, 273–284. [Google Scholar] [CrossRef]
- Gu, P.; Mendonca, O.; Carter, D.; Dube, S.; Wang, P.; Huang, X.; Li, D.; Moore, J.H.; McGovern, D.P.B. AI-luminating Artificial Intelligence in Inflammatory Bowel Diseases: A Narrative Review on the Role of AI in Endoscopy, Histology, and Imaging for IBD. Inflamm. Bowel Dis. 2024, 30, 2467–2485. [Google Scholar] [CrossRef]
- Kaplan, G.G.; Windsor, J.W. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 56–66. [Google Scholar] [CrossRef]
- Watermeyer, G.; Katsidzira, L.; Setshedi, M.; Devani, S.; Mudombi, W.; Kassianides, C. Gastroenterology and Hepatology Association of sub-Saharan Africa (GHASSA). Inflammatory bowel disease in sub-Saharan Africa: Epidemiology, risk factors, and challenges in diagnosis. Lancet Gastroenterol. Hepatol. 2022, 7, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Stidham, R.W.; Takenaka, K. Artificial Intelligence for Disease Assessment in Inflammatory Bowel Disease: How Will it Change Our Practice? Gastroenterology 2022, 162, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Da Rio, L.; Spadaccini, M.; Parigi, T.L.; Gabbiadini, R.; Dal Buono, A.; Busacca, A.; Maselli, R.; Fugazza, A.; Colombo, M.; Carrara, S.; et al. Artificial intelligence and inflammatory bowel disease: Where are we going? World J. Gastroenterol. 2023, 29, 508–520. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug. Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, H.A.; East, J.E.; Panaccione, R.; Travis, S.; Canavan, J.B.; Usiskin, K.; Byrne, M.F. Artificial Intelligence in Inflammatory Bowel Disease Endoscopy: Implications for Clinical Trials. J. Crohns Colitis. 2023, 17, 1342–1353. [Google Scholar] [CrossRef]
- Ahmed, M.; Stone, M.L.; Stidham, R.W. Artificial Intelligence and IBD: Where are We Now and Where Will We Be in the Future? Curr. Gastroenterol. Rep. 2024, 26, 137–144. [Google Scholar] [CrossRef]
- Yang, Z.R. Biological applications of support vector machines. Brief. Bioinform. 2004, 5, 328–338. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef]
- Sasaki, Y.; Hada, R.; Munakata, A. Computer-aided grading system for endoscopic severity in patients with ulcerative colitis. Dig. Endosc. 2003, 15, 206–209. [Google Scholar] [CrossRef]
- Kraszewski, S.; Szczurek, W.; Szymczak, J.; Reguła, M.; Neubauer, K. Machine Learning Prediction Model for Inflammatory Bowel Disease Based on Laboratory Markers. Working Model in a Discovery Cohort Study. J. Clin. Med. 2021, 10, 4745. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.; Nakase, H.; Vermeire, S.; de Hertogh, G.; Eelbode, T.; Ferrante, M.; Hasegawa, T.; Willekens, H.; Ikemoto, Y.; Makino, T.; et al. Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut 2020, 69, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Stidham, R.W.; Liu, W.; Bishu, S.; Rice, M.D.; Higgins, P.D.R.; Zhu, J.; Nallamothu, B.K.; Waljee, A.K. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients With Ulcerative Colitis. JAMA Netw. Open. 2019, 2, e193963. [Google Scholar] [CrossRef]
- Fan, Y.; Mu, R.; Xu, H.; Xie, C.; Zhang, Y.; Liu, L.; Wang, L.; Shi, H.; Hu, Y.; Ren, J.; et al. Novel deep learning-based computer-aided diagnosis system for predicting inflammatory activity in ulcerative colitis. Gastrointest. Endosc. 2023, 97, 335–346. [Google Scholar] [CrossRef]
- Ozawa, T.; Ishihara, S.; Fujishiro, M.; Saito, H.; Kumagai, Y.; Shichijo, S.; Aoyama, K.; Tada, T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest. Endosc. 2019, 89, 416–421.e1. [Google Scholar] [CrossRef]
- Takabayashi, K.; Kobayashi, T.; Matsuoka, K.; Levesque, B.G.; Kawamura, T.; Tanaka, K.; Kadota, T.; Bise, R.; Uchida, S.; Kanai, T.; et al. Artificial intelligence quantifying endoscopic severity of ulcerative colitis in gradation scale. Dig. Endosc. Off. J. Jpn. Gastroenterol. Endosc. Soc. 2024, 36, 582–590. [Google Scholar] [CrossRef]
- Takenaka, K.; Ohtsuka, K.; Fujii, T.; Negi, M.; Suzuki, K.; Shimizu, H.; Oshima, S.; Akiyama, S.; Motobayashi, M.; Nagahori, M.; et al. Development and Validation of a Deep Neural Network for Accurate Evaluation of Endoscopic Images From Patients With Ulcerative Colitis. Gastroenterology 2020, 158, 2150–2157. [Google Scholar] [CrossRef]
- Takenaka, K.; Ohtsuka, K.; Fujii, T.; Oshima, S.; Okamoto, R.; Watanabe, M. Deep Neural Network Accurately Predicts Prognosis of Ulcerative Colitis Using Endoscopic Images. Gastroenterology 2021, 160, 2175–2177.e3. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Najarian, K.; Gryak, J.; Bishu, S.; Rice, M.D.; Waljee, A.K.; Wilkins, H.J.; Stidham, R.W. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest. Endosc. 2021, 93, 728–736.e1. [Google Scholar] [CrossRef]
- Gottlieb, K.; Requa, J.; Karnes, W.; Chandra Gudivada, R.; Shen, J.; Rael, E.; Arora, V.; Dao, T.; Ninh, A.; McGill, J. Central Reading of Ulcerative Colitis Clinical Trial Videos Using Neural Networks. Gastroenterology 2021, 160, 710–719.e2. [Google Scholar] [CrossRef]
- Iacucci, M.; Cannatelli, R.; Parigi, T.L.; Nardone, O.M.; Tontini, G.E.; Labarile, N.; Buda, A.; Rimondi, A.; Bazarova, A.; Bisschops, R.; et al. A virtual chromoendoscopy artificial intelligence system to detect endoscopic and histologic activity/remission and predict clinical outcomes in ulcerative colitis. Endoscopy 2023, 55, 332–341. [Google Scholar] [CrossRef]
- Charisis, V.S.; Hadjileontiadis, L.J. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J. Gastroenterol. 2016, 22, 8641–8657. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Xu, L.; Fan, Y.; Wei, K.; Li, L. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys. Med. Biol. 2018, 63, 165001. [Google Scholar] [CrossRef] [PubMed]
- Afonso, J.; Saraiva, M.M.; Ferreira, J.P.S.; Cardoso, H.; Ribeiro, T.; Andrade, P.; Parente, M.; Jorge, R.N.; Macedo, G. Automated detection of ulcers and erosions in capsule endoscopy images using a convolutional neuralnetwork. Med. Biol. Eng. Comput. 2022, 60, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest. Endosc. 2019, 89, 357–363.e2. [Google Scholar] [CrossRef]
- Ferreira, J.P.S.; de Mascarenhas Saraiva, M.J.Q.E.C.; Afonso, J.P.L.; Ribeiro, T.F.C.; Cardoso, H.M.C.; Ribeiro Andrade, A.P.; de Mascarenhas Saraiva, M.N.G.; Parente, M.P.L.; Natal Jorge, R.; Lopes, S.I.O.; et al. Identification of Ulcers and Erosions by the Novel PillcamTM Crohn’s Capsule Using a Convolutional Neural Network: A Multicentre Pilot Study. J. Crohns Colitis. 2022, 16, 169–172. [Google Scholar] [CrossRef]
- Kratter, T.; Shapira, N.; Lev, Y.; Mauda, O.; Moshkovitz, Y.; Shitrit, R.; Konyo, S.; Ukashi, O.; Dar, L.; Shlomi, O.; et al. Deep Learning Multi-Domain Model Provides Accurate Detection and Grading of Mucosal Ulcers in Different Capsule Endoscopy Types. Diagnostic 2022, 12, 2490. [Google Scholar] [CrossRef]
- Brodersen, J.B.; Jensen, M.D.; Leenhardt, R.; Kjeldsen, J.; Histace, A.; Knudsen, T.; Dray, X. Artificial Intelligence-assisted Analysis of Pan-enteric Capsule Endoscopy in Patients with Suspected Crohn’s Disease: A Study on Diagnostic Performance. J. Crohns Colitis. 2024, 18, 75–81. [Google Scholar] [CrossRef]
- Klang, E.; Grinman, A.; Soffer, S.; Margalit Yehuda, R.; Barzilay, O.; Amitai, M.M.; Konen, E.; Ben-Horin, S.; Eliakim, R.; Barash, Y.; et al. Automated Detection of Crohn’s Disease Intestinal Strictures on Capsule Endoscopy Images Using Deep Neural Networks. J. Crohns Colitis. 2021, 15, 749–756. [Google Scholar] [CrossRef]
- Barash, Y.; Azaria, L.; Soffer, S.; Margalit Yehuda, R.; Shlomi, O.; Ben-Horin, S.; Eliakim, R.; Klang, E.; Kopylov, U. Ulcer severity grading in video capsule images of patients with Crohn’s disease: An ordinal neural network solution. Gastrointest. Endosc. 2021, 93, 187–192. [Google Scholar] [CrossRef]
- Ding, Z.; Shi, H.; Zhang, H.; Meng, L.; Fan, M.; Han, C.; Zhang, K.; Ming, F.; Xie, X.; Liu, H.; et al. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology 2019, 157, 1044–1054.e5. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Yamada, A.; Aoyama, K.; Saito, H.; Fujisawa, G.; Odawara, N.; Kondo, R.; Tsuboi, A.; Ishibashi, R.; Nakada, A.; et al. Clinical usefulness of a deep learning-based system as the first screening on small-bowel capsule endoscopy reading. Dig. Endosc. Off. J. Jpn. Gastroenterol. Endosc. Soc. 2020, 32, 585–591. [Google Scholar] [CrossRef]
- Quénéhervé, L.; David, G.; Bourreille, A.; Hardouin, J.B.; Rahmi, G.; Neunlist, M.; Brégeon, J.; Coron, E. Quantitative assessment of mucosal architecture using computer-based analysis of confocal laser endomicroscopy in inflammatory bowel diseases. Gastrointest. Endosc. 2019, 89, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kudo, S.E.; Mori, Y.; Misawa, M.; Ogata, N.; Sasanuma, S.; Wakamura, K.; Oda, M.; Mori, K.; Ohtsuka, K. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest. Endosc. 2019, 89, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.; De Hertogh, G.; Eelbode, T.; Vermeire, S.; Bisschops, R. Computer-Aided Diagnosis With Monochromatic Light Endoscopy for Scoring Histologic Remission in Ulcerative Colitis. Gastroenterolog 2021, 160, 23–25. [Google Scholar] [CrossRef]
- Sinonquel, P.; Lenfant, M.; Eelbode, T.; Watanabe, H.; Callaerts, B.; Bossuyt, P.; Verstockt, B.; Sabino, J.P.G.; De Hertogh, G.; Maes, F.; et al. Development of an Automated Tool for the Estimation of Histological Remission in Ulcerative Colitis Using Single-Wavelength Endoscopy Technology. J. Crohns Colitis. 2024, 19, jjae180. [Google Scholar] [CrossRef]
- Con, D.; van Langenberg, D.R.; Vasudevan, A. Deep learning vs conventional learning algorithms for clinical prediction in Crohn’s disease: A proof-of-concept study. World J. Gastroenterol. 2021, 27, 6476–6488. [Google Scholar] [CrossRef] [PubMed]
- Popa, I.V.; Burlacu, A.; Mihai, C.; Prelipcean, C.C. A Machine Learning Model Accurately Predicts Ulcerative Colitis Activity at One Year in Patients Treated with Anti-Tumour Necrosis Factor α Agents. Med. Kaunas. Lith. 2020, 56, 628. [Google Scholar] [CrossRef]
- Park, S.K.; Kim, Y.B.; Kim, S.; Lee, C.W.; Choi, C.H.; Kang, S.B.; Kim, T.O.; Bang, K.B.; Chun, J.; Cha, J.M.; et al. Development of a Machine Learning Model to Predict Non-Durable Response to Anti-TNF Therapy in Crohn’s Disease Using Transcriptome Imputed from Genotypes. J. Pers. Med. 2022, 12, 947. [Google Scholar] [CrossRef]
- Waljee, A.K.; Wallace, B.I.; Cohen-Mekelburg, S.; Liu, Y.; Liu, B.; Sauder, K.; Stidham, R.W.; Zhu, J.; Higgins, P.D.R. Development and Validation of Machine Learning Models in Prediction of Remission in Patients With Moderate to Severe Crohn Disease. JAMA Netw. Open. 2019, 2, e193721. [Google Scholar] [CrossRef]
- He, M.; Li, C.; Tang, W.; Kang, Y.; Zuo, Y.; Wang, Y. Machine learning gene expression predicting model for ustekinumab response in patients with Crohn’s disease. Immun. Inflamm. Dis. 2021, 9, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- Waljee, A.K.; Sauder, K.; Patel, A.; Segar, S.; Liu, B.; Zhang, Y.; Zhu, J.; Stidham, R.W.; Balis, U.; Higgins, P.D.R. Machine Learning Algorithms for Objective Remission and Clinical Outcomes with Thiopurines. J. Crohns Colitis. 2017, 11, 801–810. [Google Scholar] [CrossRef]
- Waljee, A.K.; Liu, B.; Sauder, K.; Zhu, J.; Govani, S.M.; Stidham, R.W.; Higgins, P.D.R. Predicting Corticosteroid-Free Biologic Remission with Vedolizumab in Crohn’s Disease. Inflamm. Bowel Dis. 2018, 24, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Dulai, P.S.; Amiot, A.; Peyrin-Biroulet, L.; Jairath, V.; Serrero, M.; Filippi, J.; Singh, S.; Pariente, B.; Loftus, E.V.; Roblin, X.; et al. A clinical decision support tool may help to optimise vedolizumab therapy in Crohn’s disease. Aliment. Pharmacol. Ther. 2020, 51, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Dulai, P.S.; Wan, Y.; Huang, Z.; Luo, M. Probability of Response as Defined by a Clinical Decision Support Tool Is Associated With Lower Healthcare Resource Utilization in Vedolizumab-Treated Patients With Crohn’s Disease. Crohns. Colitis 360 2022, 4, otac048. [Google Scholar] [CrossRef]
- Venkatapurapu, S.P.; Iwakiri, R.; Udagawa, E.; Patidar, N.; Qi, Z.; Takayama, R.; Kumar, K.; Sato, Y.; Behar, M.; Offner, P.; et al. A Computational Platform Integrating a Mechanistic Model of Crohn’s Disease for Predicting Temporal Progression of Mucosal Damage and Healing. Adv. Ther. 2022, 39, 3225–3247. [Google Scholar] [CrossRef]
- Iacucci, M.; Jeffery, L.; Acharjee, A.; Grisan, E.; Buda, A.; Nardone, O.M.; Smith, S.C.L.; Labarile, N.; Zardo, D.; Ungar, B.; et al. Computer-Aided Imaging Analysis of Probe-Based Confocal Laser Endomicroscopy With Molecular Labeling and Gene Expression Identifies Markers of Response to Biological Therapy in IBD Patients: The Endo-Omics Study. Inflamm. Bowel Dis. 2023, 29, 1409–1420. [Google Scholar] [CrossRef]
- Vande Casteele, N.; Leighton, J.A.; Pasha, S.F.; Cusimano, F.; Mookhoek, A.; Hagen, C.E.; Rosty, C.; Pai, R.K.; Pai, R.K. Utilizing Deep Learning to Analyze Whole Slide Images of Colonic Biopsies for Associations Between Eosinophil Density and Clinicopathologic Features in Active Ulcerative Colitis. Inflamm. Bowel Dis. 2022, 28, 539–546. [Google Scholar] [CrossRef]
- Reigle, J.; Lopez-Nunez, O.; Drysdale, E.; Abuquteish, D.; Liu, X.; Putra, J.; Erdman, L.; Griffiths, A.M.; Prasath, S.; Siddiqui, I.; et al. Using Deep Learning to Automate Eosinophil Counting in Pediatric Ulcerative Colitis Histopathological Images. MedRxiv. 2024. [Google Scholar] [CrossRef]
- Ohara, J.; Nemoto, T.; Maeda, Y.; Ogata, N.; Kudo, S.E.; Yamochi, T. Deep learning-based automated quantification of goblet cell mucus using histological images as a predictor of clinical relapse of ulcerative colitis with endoscopic remission. J. Gastroenterol. 2022, 57, 962–970. [Google Scholar] [CrossRef]
- Iacucci, M.; Parigi, T.L.; Del Amor, R.; Meseguer, P.; Mandelli, G.; Bozzola, A.; Bazarova, A.; Bhandari, P.; Bisschops, R.; Danese, S.; et al. Artificial Intelligence Enabled Histological Prediction of Remission or Activity and Clinical Outcomes in Ulcerative Colitis. Gastroenterology 2023, 164, 1180–1188.e2. [Google Scholar] [CrossRef]
- Gui, X.; Bazarova, A.; Del Amor, R.; Vieth, M.; de Hertogh, G.; Villanacci, V.; Zardo, D.; Parigi, T.L.; Røyset, E.S.; Shivaji, U.N.; et al. PICaSSO Histologic Remission Index (PHRI) in ulcerative colitis: Development of a novel simplified histological score for monitoring mucosal healing and predicting clinical outcomes and its applicability in an artificial intelligence system. Gut 2022, 71, 889–898. [Google Scholar] [CrossRef]
- Zand, A.; Sharma, A.; Stokes, Z.; Reynolds, C.; Montilla, A.; Sauk, J.; Hommes, D. An Exploration Into the Use of a Chatbot for Patients With Inflammatory Bowel Diseases: Retrospective Cohort Study. J. Med. Internet. Res. 2020, 22, e15589. [Google Scholar] [CrossRef] [PubMed]
- Biasci, D.; Lee, J.C.; Noor, N.M.; Pombal, D.R.; Hou, M.; Lewis, N.; Ahmad, T.; Hart, A.; Parkes, M.; McKinney, E.F.; et al. A blood-based prognostic biomarker in IBD. Gut 2019, 68, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Cushing, K.C.; Mclean, R.; McDonald, K.G.; Gustafsson, J.K.; Knoop, K.A.; Kulkarni, D.H.; Sartor, R.B.; Newberry, R.D. Predicting Risk of Postoperative Disease Recurrence in Crohn’s Disease: Patients With Indolent Crohn’s Disease Have Distinct Whole Transcriptome Profiles at the Time of First Surgery. Inflamm. Bowel Dis. 2019, 25, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xu, L.; Fan, Y.; Xiang, P.; Gao, X.; Chen, Y.; Zhang, W.; Ge, Q. A novel surgical predictive model for Chinese Crohn’s disease patients. Medicine 2019, 98, e17510. [Google Scholar] [CrossRef]
- Morilla, I.; Uzzan, M.; Laharie, D.; Cazals-Hatem, D.; Denost, Q.; Daniel, F.; Belleannee, G.; Bouhnik, Y.; Wainrib, G.; Panis, Y.; et al. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2019, 17, 905–913. [Google Scholar] [CrossRef]
- Maeda, Y.; Kudo, S.E.; Ogata, N.; Misawa, M.; Mori, Y.; Mori, K.; Ohtsuka, K. Can artificial intelligence help to detect dysplasia in patients with ulcerative colitis? Endoscopy 2021, 53, E273–E274. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Hotta, K.; Ohtsuka, K.; Matsuda, T.; Saito, S.; Kudo, T.; Baba, T.; Ishida, F.; et al. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointest. Endosc. 2021, 93, 960–967.e3. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kinugasa, H.; Hamada, K.; Tomiya, M.; Tanimoto, T.; Ohto, A.; Toda, A.; Takei, D.; Matsubara, M.; Suzuki, S.; et al. The diagnostic ability to classify neoplasias occurring in inflammatory bowel disease by artificial intelligence and endoscopists: A pilot study. J. Gastroenterol. Hepatol. 2022, 37, 1610–1616. [Google Scholar] [CrossRef]
- Rymarczyk, D.; Schultz, W.; Borowa, A.; Friedman, J.R.; Danel, T.; Branigan, P.; Chałupczak, M.; Bracha, A.; Krawiec, T.; Warchoł, M.; et al. Deep Learning Models Capture Histological Disease Activity in Crohn’s Disease and Ulcerative Colitis with High Fidelity. J. Crohns Colitis. 2024, 18, 604–614. [Google Scholar] [CrossRef]
- Matalka, I.I.; Al-Omari, F.A.; Salama, R.M.; Mohtaseb, A.H. A novel approach for quantitative assessment of mucosal damage in inflammatory bowel disease. Diagn. Pathol. 2013, 8, 156. [Google Scholar] [CrossRef]
- Ohara, J.; Maeda, Y.; Ogata, N.; Kuroki, T.; Misawa, M.; Kudo, S.E.; Nemoto, T.; Yamochi, T.; Iacucci, M. Automated Neutrophil Quantification and Histological Score Estimation in Ulcerative Colitis. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2024, 23, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Del Amor, R.; Meseguer, P.; Parigi, T.L.; Villanacci, V.; Colomer, A.; Launet, L.; Bazarova, A.; Tontini, G.E.; Bisschops, R.; de Hertogh, G.; et al. Constrained multiple instance learning for ulcerative colitis prediction using histological images. Comput. Methods Programs Biomed. 2022, 224, 107012. [Google Scholar] [CrossRef] [PubMed]
- Peyrin-Biroulet, L.; Adsul, S.; Stancati, A.; Dehmeshki, J.; Kubassova, O. An artificial intelligence-driven scoring system to measure histological disease activity in ulcerative colitis. United Eur. Gastroenterol. J. 2024, 12, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Najdawi, F.; Sucipto, K.; Mistry, P.; Hennek, S.; Jayson, C.K.B.; Lin, M.; Fahy, D.; Kinsey, S.; Wapinski, I.; Beck, A.H.; et al. Artificial Intelligence Enables Quantitative Assessment of Ulcerative Colitis Histology. Mod. Pathol. Off. 2023, 36, 100124. [Google Scholar] [CrossRef]
- Kiyokawa, H.; Abe, M.; Matsui, T.; Kurashige, M.; Ohshima, K.; Tahara, S.; Nojima, S.; Ogino, T.; Sekido, Y.; Mizushima, T.; et al. Deep Learning Analysis of Histologic Images from Intestinal Specimen Reveals Adipocyte Shrinkage and Mast Cell Infiltration to Predict Postoperative Crohn Disease. Am. J. Pathol. 2022, 192, 904–916. [Google Scholar] [CrossRef]
- Rubin, D.T.; Kubassova, O.; Weber, C.R.; Adsul, S.; Freire, M.; Biedermann, L.; Koelzer, V.H.; Bressler, B.; Xiong, W.; Niess, J.H.; et al. Deployment of an Artificial Intelligence Histology Tool to Aid Qualitative Assessment of Histopathology Using the Nancy Histopathology Index in Ulcerative Colitis. Inflamm. Bowel Dis. 2024, 31, izae204. [Google Scholar] [CrossRef] [PubMed]
- Guerrero Vinsard, D.; Fetzer, J.R.; Agrawal, U.; Singh, J.; Damani, D.N.; Sivasubramaniam, P.; Poigai Arunachalam, S.; Leggett, C.; Raffals, L.E.; Coelho-Prabhu, N. Development of an artificial intelligence tool for detecting colorectal lesions in inflammatory bowel disease. iGIE 2023, 2, 91–101.e6. [Google Scholar] [CrossRef]
- Abdelrahim, M.; Siggens, K.; Iwadate, Y.; Maeda, N.; Htet, H.; Bhandari, P. New AI model for neoplasia detection and characterisation in inflammatory bowel disease. Gut 2024, 73, 725–728. [Google Scholar] [CrossRef]
- Stidham, R.W.; Liu, Y.; Enchakalody, B.; Van, T.; Krishnamurthy, V.; Su, G.L.; Zhu, J.; Waljee, A.K. The Use of Readily Available Longitudinal Data to Predict the Likelihood of Surgery in Crohn Disease. Inflamm. Bowel Dis. 2021, 27, 1328–1334. [Google Scholar] [CrossRef]
- Majumder, S.; Santacroce, G.; Maeda, Y.; Zammarchi, I.; Puga-Tejada, M.; Ditonno, I.; Hayes, B.; Crotty, R.; Fennell, E.; Shivaji, U.N.; et al. Endocytoscopy with automated multispectral intestinal barrier pathology imaging for assessment of deep healing to predict outcomes in ulcerative colitis. Gut 2024, 73, 1603–1606. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kudo, S.E.; Ogata, N.; Misawa, M.; Iacucci, M.; Homma, M.; Nemoto, T.; Takishima, K.; Mochida, K.; Miyachi, H.; et al. Evaluation in real-time use of artificial intelligence during colonoscopy to predict relapse of ulcerative colitis: A prospective study. Gastrointest. Endosc. 2022, 95, 747–756.e2. [Google Scholar] [CrossRef]
- Omori, T.; Yamamoto, T.; Murasugi, S.; Koroku, M.; Yonezawa, M.; Nonaka, K.; Nagashima, Y.; Nakamura, S.; Tokushige, K. Comparison of Endoscopic and Artificial Intelligence Diagnoses for Predicting the Histological Healing of Ulcerative Colitis in a Real-World Clinical Setting. Crohns Colitis. 2024, 6, otae005. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, T.; Maeda, Y.; Kudo, S.E.; Ogata, N.; Iacucci, M.; Takishima, K.; Ide, Y.; Shibuya, T.; Semba, S.; Kawashima, J.; et al. A novel artificial intelligence-assisted “vascular healing” diagnosis for prediction of future clinical relapse in patients with ulcerative colitis: A prospective cohort study (with video). Gastrointest. Endosc. 2024, 100, 97–108. [Google Scholar] [CrossRef]
- Turner, D.; Ricciuto, A.; Lewis, A.; D’Amico, F.; Dhaliwal, J.; Griffiths, A.M.; Bettenworth, D.; Sandborn, W.J.; Sands, B.E.; Reinisch, W.; et al. International Organization for the Study of IBD. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology 2021, 160, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Hashash, J.G.; Yu Ci Ng, F.; Farraye, F.A.; Wang, Y.; Colucci, D.R.; Baxi, S.; Muneer, S.; Reddan, M.; Shingru, P.; Melmed, G.Y. Inter- and Intraobserver Variability on Endoscopic Scoring Systems in Crohn’s Disease and Ulcerative Colitis: A Systematic Review and Meta-Analysis. Inflamm. Bowel Dis. 2024, 30, 2217–2226. [Google Scholar] [CrossRef]
- Stidham, R.W.; Cai, L.; Cheng, S.; Rajaei, F.; Hiatt, T.; Wittrup, E.; Rice, M.D.; Bishu, S.; Wehkamp, J.; Schultz, W.; et al. Using Computer Vision to Improve Endoscopic Disease Quantification in Therapeutic Clinical Trials of Ulcerative Colitis. Gastroenterology 2024, 166, 155–167.e2. [Google Scholar] [CrossRef]
- Takenaka, K.; Fujii, T.; Kawamoto, A.; Suzuki, K.; Shimizu, H.; Maeyashiki, C.; Yamaji, O.; Motobayashi, M.; Igarashi, A.; Hanazawa, R.; et al. Deep neural network for video colonoscopy of ulcerative colitis: A cross-sectional study. Lancet Gastroenterol. Hepatol. 2022, 7, 230–237. [Google Scholar] [CrossRef]
- Rimondi, A.; Gottlieb, K.; Despott, E.J.; Iacucci, M.; Murino, A.; Tontini, G.E. Can artificial intelligence replace endoscopists when assessing mucosal healing in ulcerative colitis? A systematic review and diagnostic test accuracy meta-analysis. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2024, 56, 1164–1172. [Google Scholar] [CrossRef]
- Lv, B.; Ma, L.; Shi, Y.; Tao, T.; Shi, Y. A systematic review and meta-analysis of artificial intelligence-diagnosed endoscopic remission in ulcerative colitis. iScience 2023, 26, 108120. [Google Scholar] [CrossRef] [PubMed]
- Jahagirdar, V.; Bapaye, J.; Chandan, S.; Ponnada, S.; Kochhar, G.S.; Navaneethan, U.; Mohan, B.P. Diagnostic accuracy of convolutional neural network-based machine learning algorithms in endoscopic severity prediction of ulcerative colitis: A systematic review and meta-analysis. Gastrointest. Endosc. 2023, 98, 145–154.e8. [Google Scholar] [CrossRef] [PubMed]
- Sipponen, T.; Nuutinen, H.; Turunen, U.; Färkkilä, M. Endoscopic evaluation of Crohn’s disease activity: Comparison of the CDEIS and the SES-CD. Inflamm. Bowel Dis. 2010, 16, 2131–2136. [Google Scholar] [CrossRef]
- Leenhardt, R.; Buisson, A.; Bourreille, A.; Marteau, P.; Koulaouzidis, A.; Li, C.; Keuchel, M.; Rondonotti, E.; Toth, E.; Plevris, J.N.; et al. Nomenclature and semantic descriptions of ulcerative and inflammatory lesions seen in Crohn’s disease in small bowel capsule endoscopy: An international Delphi consensus statement. United Eur. Gastroenterol. J. 2020, 8, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Mohan, B.P.; Khan, S.R.; Kassab, L.L.; Ponnada, S.; Chandan, S.; Ali, T.; Dulai, P.S.; Adler, D.G.; Kochhar, G.S. High pooled performance of convolutional neural networks in computer-aided diagnosis of GI ulcers and/or hemorrhage on wireless capsule endoscopy images: A systematic review and meta-analysis. Gastrointest. Endosc. 2021, 93, 356–364.e4. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Picetti, D.; Dulai, P.S.; Jairath, V.; Sandborn, W.J.; Ohno-Machado, L.; Chen, P.L.; Singh, S. Machine Learning-based Prediction Models for Diagnosis and Prognosis in Inflammatory Bowel Diseases: A Systematic Review. J. Crohns Colitis. 2022, 16, 398–413. [Google Scholar] [CrossRef]
- Gordon, H.; Minozzi, S.; Kopylov, U.; Verstockt, B.; Chaparro, M.; Buskens, C.; Warusavitarne, J.; Agrawal, M.; Allocca, M.; Atreya, R.; et al. ECCO Guidelines on Therapeutics in Crohn’s Disease: Medical Treatment. J. Crohns Colitis. 2024, 18, 1531–1555. [Google Scholar] [CrossRef]
- Brooks-Warburton, J.; Ashton, J.; Dhar, A.; Tham, T.; Allen, P.B.; Hoque, S.; Lovat, L.B.; Sebastian, S. Artificial intelligence and inflammatory bowel disease: Practicalities and future prospects. Frontline Gastroenterol. 2022, 13, 325–331. [Google Scholar] [CrossRef]
- Yin, A.L.; Hachuel, D.; Pollak, J.P.; Scherl, E.J.; Estrin, D. Digital Health Apps in the Clinical Care of Inflammatory Bowel Disease: Scoping Review. J. Med. Internet. Res. 2019, 21, e14630. [Google Scholar] [CrossRef]
- Jagannath, B.; Muthukumar, S.; Prasad, S. Wearable Sweat Sensing Device For Detection Of Ibd Biomarkers. Inflamm. Bowel Dis. 2021, 27, S12. [Google Scholar] [CrossRef]
- Cross, R.K.; Langenberg, P.; Regueiro, M.; Schwartz, D.A.; Tracy, J.K.; Collins, J.F.; Katz, J.; Ghazi, L.; Patil, S.A.; Quezada, S.M.; et al. A Randomized Controlled Trial of TELEmedicine for Patients with Inflammatory Bowel Disease (TELE-IBD). Am. J. Gastroenterol. 2019, 114, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Puga-Tejada, M.; Majumder, S.; Maeda, Y.; Zammarchi, I.; Ditonno, I.; Santacroce, G.; Capobianco, I.; Robles-Medranda, C.; Ghosh, S.; Iacucci, M. Artificial intelligence–enabled histology exhibits comparable accuracy to pathologists in assessing histological remission in ulcerative colitis: A systematic review, meta-analysis, and meta-regression. J. Crohns Colitis. 2025, 19, jjae198. [Google Scholar] [CrossRef]
- Bajwa, J.; Munir, U.; Nori, A.; Williams, B. Artificial intelligence in healthcare: Transforming the practice of medicine. Futur. Healthc. J. 2021, 8, e188–e194. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health. 2019, 1, e271–e297. [Google Scholar] [CrossRef] [PubMed]
- Kröner, P.T.; Engels, M.M.; Glicksberg, B.S.; Johnson, K.W.; Mzaik, O.; van Hooft, J.E.; Wallace, M.B.; El-Serag, H.B.; Krittanawong, C. Artificial intelligence in gastroenterology: A state-of-the-art review. World J. Gastroenterol. 2021, 27, 6794–6824. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef]
- Tontini, G.E.; Rimondi, A.; Vernero, M.; Neumann, H.; Vecchi, M.; Bezzio, C.; Cavallaro, F. Artificial intelligence in gastrointestinal endoscopy for inflammatory bowel disease: A systematic review and new horizons. Ther. Adv. Gastroenterol. 2021, 14, 17562848211017730. [Google Scholar] [CrossRef]
- Sedano, R.; Hogan, M.; McDonald, C.; Aswani-Omprakash, T.; Ma, C.; Jairath, V. Underrepresentation of Minorities and Lack of Race Reporting in Ulcerative Colitis Drug Development Clinical Trials. Inflamm. Bowel Dis. 2022, 28, 1293–1295. [Google Scholar] [CrossRef]
- Sedano, R.; Hogan, M.; Mcdonald, C.; Aswani-Omprakash, T.; Ma, C.; Jairath, V. Underrepresentation of Minorities and Underreporting of Race and Ethnicity in Crohn’s Disease Clinical Trials. Gastroenterology 2022, 162, 338–340.e2. [Google Scholar] [CrossRef]
- Shah, S.; Shillington, A.C.; Kabagambe, E.K.; Deering, K.L.; Babin, S.; Capelouto, J.; Pulliam, C.; Patel, A.; LaChappelle, B.; Liu, J. Racial and Ethnic Disparities in Patients With Inflammatory Bowel Disease: An Online Survey. Inflamm. Bowel Dis. 2024, 30, 1467–1474. [Google Scholar] [CrossRef]
- Liu, X.; Cruz Rivera, S.; Moher, D.; Calvert, M.J.; Denniston, A.K.; SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: The CONSORT-AI extension. Lancet Digit. Health. 2020, 2, e537–e548. [Google Scholar] [CrossRef] [PubMed]
- Ehsani-Moghaddam, B.; Martin, K.; Queenan, J.A. Data quality in healthcare: A report of practical experience with the Canadian Primary Care Sentinel Surveillance Network data. Health Inf. Manag. J. Health Inf. Manag. Assoc. Aust. 2021, 50, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Nazer, L.H.; Zatarah, R.; Waldrip, S.; Chen Ke, J.X.; Moukheiber, M.; Khanna, A.K.; Hicklen, R.S.; Moukheiber, L.; Moukheiber, D.; Ma, H.; et al. Bias in artificial intelligence algorithms and recommendations for mitigation. PLoS Digit. Health 2023, 2, e0000278. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Soltan, A.A.S.; Eyre, D.W.; Clifton, D.A. Algorithmic fairness and bias mitigation for clinical machine learning with deep reinforcement learning. Nat. Mach. Intell. 2023, 5, 884–894. [Google Scholar] [CrossRef]
- Health C for D and, R. Artificial Intelligence and Machine Learning in Software as a Medical Device; FDA: Silver Spring, MD, USA, 2025.
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef]
- Fraser, A.G.; Biasin, E.; Bijnens, B.; Bruining, N.; Caiani, E.G.; Cobbaert, K.; Davies, R.H.; Gilbert, S.H.; Hovestandt, L.; Kamenjasevic, E.; et al. Artificial intelligence in medical device software and high-risk medical devices-a review of definitions, expert recommendations and regulatory initiatives. Expert. Rev. Med. Devices. 2023, 20, 467–491. [Google Scholar] [CrossRef]
- Sedano, R.; Solitano, V.; Vuyyuru, S.K.; Yuan, Y.; Hanžel, J.; Ma, C.; Nardone, O.M.; Jairath, V. Artificial intelligence to revolutionize IBD clinical trials: A comprehensive review. Ther. Adv. Gastroenterol. 2025, 18, 17562848251321915. [Google Scholar] [CrossRef]
| Study | Field of Application | AIM | Outcome |
|---|---|---|---|
| Sasaki et al., 2003 [20] | Endoscopic activity | Matts score was characterised using mucosal redness parameters, considered proportional to the histological microvascular bed and to disease activity | The algorithm was able to differentiate Matts 1 from Matts 2, Matts 2 from Matts 3, and Matts 3 from Matts 4 with high sensitivity and specificity |
| WLE | |||
| Kraszewski et al., 2021 [21] | Diagnosis | ML model based on routinely performed laboratory blood, urine, and faecal tests to diagnose IBD | The model could diagnose CD and UC with an average accuracy of 97% and 91%, respectively |
| Bossuyt et al., 2020 [22] | Endoscopic activity | Application of a new algorithm (Red Density) based on the red channel and vessel pattern detection on UC patients | The algorithm significantly correlated with MES, UCEIS and RHI (r 0.76, 0.74, and 0.74, p < 0.01, respectively) |
| WLE | |||
| Stidham et al., 2019 [23] | Endoscopic activity | Analysis of DL for distinguishing moderate to severe UC from remission compared with multiple expert reviewers | The CNN demonstrated excellent performance in distinguishing MES 0–1 from MES 2–3 and good agreement between expert reviewers (κ = 0.86) |
| WLE | |||
| Fan et al., 2023 [24] | Endoscopic activity | Application of DL for objective scoring of endoscopic images and videos in UC patients | The CNN exhibited good accuracy for MES and UCEIS, with a very good agreement (k 0.8) with endoscopists’ scores |
| WLE | |||
| Ozawa et al., 2019 [25] | Endoscopic activity | Application of a CNN to evaluate MES in endoscopic pictures from UC patients | The CNN demonstrated a high level of performance with an AUROC of 0.86 and 0.98 for identifying Mayo 0 and 0–1 |
| WLE | |||
| Takabayashi et al., 2024 [26] | Endoscopic activity | Application of an AI system to assess endoscopic severity of UC | The correlation coefficients between IBD expert endoscopists and the AI of the evaluation results were all higher than 0.95 |
| WLE | |||
| Takenaka et al., 2020 [27] | Endoscopic/histological activity | Application of a DNN to assess both endoscopic (UCEIS) and histopathological (Geboes score) disease activity | The DNN showed 90% and 93% accuracy for endoscopic and histological remission, respectively; the intraclass correlation coefficients between DNN and experienced endoscopists were 0.917 and 0.859, respectively |
| WLE | |||
| Takenaka et al., 2022 [28] | Endoscopic/histological activity | The same group refined the previous algorithm to assess disease activity directly on videos | For predicting histological remission, the DNUC had a sensitivity of 97.9% and a specificity of 94.6%. The intraclass correlation coefficient between DNUC and experts for endoscopic scoring was 0.927 |
| WLE | |||
| Yao et al., 2021 [29] | Endoscopic activity | To pilot a fully automated video analysis system for grading UC endoscopic disease | The CNN performed better in automatically scoring the local high-resolution video (κ = 0.84) but less well in the unadjusted analysis of the external patient cohort |
| WLE | |||
| Gottlieb et al., 2021 [30] | Endoscopic activity | Application of a CNN system to assess mucosal activity according to MES and UCEIS on videos | Agreement with expert readers was excellent for both MES and UCEIS (0.844 and 0.855, respectively). Model performance was best for MES scores 0 and 3 and worst for MES scores 1 and 2 |
| WLE | |||
| Iacucci et al., 2023 [31] | Endoscopic/histological activity | Application of a new CNN to evaluate endoscopic and histological activity on videos in WLE and VCE of the multicentre Picasso study | The algorithm showed a sensitivity, specificity and AUROC of 72%, 87% and 0.85 for WLE, and of 79%, 95% and 0.94 for VCE |
| WLE-advanced imaging | |||
| Charisis and Hadjileontiadis, 2016 [32] | CE | Application of an AI system (HFA DLac) for describing and detecting CD-associated lesions in CE | The accuracy ranged from 81.2% in mild lesions to 93.8% in severe lesions (total 90.5%) |
| Fan et al., 2018 [33] | CE | Application of a CNN to detect small intestinal ulcer and erosion in CE | Ulcer and erosion detection reached a high accuracy of 95.16% and 95.34%, sensitivity of 96.80% and 93.67%, and specificity of 94.79% and 95.98%, correspondingly, an AUROC of 0.98 in both of the network |
| Afonso et al., 2022 [34] | CE | A CNN for the automatic identification of small intestinal ulcers and erosions | The model was able to detect and distinguish ulcers and erosions with an accuracy of 95.6%, sensitivity of 90.8%, and a specificity of 97.1% |
| Aoki et al., 2019 [35] | CE | Application of a CNN created to detect CD ulcers or erosions on CE images | The evaluation was completed in just under 4 min with a sensitivity of 88%, a specificity of 99%, and an overall AUROC of 0.99 |
| Ferreira et al., 2022 [36] | CE | An AI algorithm for the automatic detection of ulcers and erosions of the small intestine and colon in PillCam™ Crohn’s Capsule images | The model had a sensitivity of 98.0% and a specificity of 99.0%. The overall accuracy of the network was 98.8%. The AUROC for detection of ulcers and erosions in PCC images was 1.00 |
| Kratter et al., 2022 [37] | CE | Application of a combined model for two different capsules | The combined model achieved an average AUC of 0.99 and average mean patient accuracy of 0.974 |
| Brodersen, 2024 [38] | CE | Application of the deep learning solution AXARO on panenteric capsules endoscopy | AXARO reduced the initial review time maintaining high diagnostic accuracy |
| Klang et al., 2020 [37] | CE | Evaluation of a DL algorithm for the automated detection of small-bowel ulcers in CD on CE | ANNs trained on CE images can detect small bowel ulcers with approximately 95% accuracy |
| Klang et al., 2021 [39] | CE | DL applied on CE images for identification of CD intestinal strictures | DL provided excellent differentiation between strictures vs. normal mucosa, and strictures vs. ulcers |
| Barash et al., 2021 [40] | CE | Development of DL algorithm for automated grading of CD ulcers on CE | CNN-assisted CE readings have high potential in classifying ulcers in CD |
| Ding et al., 2019 [41] | CE | Development of a CNN-based algorithm to assist in the evaluation of CE images | The CNN identified abnormalities with 99.90% sensitivity. The mean reading time per patient was 96.6 ± 22.53 min by conventional reading vs. 5.9 ± 2.23 min by CNN |
| Aoki et al., 2020 [42] | CE | To examine if AI systems can reduce the reading time of endoscopists without decreasing the detection rate of mucosal breaks | AI reduced reading time from 12.2 min to 3.1 for experienced examiners and from 20.7 to 5.2 for trainees, without affecting overall accuracy |
| Quénéhervé et al., 2019 [43] | CE | Evaluation of the potential of AI-guided CE diagnosis in a retrospective analysis of IBD patient | Excellent accuracy was obtained for the diagnosis of IBD (sensitivity and specificity of 100%) and for the differentiation of UC from CD (sensitivity of 92%, specificity of 91%) |
| Maeda et al., 2019 [44] | Histological activity | Evaluation of a CAD system to predict persistent histologic inflammation from endocytoscopy, validated on UC patients | The CAD system showed a sensitivity, specificity, and accuracy of 74%, 97%, and 91%, respectively; it also predicted clinical recurrence at 12 months, finding that this was at a higher rate in the AI-histologically active group (28.4 vs. 4.9%, p < 0.001) |
| Advanced imaging | |||
| Bossuyt et al., 2021 [45] | Histological activity | Application of a CAD technique to assess histologic remission on images obtained from a Single-wavelength endoscope | The CAD algorithm successfully predicted histologic remission of UC with high accuracy (86%) |
| Advanced imaging | |||
| Sinonquel et al., 2024 [46] | Histological activity | Evaluation of histological activity using a CAD system based on either WLE or SWE. | SWE-CAD exceeded the accuracy of WLE-CAD; it showed an accuracy of 95.2%, sensitivity of 96.4%, and specificity of 92.9% |
| WLE-advanced imaging | |||
| Con et al., 2021 [47] | Response to therapy | A deep learning model developed to predict response to anti-TNF therapy in CD patients, using the CRP biomarker | ML methods showed stronger predictive performance than the conventional statistical model (AuROC; 0.754 [95% CI: 0.674–0.834] vs. 0.659 [95% CI: 0.562–0.756]; p = 0.036) |
| Popa et al., 2020 [48] | Response to therapy | AI algorithm to predict clinical remission in UC patients on anti-TNF therapy, using clinical and endoscopic data | This system showed a well-performing ROC curve (PPV 100%, NPV 100%; p < 0.001), with ability to differentiate those who will achieve clinical remission from those who will have active disease |
| WLE | |||
| Park et al., 2022 [49] | Response to therapy | A ML model using transcriptome imputed from genotypes to predict non-durable response to anti-TNF treatment in CD | Imputed gene expression characteristics in machine learning models successfully predicted a non-durable response to anti-TNF |
| Waljee et al., 2019 [50] | Response to therapy | A ML models in prediction response to ustekinumab in CD patients | Predictions of remission at week 42 had a sensitivity and specificity of 0.79 and 0.67 using week 8 post-treatment data |
| He et al., 2021 [51] | Response to therapy | A ML model based on the expression of four gene to predict response to ustekinumab in CD patients | The AuROC of the model for the training and testing datasets was 0.746 and 0.734 respectively |
| Waljee et al., 2017 [52] | Response to therapy | ML model to predict response to thiopurines | The AuROC for remission predicted by the algorithm was 0.79 |
| Waljee et al., 2018 [53] | Response to therapy | ML models for UC patients to predict clinical remission to vedolizumab at week 52 | The model showed a sensitivity and specificity of 0.76 and 0.71, respectively. Furthermore, the model was also able to predict therapeutic failure in 95.3% of patients using week 6 data and in 88% of cases using only pre-treatment data |
| Dulai et al., 2020 [54] | Response to therapy | A CDST was created for predicting treatment effectiveness of vedolizumab in CD using data from GEMINI 2 study | A linear relationship existed between CDST-defined groups, measured vedolizumab exposure, rapidity of onset of action and efficacy in GEMINI through week 52 |
| Dulai et al., 2022 [55] | Response to therapy | A CDST identified patients with CD most likely to respond to vedolizumab and to predict real-world healthcare resource utilisation (HRU) | CDSTs identified lower rates of surgery or hospitalisation in CD patients with higher probability of vedolizumab response |
| Venkatapurapu et al., 2022 [56] | Response to therapy | A platform to predict endoscopic remission and mucosal healing after vedolizumab treatment | The model predicted endoscopic remission and mucosal healing for treatment with vedolizumab over 26 weeks, with overall sensitivities of 80% and 75% and overall specificities of 69% and 70%, respectively |
| Iacucci et al., 2023 [57] | Response to therapy | A model to predict response to biologics in IBD using pCLE in vivo and assess the binding of fluorescent-labelled biologics ex vivo | Higher mucosal binding of the drug target is associated with response to therapy in UC. In vivo, mucosal and microvascular changes detected by pCLE are associated with response to biologics in inflammatory bowel disease |
| Advanced imaging | |||
| Takenaka et al., 2021 [28] | Histological activity | In a previous study a deep neural network system based on endoscopic images of UC (DNUC) predicted histologic remission. In this follow-up study, it was evaluated if DNUC could predict patient prognosis | The DNUC could predict patient prognosis, and its predictive value was comparable with that of assessments by experts |
| WLE | |||
| Vande Casteele et al., 2022 [58] | Histological activity | A DL algorithm to quantify eosinophils in colonic biopsies | The model had sensitivity 0.86, specificity 0.91, accuracy 0.89 |
| WLE | |||
| Reigle et al., 2024 [59] | Histological activity | Application of a DL to automate eosinophil counting | The inter-rater reliability was 0.96 (95% CI: 0.93–0.97). The correlation between two pathologists and the algorithm was 0.89 (95% CI: 0.82–0.94) and 0.88 (95% CI: 0.80–0.94), respectively |
| Ohara et al., 2022 [60] | Histological activity | DL-based models were trained to detect goblet cell mucus area from whole slide images of biopsy specimens | The model had sensitivity 0.83, specificity 0.99, accuracy 0.97 |
| WLE | |||
| Iacucci et al., 2023 [61] | Histological activity | The PHRI was applied to a computer-assisted diagnostic system | When comparing the AI-generated assessment results with those generated by pathologists, the AI model was found to be highly sensitive and specific in determining the presence of neutrophils |
| WLE-advanced imaging | |||
| Gui et al., 2022 [62] | Histological activity | Evaluation of the applicability of the PHRI in an AI system on 614 biopsies from 307 UC patients | The algorithm showed a sensitivity of 78%, a specificity of 91.7%, and an accuracy of 86% in determining the presence or absence of neutrophils |
| WLE-advanced imaging | |||
| Zand et al., 2020 [63] | Response to therapy | Evaluation of a natural language processing (NLP)-based chatbot to categorise IBD patients’ electronic messages into various categories | The agreement between the algorithm and clinicians was 95% |
| Biasci et al., 2019 [64] | Risk stratification | ML models using RNA expression levels from whole blood samples to perform risk stratification | The ML model identified high- and low-risk groups for future dose escalations in both CD (75% vs. 35%) and UC (60% vs. 20%) |
| Cushing et al., 2019 [65] | Risk stratification | ML model created to predict 1-year relapse risk in CD after surgery, using non-invasive markers | Anti-TNF exposed patients with indolent postoperative courses were found to have a transcriptome signature distinct from those with aggressive disease |
| Stidham et al., 2021 [13] | Risk stratification | ML model developed to predict the risk of surgery in patients with CD | Anti-TNF therapy is the strongest predictor associated with a lower risk of surgery within 1 year |
| Dong et al., 2019 [66] | Risk stratification | ML model developed to predict the risk of surgery in patients with CD | Using variables such as age, sex, smoking status, perianal disease, previous surgical resection, the ML model showed higher accuracy and precision than the statistical model |
| Morilla et al., 2019 [67] | Response to therapy | A neural network was created that combining data from a pool of 3391 miRNA candidates and clinical factors in patients with ASUC | It effectively distinguished medical responders from non-responders with 97% accuracy; the miRNA-only model had 94% accuracy |
| Maeda et al., 2020 [68] | Detection of colonic neoplasm | Evaluation of an AI-assisted detection of colitis-associated neoplasms | The first case report in which an AI system detected colitis-associated neoplasms |
| Advanced imaging | |||
| Misawa et al., 2021 [69] | Detection of colonic neoplasm | Applications of EndoBrain-EYE in IBD patients | Two flat lesions with low-grade dysplasia were clearly highlighted by EndoBRAIN-EYE |
| WLE | |||
| Yamamoto et al., 2022 [70] | Detection of colonic neoplasm | Evaluation of an AI system for characterising neoplasia occurring in IBD | AI diagnostic accuracy of the AI model was higher than experts and non-experts (nonexperts, 77.8%; experts, 75.8%; AI model, 79.0%) |
| WLE | |||
| Rymarczyk et al., 2023 [71] | Histological activity | Evaluation of a DL models for automating histological assessments in IBD | AI-modelled GHAS and Geboes subgrades matched central readings with moderate to substantial agreement, with accuracies ranging from 65% to 89% |
| Matalka et al., 2013 [72] | Histological activity | Evaluation of a novel automated system to assess mucosal damage and architectural distortion in IBD | The developed system achieved an overall precision of 98.31% |
| Ohara et al., 2024 [73] | Histological activity | Evaluation of an AI system to detect neutrophils in UC biopsy specimens | The model achieved a performance of 0.77, 0.81, and 0.79 for precision, recall, and F-score, respectively |
| Del Amor et al., 2022 [74] | Histological activity | Evaluation of a novel MIL framework with location constraints able to determine the presence of UC activity based on neutrophils detection using WSI | In comparison with prior multiple instance learning settings, this method allowed for 10% improvements in accuracy |
| Peyrin Biroulet et al., 2024 [75] | Histological activity | Evaluation of an AI system to measure histological disease activity based on the Nancy index | The average ICC among the histopathologists was 89.3 and the average ICC between histopathologists and the AI tool was 87.2 |
| Najdawi et al., 2023 [76] | Histological activity | Validation of CNN models that quantify histologic features in UC, directly from haematoxylin and eosin-stained whole slide images | The model accurately predicted Nancy histological index scores (⍴ = 0.89, p < 0.001) when compared with pathologist consensus Nancy histological index scores. It also predicted histologic remission with a high accuracy of 0.97 |
| Kiyokawa et al., 2022 [77] | Histological activity | Evaluation of a DL model to predict postoperative recurrence of CD by computational analysis of histopathologic images and to extract histologic characteristics associated with recurrence | The model achieved a highly accurate prediction of recurrence (area under the curve, 0.995 |
| Rubin et al., 2024 [78] | Histological activity | Application of an AI tool based on DL to streamline the quantitative assessment of histopathology using the Nancy Index in UC | Confusion matrix analysis demonstrated an 80% correlation between predicted and true labels for Nancy scores of 0 or 4; a 96% correlation for a true score of 0 being predicted as 0 or 1; and a 100% correlation for a true score of 2 being predicted as 2 or 3 |
| Vinsard et al., 2023 [79] | Detection of colonic neoplasm | Application of a CADe model of colorectal lesions in patients with IBD | IBD-CADe model on HDWLE had sensitivity, 95.1%; specificity, 98.8% and accuracy, 96.8%; and area under the curve, 0.85. IBD-CADe for chromoendoscopy images showed a sensitivity of 67.4%, specificity of 88.0%, accuracy of 77.8%, and area under the curve of 0.65 |
| WLE | |||
| Abdelrahim et al., 2024 [80] | Detection of colonic neoplasm | AI model for lesion detection in IBD | The AI model had lesion detection rate, lesion per colonoscopy and neoplasia per colonoscopy of 90.4%, 4.6% and 0.96., respectively. The sensitivity and specificity of lesion characterisation were 87.5% and 80.6%, respectively |
| WLE | |||
| Stidham et al., 2021 [81] | Risk stratification | Evaluation of a ML models incorporating routinely collected laboratory studies to predict surgical outcomes in CD | The model achieved a mean area under the receiver operating characteristic of 0.78 (SD, 0.002). Anti-tumour necrosis factor use was the most influential predictor |
| Majumder et al., 2024 [82] | Risk stratification | This study aims to combine endocytoscope with intestinal barrier proteins assessment through ML-enabled multispectral spatial imaging to predict MAOs | The combination of endocytoscopy with Claudin-2 expression showed promise in accurately predicting MAOs over 12 months |
| Advanced imaging | |||
| Maeda et al., 2022 [83] | Risk stratification | Application of AI to predict clinical relapse of UC in clinical remission | The relapse rate was higher in the AI-Active group (28.4% [21/74]; 95% confidence interval, 18.5–40.1%) than in the AI-Healing group (4.9% [3/61]; 95% confidence interval, 1.0–13.7%; p < 0.001) |
| Advanced imaging | |||
| Omori et al., 2024 [84] | Risk stratification | Comparison between AI-assisted ultra-magnifying colonoscopy system for histological healing in UC and conventional light non-magnifying endoscopy | EndoBRAIN-UC showed a sensitivity of 74.2% and a specificity of 93.8% for histological diagnosis of remission |
| WLE-advanced imaging | |||
| Kuroki et al., 2024 [85] | Risk stratification | Evaluation of an AI-based system to diagnose “vascular-healing” | The clinical relapse rate was significantly higher in the AI-based vascular-active group (23.9% [16/67]) compared with the AI-based vascular-healing group (3.0% [1/33)]; p = 0.01) |
| WLE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labarile, N.; Vitello, A.; Sinagra, E.; Nardone, O.M.; Calabrese, G.; Bonomo, F.; Maida, M.; Iacucci, M. Artificial Intelligence in Advancing Inflammatory Bowel Disease Management: Setting New Standards. Cancers 2025, 17, 2337. https://doi.org/10.3390/cancers17142337
Labarile N, Vitello A, Sinagra E, Nardone OM, Calabrese G, Bonomo F, Maida M, Iacucci M. Artificial Intelligence in Advancing Inflammatory Bowel Disease Management: Setting New Standards. Cancers. 2025; 17(14):2337. https://doi.org/10.3390/cancers17142337
Chicago/Turabian StyleLabarile, Nunzia, Alessandro Vitello, Emanuele Sinagra, Olga Maria Nardone, Giulio Calabrese, Federico Bonomo, Marcello Maida, and Marietta Iacucci. 2025. "Artificial Intelligence in Advancing Inflammatory Bowel Disease Management: Setting New Standards" Cancers 17, no. 14: 2337. https://doi.org/10.3390/cancers17142337
APA StyleLabarile, N., Vitello, A., Sinagra, E., Nardone, O. M., Calabrese, G., Bonomo, F., Maida, M., & Iacucci, M. (2025). Artificial Intelligence in Advancing Inflammatory Bowel Disease Management: Setting New Standards. Cancers, 17(14), 2337. https://doi.org/10.3390/cancers17142337






